Abstract
Wolfram syndrome is an autosomal-recessive disorder characterized by insulin-dependent diabetes mellitus, caused by nonautoimmune loss of β cells, and neurological dysfunctions. We have previously shown that mutations in the Wolfram syndrome 1 (WFS1) gene cause Wolfram syndrome and that WFS1 has a protective function against ER stress. However, it remained to be determined how WFS1 mitigates ER stress. Here we have shown in rodent and human cell lines that WFS1 negatively regulates a key transcription factor involved in ER stress signaling, activating transcription factor 6α (ATF6α), through the ubiquitin-proteasome pathway. WFS1 suppressed expression of ATF6α target genes and repressed ATF6α-mediated activation of the ER stress response element (ERSE) promoter. Moreover, WFS1 stabilized the E3 ubiquitin ligase HRD1, brought ATF6α to the proteasome, and enhanced its ubiquitination and proteasome-mediated degradation, leading to suppression of ER stress signaling. Consistent with these data, β cells from WFS1-deficient mice and lymphocytes from patients with Wolfram syndrome exhibited dysregulated ER stress signaling through upregulation of ATF6α and downregulation of HRD1. These results reveal a role for WFS1 in the negative regulation of ER stress signaling and in the pathogenesis of diseases involving chronic, unresolvable ER stress, such as pancreatic β cell death in diabetes.
Introduction
Productive folding of secretory proteins and degradation of misfolded proteins are essential to ensure normal cell function. Both these processes occur in the ER. Perturbations in ER function cause an imbalance between these processes, leading to accumulation of misfolded and unfolded proteins in the organelle, a state called ER stress. Cells cope with ER stress by activating an ER stress signaling network, also called the unfolded protein response (UPR). Activation of the UPR not only results in the upregulation of gene expression for molecular chaperones, but expands the size of the ER, decreases general protein translation to reduce the ER workload, and degrades abnormal proteins accumulated in the ER (1, 2). As long as the UPR can mitigate ER stress, cells can produce proper amounts of proteins in response to the need for them and perform their normal functions.
Activating transcription factor 6α (ATF6α) is 1 of the 3 master regulators of the UPR (1). ATF6 encodes a bZIP-containing transcription factor localized to the ER membrane (3). Under ER stress, the N-terminal DNA binding domain of ATF6α is cleaved and released from the ER (3–5). The bZIP domain of ATF6α then translocates into the nucleus and upregulates downstream target genes, such as BiP and XBP-1, that function in protein folding and processing (3, 4, 6). Therefore, deletion of ATF6α compromises the secretory pathway during ER stress (7, 8). It has been reported that the noncleaved form of ATF6α is unstable and quickly degraded by the ubiquitin-proteasome pathway to prevent hyperactivation of the UPR (9). However, the mechanism underlying this phenomenon has yet to be elucidated.
WFS1, a transmembrane protein localized to the ER (10), has previously been shown to be a UPR component that mitigates ER stress response in cells (11). Mutations in the gene encoding WFS1 cause Wolfram syndrome, a genetic form of diabetes, optic atrophy, neurodegeneration, and psychiatric illness (12, 13). Recent reports also indicated that WFS1 polymorphisms are associated with type 2 diabetes (14–16). Accumulating evidence indicates that β cell death and neuronal cell dysfunction in Wolfram syndrome are attributed to high levels of ER stress signaling in affected cells (11, 17–19). However, the function of WFS1 in the UPR has been unclear. Here we showed that WFS1 controls a regulatory feedback loop of the ER stress signaling network. Activation of the ER stress response element (ERSE) by ATF6α was attenuated by WFS1 expression. WFS1 recruited ATF6α to an E3 ligase, HRD1, and the proteasome, where it enhanced ATF6α degradation, thus suppressing the UPR. Inducible overexpression of WFS1 thereby decreased expression levels of ATF6α target genes, such as BiP and XBP-1. These results indicate that WFS1 has an important function in the negative regulation of a feedback loop of the ER stress signaling network and prevents secretory cells from death caused by dysregulation of this signaling pathway.
Results
WFS1 forms an ER stress–mediated complex with ATF6α and suppresses its activity.
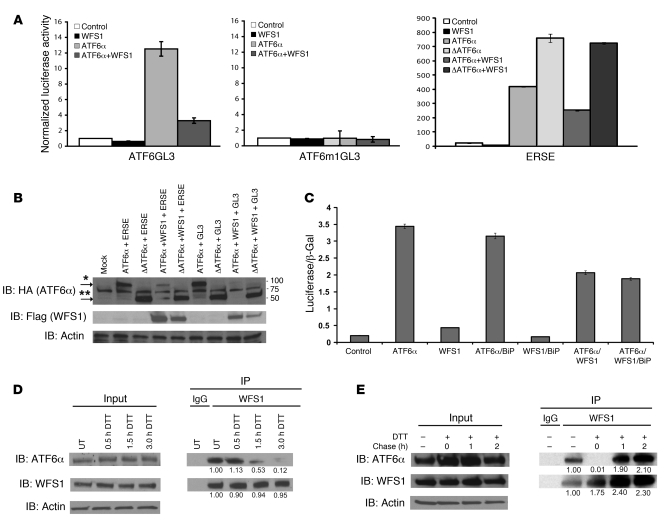
In order to further define the role of WFS1 in the UPR, we assessed whether WFS1 expression affects the function of UPR components. Transcriptional activity of a transmembrane transcription factor and master regulator of the UPR, ATF6α, is attenuated by WFS1 expression. Under ER stress, the N-terminal DNA binding domain of ATF6α is cleaved and released from the ER to upregulate UPR target genes in the nucleus (3–5). As expected, when full-length ATF6α was transfected with the ATF6α binding site reporter gene ATF6GL3, this reporter was induced 12-fold by ATF6α (20), an induction reduced to 3-fold by cotransfection with WFS1 (Figure 1A). ATF6α has also been shown to strongly activate the BiP/GRP78 promoter (4). To confirm that WFS1 regulates ATF6α transcriptional activity on the BiP/GRP78 promoter, full-length ATF6α or cleaved ATF6α (ΔATF6α) was cotransfected with WFS1 and a rat GRP78 promoter reporter gene containing the ER stress response element (ERSE). This reporter was induced by both full-length ATF6α and ΔATF6α; however, only full-length ATF6α activity was suppressed by WFS1 expression (Figure 1A). In addition, full-length ATF6α protein expression decreased when it was coexpressed with WFS1 (Figure 1B). BiP has previously been shown to anchor full-length ATF6α to the ER membrane and prevent ATF6α activation (6, 21). To compare the ability of WFS1 to suppress ATF6α with that of BiP, the GRP78 promoter reporter was cotransfected with full-length ATF6α and BiP, with full-length ATF6α and WFS1, or with full-length ATF6α, BiP, and WFS1. Suppression of ATF6α activity by WFS1 was stronger than that by BiP (Figure 1C). Collectively, these results indicate that WFS1 suppresses ATF6α transcriptional activity before its translocation to the nucleus.
Figure 1. WFS1 interacts with ATF6α in an ER stress–dependent manner and suppresses ATF6α transcriptional activation.
(A) COS7 cells were transfected with a full-length ATF6α expression plasmid or ΔATF6α with a WFS1 plasmid together with the following luciferase reporter genes: ATF6α binding site reporter gene ATF6GL3, ATF6α mutant site reporter ATF6m1GL3, and GRP78 promoter reporter gene ERSE. Relative intensity of luciferase was then measured (n = 3). (B) Protein lysates from the luciferase assay were analyzed by IB using anti-HA (ATF6α), anti-Flag (WFS1), and anti-actin antibodies. ATF6α and ΔATF6α are denoted by single and double asterisks, respectively. (C) COS7 cells were transfected with a full-length ATF6α expression plasmid with a BiP expression plasmid, WFS1 expression plasmid, or WFS1 and BiP expression plasmid together with the GRP78 reporter gene (n = 3). (D) An anti-WFS1 antibody was used to IP WFS1 protein from INS1 832/13 cells untreated (UT) or treated with the ER stress inducer DTT (1 mM) for 0.5, 1.5, or 3 hours. IPs were then subject to IB analysis using anti-ATF6α, anti-WFS1, and anti-actin antibodies (n = 3). (E) INS1 832/13 cells were treated with DTT (1 mM) for 2 hours and then chased in normal media for 0, 1, or 2 hours. WFS1 was subjected to IP from cell lysates, and IPs were analyzed by IB using anti-ATF6α, anti-WFS1, and anti-actin antibodies (n = 3).
Both WFS1 and ATF6α are transmembrane proteins localized to the ER (3, 10), raising the possibility that the suppression of the ATF6α reporter by WFS1 might be mediated by direct interaction between the WFS1 and ATF6α proteins. To confirm this, the association of WFS1 with ATF6α was examined in the pancreatic β cell line INS-1 832/13. WFS1 associated with ATF6α under nonstress conditions (Figure 1D). To examine whether this interaction was maintained during ER stress conditions, the cells were treated with the ER stress inducer dithiothreitol (DTT), which caused a dissociation of ATF6α from WFS1 in a time-dependent manner, with almost complete dissociation 3 hours after treatment (Figure 1D). This ER stress–dependent interaction was also observed in cells treated with another ER stress inducer, thapsigargin (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI39678DS1). To confirm that this interaction was recovered after stress, cells were treated for 2 hours with DTT and then chased in normal media. As expected, the interaction of ATF6 and WFS1 began to recover after a 1-hour chase (Figure 1E). This interaction was also seen in the neuronal cell line Neuro2A (Supplemental Figure 2). Together, these data suggest that ATF6 is released from WFS1 under stress in order to activate its target UPR genes.
WFS1 has a function in the degradation of ATF6α through the ubiquitin-proteasome pathway.
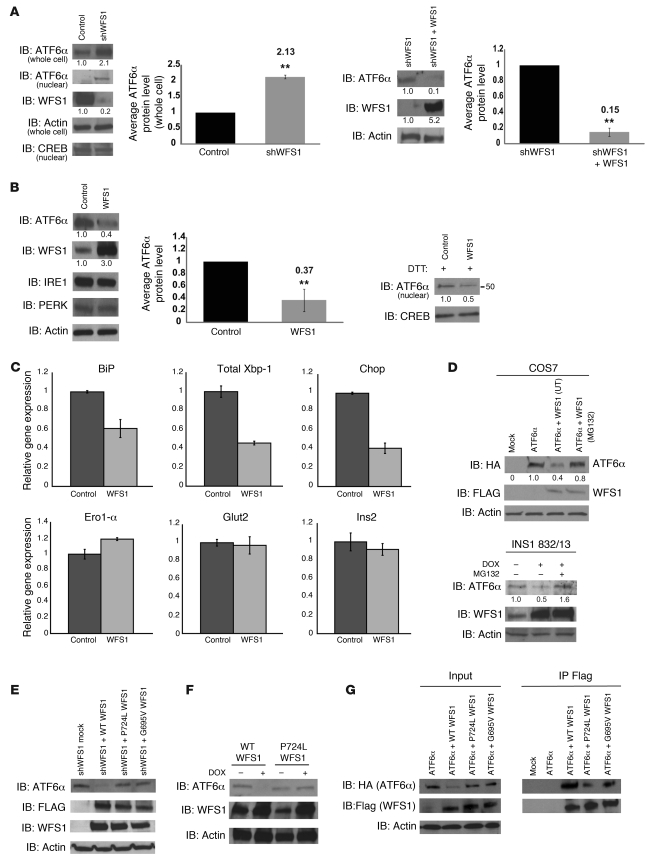
Suppression of ATF6α transcriptional activity by WFS1 and the formation of an ATF6α-WFS1 complex led to the prediction that WFS1 regulates ATF6α function at the posttranslational level. To test this prediction, we derived a pancreatic β cell line, MIN6 cells, stably expressing a shRNA directed against WFS1. Full-length as well as nuclear ATF6α protein levels increased approximately 2-fold compared with control cells (Figure 2A). To confirm that upregulation of ATF6α protein is directly regulated by WFS1, we reintroduced a lentivirus expressing WFS1 into the cells expressing shRNA directed against WFS1; ATF6 protein expression levels were again reduced when WFS1 was reintroduced (Figure 2A).
Figure 2. WFS1 regulates ATF6α protein levels.
(A) IB analysis measured ATF6α and WFS1 levels in MIN6 cells expressing shGFP (control) or shWFS1, as well as in MIN6 cells expressing shWFS1 or expressing shWFS1 and rescued with WFS1 (n = 3). (B) IB analysis measuring ATF6α, WFS1, IRE1α, and PERK levels in INS1 832/13 cells (treated with 2 mM DTT for 3 hours) overexpressing GFP (control) or WFS1 (n = 3). (C) Quantitative real-time PCR analysis of BiP, total Xbp-1, Chop, Ero1-α, Glut2, and Ins2 mRNA levels in INS1 832/13 cells overexpressing GFP (control) or WFS1 (n = 3). (D) IB analysis of ATF6α and WFS1 in COS7 cells transfected with ATF6α-HA or ATF6α-HA and WFS1-FLAG at 2 different ratios, and in INS1 832/13 cells expressing inducible WFS1 and treated with or without MG132. (E) IB analysis of ATF6α and WFS1 in MIN6 cells expressing shWFS1 and transfected with WT WFS1-FLAG or mutant P724L WFS1-FLAG and G695V WFS1-FLAG (n = 3). (F) IB analysis measuring ATF6α and WFS1 levels in INS1 832/13 cells expressing WT WFS1 or P724L WFS1 (n = 3). (G) WFS1 was subjected to IP from COS7 cells expressing ATF6α-HA or ATF6α-HA with WT, P724L, or G695V WFS1-Flag using an anti-Flag antibody. IPs and input proteins were analyzed using anti-HA and anti-Flag antibodies. **P < 0.01.
ATF6α mRNA was unchanged in the WFS1-knockdown INS-1 832/13 cells, but ATF6α target genes, such as p58IPK and BiP (7, 8), were upregulated as predicted (Supplemental Figure 3). To further confirm that this upregulation is directly regulated by ATF6α, we suppressed ATF6α expression by siRNA in WFS1 knockdown INS-1 832/13 cells and then measured expression levels of its major target, BiP. Upregulation of BiP by WFS1 inhibition was cancelled out by ATF6α inhibition (Supplemental Figure 4).
ATF6α protein levels were also measured in INS-1 832/13 cells overexpressing WFS1. Full-length and nuclear ATF6α protein levels were suppressed in these cells, whereas there was no significant change in protein levels of the other 2 master regulators of the UPR, IRE1 and PERK (Figure 2B). IRE1 and PERK protein expression levels were not decreased even with higher levels of WFS1 expression (Supplemental Figure 5). Suppression of ATF6α protein expression was also seen in a neuronal cell line (Supplemental Figure 6). ATF6 target gene mRNA levels were also suppressed in β cells overexpressing WFS1 (Figure 2C). The relationship of WFS1 and ATF6 protein expression was found to be dose-dependent: increased expression of WFS1 leads to a decrease in ATF6 protein expression (Supplemental Figure 7). We asked whether this relationship was proteasome dependent. Treatment of 2 WFS1-overexpressing cell lines with the proteasome inhibitor MG132 rescued ATF6α protein levels (Figure 2D). We cloned 2 missense mutants, WFS1 P724L and WFS1 G695V, and 1 inactivating mutant, WFS1 ins483fs/ter544, from patient samples (13). Mutant variants of WFS1 did not affect ATF6α protein levels in MIN6 cells expressing shRNA directed against WFS1 (Figure 2E and Supplemental Figure 8). This was also confirmed in INS-1 832/13 cells expressing the missense mutant WFS1 P724L (Figure 2F) and in neuronal cells expressing the missense mutant WFS1 G695V (Supplemental Figure 9). Although ATF6α weakly interacted WFS1 P724L and WFS1 G695V, there was no significant decrease in ATF6α protein levels in these cells (Figure 2G).
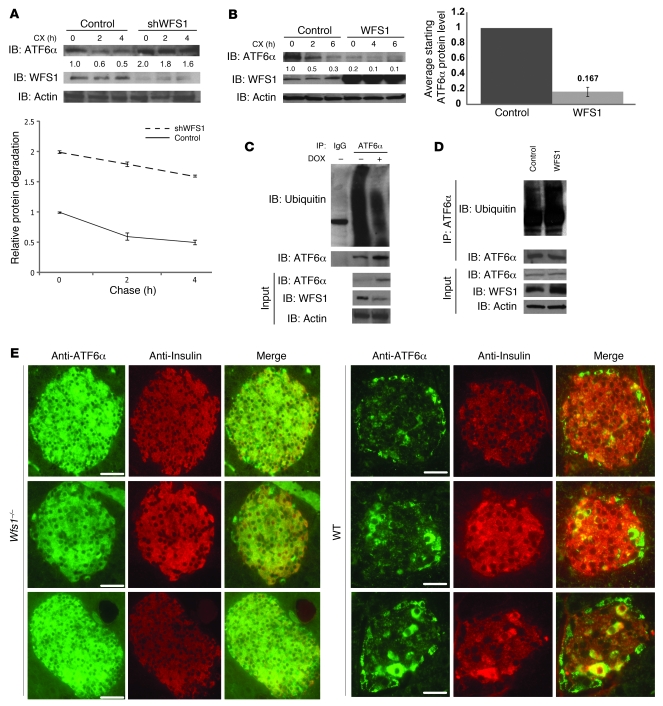
To assess the impact of WFS1 on ATF6α protein degradation, cycloheximide experiments were performed. In MIN6 cells expressing shRNA directed against WFS1, there was a block in ATF6α protein degradation, whereas in cells overexpressing WFS1, there was minimal ATF6α protein expression (Figure 3, A and B). WFS1 could not enhance the degradation of 2 other ER proteins susceptible to misfolding, TCRα and mutant alpha-1-antitrypsin NHK3 (refs. 22–24 and Supplemental Figure 10), which indicates that WFS1 specifically degrades ATF6α protein. WFS1 also enhanced the ubiquitination of ATF6α. In cells expressing shRNA directed against WFS1, there was a decrease in ATF6α ubiquitination after blocking proteasome activity (Figure 3C), whereas in cells overexpressing WFS1, there was an enhancement of ATF6α ubiquitination (Figure 3D). In Wfs1–/– mouse pancreata, ATF6α protein expression was strikingly high compared with control littermate pancreata (Figure 3E), indicating that WFS1 functions in ATF6α protein expression in vivo. In samples from patients with WFS1 mutations, there was a higher expression of ATF6α protein compared with control samples (Supplemental Figure 11). Together, these results indicate that WFS1 is important for regulating ATF6α protein expression. When WFS1 was not present, there was increased expression of ATF6α protein and hyperactivation of its downstream effectors. This suggests that in response to ER stress, ATF6α escapes from WFS1-dependent degradation, is cleaved in the Golgi to its active form, and then translocates to the nucleus to upregulate its UPR target genes.
Figure 3. WFS1 enhances ATF6α ubiquitination and degradation.
(A) IB analysis measuring ATF6α, WFS1, and actin levels in MIN6 cells stably expressing shGFP (control) or shWFS1 treated with 40 μM cycloheximide (CX) for 0, 2, and 4 hours (n = 3). (B) IB analysis measuring ATF6α, WFS1, and actin levels in INS1 832/13 cells expressing GFP (control) or WFS1 treated with 40 μM cycloheximide for 0, 2, and 6 hours (n = 3). (C) ATF6α was subjected to IP using an anti-ATF6α antibody from an INS1 832/13 cells inducibly expressing shWFS1 (treated for 48 hours with 2 μM doxycycline) and treated with MG132 (20 μM) for 3 hours. IPs were then subjected to IB with anti-ubiquitin and anti-ATF6α antibodies, and input lysates were blotted with anti-ATF6α, anti-WFS1, and anti-actin antibodies (n = 3). (D) ATF6α was subjected to IP using an anti-ATF6α antibody, from INS1 832/13 cells overexpressing GFP (control) or WFS1, then treated with MG132 (0.1 μM) overnight. IPs were subjected to IB with anti-ubiquitin and anti-ATF6α antibodies. Input lysates were subjected to IB with anti-ATF6α, anti-WFS1, and anti-actin antibodies (n = 3). (E) Wfs1–/– and WT littermate mouse pancreata were analyzed by immunohistochemistry using anti-ATF6α and anti-insulin antibodies. Scale bars: 50 μm.
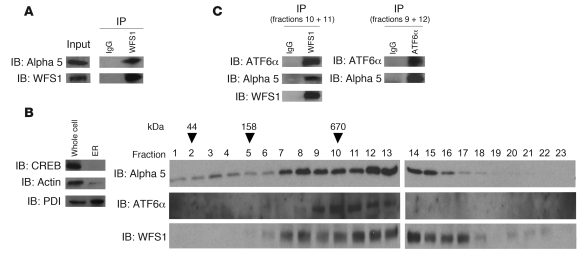
These data raised the possibility that WFS1 recruits ATF6α to the proteasome for its degradation. As we predicted, WFS1 formed a complex with the proteasome (Figure 4A). When glycerol-gradient fractionation was performed on ER-isolated lysates, the proteasome ATF6α and WFS1 comigrated in the same high–molecular weight fractions, and a complex between them was formed (Figure 4, B and C).
Figure 4. WFS1 forms a complex with the proteasome and ATF6α.
(A) WFS1 was subjected to IP from INS1 832/13 cells using an anti-WFS1 specific antibody. IPs were subjected to IB with anti–alpha 5 20S proteasome and anti-WFS1 antibodies. (B) IB analysis measuring CREB, actin, and PDI levels using whole cell lysates or ER-isolated lysates of INS1 832/13 cells. ER-isolated lysates of INS1 832/13 cells were also subjected to fractionation using a 10%–40% glycerol gradient. Fractions were analyzed by IB using anti–alpha 5 20s proteosome, anti-ATF6α, and anti-WFS1 antibodies. Lanes were run on separate gels and were not contiguous. (C) WFS1 was subjected to IP from a mixture of fractions 10 and 11 using an anti-WFS1 antibody, and IP products were subjected to IB analysis using anti-alpha 5 20s proteosome, anti-ATF6α, and anti-WFS1 antibodies. ATF6 was subjected to IP from a mixture of fractions 9 and 12, and IP products were analyzed by IB with anti–alpha 5 20s proteosome and anti-ATF6α (n = 3).
WFS1 stabilizes HRD1, which functions as an E3 ligase for ATF6α.
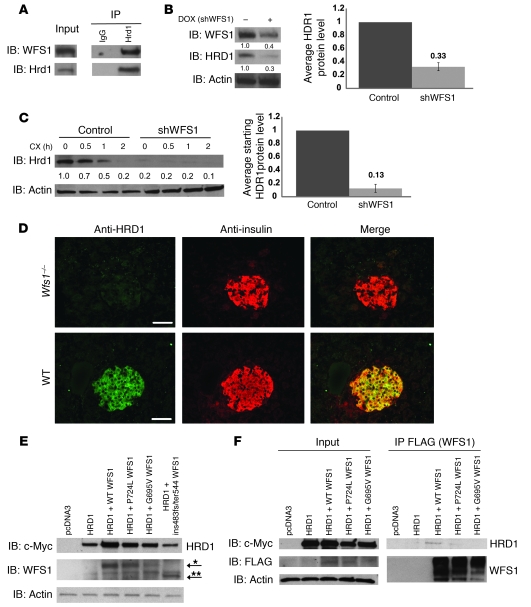
Because WFS1 is localized to the ER membrane and recruits ATF6α to the proteasome, but is not itself an E3 ligase, we searched for ER-localized E3 ligases with which WFS1 could interact. A top candidate was the ER-resident E3 ligase HRD1, which has a known role in ER stress signaling (25, 26). SEL1/HRD3, which has an important function in hydroxy-3-methylglutaryl-CoA reductase (HMG-R) degradation (27), has been shown to interact with and stabilize HRD1 (28), raising the possibility that WFS1 may also have a similar function and could interact with HRD1. Indeed, WFS1 and HRD1 formed a complex (Figure 5A). We next asked whether WFS1 also plays a role in HRD1 protein expression. Inducible suppression of WFS1 in INS-1 832/13 cells expressing shRNA directed against WFS1 suppressed HRD1 protein expression (Figures 5B). To test the effect of WFS1 on HRD1 protein stability, we performed cycloheximide experiments using MIN6 cells stably expressing shRNA directed against WFS1. HRD1 protein expression was significantly decreased in WFS1 knockdown cells compared with control cells, and it was difficult to measure the stability of HRD1 (Figure 5C).
Figure 5. WFS1 interacts with and stabilizes the E3 ligase HRD1.
(A) Hrd1 was subjected to IP from INS1 832/13 cells, and IPs were subjected to IB analysis using anti-WFS1 and anti-Hrd1 antibodies (n = 3). (B) Total lysates from INS1 832/13 cells inducibly expressing shWFS1 (treated with 2 μM doxycycline for 48 hours) were analyzed by IB using anti-WFS1, anti-Hrd1, and anti-actin antibodies (n = 3). (C) IB analysis measuring HRD1 levels in MIN6 cells stably expressing shGFP (control) or shWFS1 treated with 40 μM cycloheximide for 0, 0.5, 1, and 2 hours (n = 3). (D) Wfs1–/– and WT littermate mouse pancreata were analyzed by immunohistochemistry using anti-HRD1 and anti-insulin antibodies (n = 3). Scale bars: 100 μm. (E) COS7 cells were transfected with pcDNA3, HRD1–c-Myc, HRD1–c-Myc and WT WFS1, or HRD1–c-Myc and WFS1 mutants (P724L, G695V, and ins483fs/ter544) expression plasmids. Expression levels of HRD1–c-Myc, WFS1, and actin were measured by IB using anti–c-Myc, anti-WFS1, and anti-actin antibodies, respectively. WT and mutant WFS1 are denoted by single and double asterisks, respectively. (F) COS7 cells were transfected with pcDNA3, HRD1–c-Myc, HRD1–c-Myc and WT WFS1-Flag, HRD1–c-Myc and WFS P724L-Flag, and HRD1–c-Myc and WFS1 G695V-Flag expression plasmids. The lysates were subjected to IP with anti-Flag antibody and IB with anti–c-Myc antibody to study the interaction between HRD1 and WFS1.
We further confirmed the effects of WFS1 on HRD1 protein expression in vivo using Wfs1–/– mice. As expected from the results using β cell lines, HRD1 expression was undetectable in islets of Wfs1–/– mice (Figure 5D). In addition, in samples from patients with Wolfram syndrome, there was less HRD1 protein expression compared with control samples (Supplemental Figure 12A). HRD1 expression did not affect WFS1 protein expression (Supplemental Figure 12B).
We next sought to compare the effects of WT WFS1 and WFS1 mutants on HRD1 protein expression. Ectopic expression of WT WFS1 increased HRD1 protein expression, whereas ectopic expression of missense and inactivating WFS1 mutants did not increase or decrease HRD1 expression (Figure 5E). To determine whether WFS1 mutants interact with HRD1, comparable amounts of WT and missense mutant WFS1 proteins were expressed together with HRD1 in COS7 cells, and the interaction was monitored by co-IP. HRD1 interacted with WT WFS1, but not with WFS1 mutants (Figure 5F). Collectively, these results demonstrated that WFS1 stabilizes and enhances the function of the E3 ligase HRD1 through direct binding.
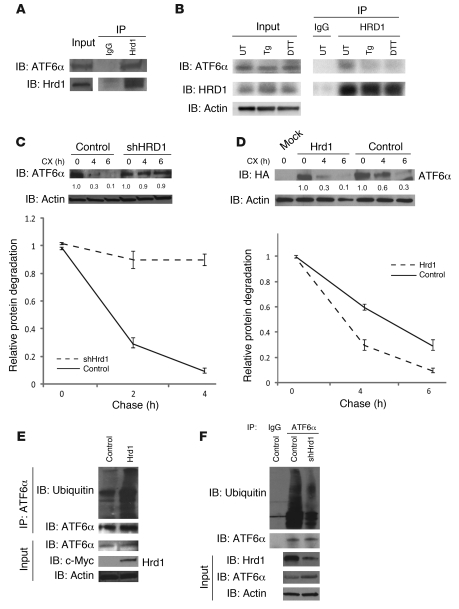
Based on the ability of WFS1 to regulate ATF6α protein, as well as its function in stabilizing HRD1, it followed that WFS1 may be recruiting ATF6α to HRD1 and that ATF6α is a substrate of HRD1. Indeed, HRD1 interacted with ATF6α (Figure 6A). In glycerol-gradient fractionation experiments of ER-isolated lysates, HRD1, ATF6α, and WFS1 were found to form a complex (Supplemental Figure 13). We next analyzed the interaction between ATF6α and HRD1 under ER stress conditions. ATF6α was released from HRD1 by DTT and thapsigargin treatments (Figure 6B), which indicates that the interaction between these proteins is disrupted by ER stress. To study the relationship between HRD1 and ATF6α protein expression levels, the stability of ATF6α protein was measured in MIN6 cells stably expressing shRNA directed against HRD1 and control cells. HRD1 suppression in cells enhanced ATF6α protein stability (Figure 6C). In contrast, overexpression of HRD1 enhanced ATF6α protein degradation (Figure 6D). HRD1 also enhanced ATF6α ubiquitination, and lack of HRD1 decreased ATF6α ubiquitination (Figure 6, E and F). Collectively, these results indicate that the WFS1-HRD1 complex enhances ATF6α ubiquitination and degradation.
Figure 6. HRD1 is an E3 ligase for ATF6α.
(A) HRD1 was subjected to IP from INS1 832/13 cells treated for 3 hours with 30 μM MG132. IPs and input proteins were analyzed by IB using anti-ATF6α and anti-HRD1 antibodies. Lanes were run on the same gel but were noncontiguous (white line). (B) An anti-HRD1 antibody was used to IP HRD1 protein from INS1 832/13 cells untreated or treated with DTT (1 mM) and thapsigargin (Tg; 1 μM) for 3 hours. IPs were then subjected to IB analysis using anti-ATF6α, anti-HRD1, and anti-actin antibodies (n = 3). (C) IB analysis measuring ATF6α levels in MIN6 cells stably expressing shGFP (control) or shRNA shHRD1 treated with 40 μM cycloheximide for 0, 4, and 6 hours (n = 3). (D) COS7 cells transfected with ATF6α-HA expression plasmid (control) or ATF6α-HA together with Hrd1-myc expression plasmids (Hrd1) were treated with 40 μM cycloheximide for 0, 4, and 6 hours. Whole cell lysates were subjected to IB with an anti-HA antibody (n = 3). (E) ATF6α was subjected to IP using an anti-ATF6α antibody from INS1 832/13 cells either mock transfected (control) or transfected with a Hrd1-Myc expression plasmid and treated with MG132 (20 μM) for 3 hours. IPs were then subjected to IB with anti-ubiquitin and anti-ATF6α antibodies, and input lysates were blotted with anti-ATF6α, anti–c-Myc, and anti-actin antibodies (n = 3). (F) ATF6 was subjected to IP using an anti-ATF6α antibody from MIN6 cells stably expressing shGFP (control) or shHRD1 and treated with MG132 (20 μM) for 3 hours. IPs were then subjected to IB with anti-ubiquitin and anti-ATF6α antibodies, and input lysates were blotted with anti-HRD1, anti-ATF6α, and anti-actin antibodies (n = 3).
Discussion
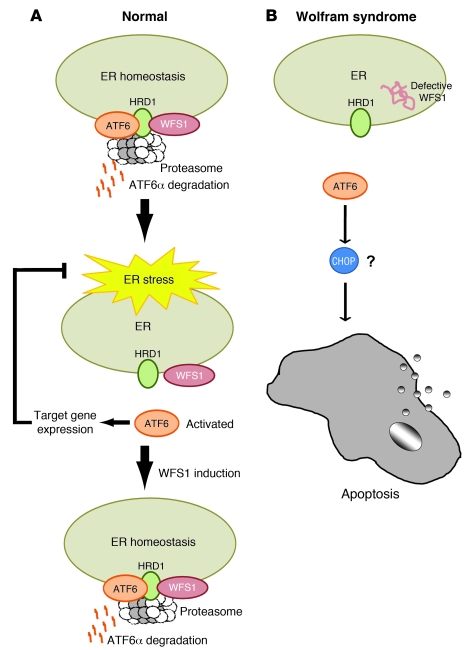
In this study, we provide evidence that WFS1 plays a crucial role in regulating ATF6α transcriptional activity through HRD1-mediated ubiquitination and proteasome-mediated degradation of ATF6α protein. Based upon the data provided, we propose a pathway for the negative-feedback regulation of the ER stress signaling network by WFS1 (Figure 7). In healthy cells, WFS1 prevents dysregulated ER stress signaling by recruiting ATF6α to HRD1 and the proteasome for ubiquitin-mediated degradation under non–ER stress conditions. When stress is applied to the ER, such as through the chemical ER stress inducer DTT, ATF6α is released from WFS1. It is then released from the ER membrane and translocates to the nucleus, where it upregulates stress signaling targets. At later time points, WFS1 is induced by ER stress, causing eventual degradation of ATF6α when ER homeostasis is established. In patients with Wolfram syndrome or Wfs1–/– mice, ATF6α escapes from this proteasome-dependent degradation, leading to dysregulated ATF6α signaling. This ATF6α hyperactivation caused by the lack of WFS1 is probably involved in β cell apoptosis.
Figure 7. WFS1 controls steady-state levels of ATF6α protein and activation.
(A) In normal cells, WFS1 recruits the ER transcription factor ATF6α to the E3 ligase Hrd1 under non–ER stress conditions. Hrd1 marks ATF6α with ubiquitin for proteasomal degradation. Under ER stress, ATF6α dissociates from WFS1 and undergoes proteolysis, and its soluble aminoportion, p60ATF6α, translocates to the nucleus, where it upregulates ER stress target genes, such as BiP, CHOP, and XBP-1. At later time points, WFS1 is induced by ER stress, which causes the eventual degradation of ATF6α. (B) In patients with Wolfram syndrome or Wfs1–/– mice, ATF6α escapes from the proteasome-dependent degradation, leading to chronic hyperactivation of ATF6α signaling. This ATF6α hyperactivation is involved in apoptosis through apoptotic effectors of the UPR, such as CHOP.
It has previously been shown that WFS1-deficient β cells are susceptible to ER stress–mediated apoptosis (11, 18, 29). We confirmed that knockdown of WFS1 made β cells sensitive to ER stress–mediated cell death (Supplemental Figure 14). In addition, we found that ectopic expression of an active form of ATF6α in β cells caused apoptosis (Supplemental Figure 15). Although this fits to our model that hyperactivation of ATF6α has a harmful effect on β cells and leads to apoptosis, it may be contrary to previous studies showing beneficial effects of ATF6α upregulation. For example, it has been shown that activation of the ATF6α pathway by a chemical compound protects neuronal cells from ER stress–mediated apoptosis (30). This effect is mediated by BiP upregulation. The induction of ATF6α has also been shown to protect cardiomyocytes from ischemia/reperfusion-mediated apoptosis (31). This effect is also mediated by BiP and GRP94 upregulation. Conversely, a recent report has shown that ATF6α upregulation attenuates diet-induced obesity and insulin resistance (32). More importantly, it has been shown that WT mice are better protected from ER stress in vivo than are ATF6α knockout mice. ATF6α knockout hepatocytes have been shown to be more sensitive to ER stress–mediated cell death compared with control hepatocytes (7). It is not surprising that previous studies found induction of ATF6α to have beneficial effects on cell function and cell survival, because ATF6α is a major regulator for BiP, a central molecular chaperone in the ER (7, 8). The main conclusion of the present study is that chronic dysregulation of the UPR, more specifically hyperactivation of ATF6α signaling, has a negative effect on β cell survival. It has been suggested that the UPR regulates both adaptive and apoptotic effectors (2, 33). The balance between these effectors depends on the nature of the ER stress, whether it is tolerable or unresolvable. Thus, the UPR acts as a binary switch between life and death. Our results demonstrated that, in patients with Wolfram syndrome and Wfs1–/– mice, unresolvable ER stress occurs in β cells and neurons, leading to a switch toward apoptosis.
ER stress is caused by both physiological and pathological stimuli that can lead to the accumulation of unfolded and misfolded proteins in the ER. Physiological ER stress can be caused by a large biosynthetic load placed on the ER, for example, during postprandial stimulation of proinsulin biosynthesis in pancreatic β cells. This stimulation leads to the activation of ER stress signaling and enhancement of insulin synthesis (34). Under physiological ER stress conditions, activation of ER stress signaling must be tightly regulated because hyperactivation or chronic activation of this signaling pathway can cause cell death. For example, when eukaryotic translation initiation factor 2α, a downstream component of ER stress signaling, is hyperphosphorylated by the compound salubrinal in pancreatic β cells, apoptosis is induced in these cells (35). Our results showed that WFS1 has an important function in the tight regulation of ER stress signaling through its interaction with a key transcription factor, ATF6α, thereby protecting cells from the damaging effects of hyperactivation of this signaling pathway.
On the basis of our present results, we believe WFS1 plays a similar role in mammals as HRD3 does in yeast: stabilizing and enhancing the activity of a key ER-resident E3 ligase, HRD1 (28). Thus, a loss of functional WFS1 may affect ER stress levels in 2 ways: (a) enhancing ATF6α signaling by increasing the pool of ATF6α, and (b) destabilizing HRD1 protein and thus its activity. The latter would independently contribute to ER stress by promoting the buildup of unfolded and misfolded proteins in the ER. In support of this is our present finding that silencing of HRD1 in β cell lines indeed led to mild ER stress (Supplemental Figure 16). It has previously been shown that HRD1 is regulated by the IRE1–XBP-1 pathway (26) and is activated at a later time point during ER stress. The ATF6α pathway, however, is activated at an earlier phase (36). Thus, WFS1 may also function as a switch from the ATF6α pathway to the IRE1–XBP-1 pathway, through the stabilization of HRD1 and consequent destruction of ATF6α protein. A previous publication has reported that WFS1 deficiency could lead to increased HRD1 expression (17), contrary to our findings. This discrepancy could be attributed to the fact that WFS1 deficiency can cause dysregulated ER stress signaling and can lead to hyperactivation of the IRE1–XBP-1 pathway under some circumstances.
It has been established that WFS1 is induced under ER stress (11). However, WFS1 increased steadily over a 24-hour time period (data not shown). ATF6α upregulation, on the other hand, occurred much more rapidly. Thus, the initial pool of WFS1 protein induced under stress may not have an inhibitory effect on ATF6α protein. In addition, we have shown that the ER-resident chaperone BiP also bound to ATF6α. The release of BiP from ATF6α when unfolded/misfolded proteins accumulate in the ER may be a key step in how ATF6α escapes WFS1-mediated proteolysis. BiP binding may be essential for the interaction of ATF6α and WFS1, and, upon release, cause a conformational change in ATF6α, leading to its consequent release from WFS1.
WFS1 is highly expressed in pancreatic β cells that are specialized for the production and regulated secretion of insulin to control blood glucose levels. In β cells, ER stress signaling needs to be tightly regulated to adapt to the frequent fluctuations of blood glucose levels and to produce the proper amount of insulin in response to the need for it (34, 37). To achieve tight regulation, mammals may have developed WFS1 as a regulator of HRD1 function in addition to SEL1. Higher expression of WFS1 in β cells, therefore, prevents hyperactivation of ER stress signaling in these cells that are particularly sensitive to disruption of ER homeostasis and dysregulation of the UPR. Therefore, WFS1 has a role in protecting β cells from death by acting as an ER stress signaling suppressor.
Mutations in the gene encoding WFS1 cause Wolfram syndrome, a genetic form of diabetes and neurodegeneration. It has been proposed that a high level of ER stress causes β cell death and neurodegeneration in this disorder. Collectively, our results suggest that a loss-of-function mutation of WFS1 causes the instability of an E3 ligase, HRD1, leading to the upregulation of ATF6α protein and hyperactivation of ATF6α signaling. Therefore, we predict that a loss-of-function or hypomorphic mutation of the WFS1, HRD1, or ATF6α genes can cause ER stress–related disorders, such as diabetes, neurodegeneration, and bipolar disorder. Indeed, it has been shown recently that common variants in WFS1 confer risk of type 2 diabetes (14–16), and there is a link between WFS1 mutations and type 1A diabetes (38, 39). It has also been shown that ATF6α polymorphisms and haplotypes are associated with impaired glucose homeostasis and type 2 diabetes (40). Excessive β cell loss is a component of both type 1 and type 2 diabetes (41); therefore, WFS1 may have a key role in the protection of these cells from apoptosis through the tight regulation of ER stress signaling, thereby suppressing the diabetes phenotype. In addition, about 60% of patients with Wolfram syndrome have some mental disturbance such as severe depression, psychosis, or organic brain syndrome, as well as impulsive verbal and physical aggression (42). The heterozygotes who do not have Wolfram syndrome are 26-fold more likely than noncarriers to have a psychiatric hospitalization (43). The relative risk of psychiatric hospitalization for depression was estimated to be 7.1 in these heterozygotes (44). Therefore, it is possible that dysregulation of a negative feedback loop of ER stress signaling may have a pathological role in psychiatric illness.
In this study, we focused on determining the physiological function of WFS1 in ER stress signaling because of its implication in diabetes, neurodegeneration, and bipolar disorder. We propose that WFS1 has a critical function in the regulation of ER stress signaling and prevents secretory cells, such as pancreatic β cells, from dysfunction and premature death caused by hyperactivation of ER stress signaling through its interaction with the transcription factor ATF6α. WFS1 could therefore be a key target for prevention and/or therapy of ER stress–mediated diseases such as diabetes, neurodegenerative diseases, and bipolar disorder.
Methods
Cell culture.
Rat insulinoma cells (INS1 832/13) were a gift from C. Newgard (Duke University Medical Center, Durham, North Carolina) and cultured in RPMI 1640 supplemented with 10% FBS. Mouse insulinoma cells (MIN6) were maintained in DMEM with 15% FBS and 1% sodium pyruvate. COS7 and Neuro2A cells were cultured in DMEM supplemented with 10% FBS. For generation of cells inducibly overexpressing WFS1 and GFP, INS-1 832/13 cells stably expressing pTetR were transduced with a lentivirus expressing human WFS1-FLAG or GFP and cultured in 2 μM doxycycline for 24 hours prior to protein/RNA isolation. For generation of cells stably suppressing WFS1 or GFP, MIN6 cells were transduced with a retrovirus expressing shRNA against mouse WFS1 or GFP. For overexpression of ATF6, Hrd1, and WFS1, COS7 cells were transfected with ATF6-HA and WFS1-FLAG expression plasmids using FuGENE 6 transfection reagent (Roche Applied Science). As a control for coexpression, equivalent amounts of pcDNA3 plasmid was used. DTT, cycloheximide, and MG132 were purchased from Sigma-Aldrich.
Plasmids.
ATF6 plasmids were provided by R. Prywes (Columbia University, New York). GRP78 reporter plasmid was provided by K. Mori (Kyoto University, Japan). Hrd1-myc plasmid was a gift from M. Kaneko and M. Nomura (Hokkaido University, Sapporo, Japan). TCRα plasmids were provided by R. Kopito (Stanford University, California), and NHK3 plasmids were a gift from K. Nagata (Kyoto University). Entry vectors, destination vectors, and viral plasmids for establishing lentiviral and retroviral cell lines were provided by E. Campeau (University of Massachusetts Medical School; ref. 45). shRNA against WFS1 and GFP were purchased from the shRNA Library Core Facility at the University of Massachusetts Medical School.
IB.
Cells were lysed in ice-cold TNE buffer (50 mM Tris HCl, pH 7.5; 150 mM NaCl; 1 mM EDTA; and 1% NP-40) containing a protease inhibitor cocktail (Sigma-Aldrich) for 15 minutes on ice. Lysates were then cleared by centrifuging the cells at 12,000 g for 20 minutes at 4°C. Lysates were normalized for total protein (30 μg/lane), separated using a 4%–20% linear gradient SDS-PAGE (BioRad), and electroblotted. Anti-WFS1 antibody was a gift from Y. Oka (Tohoku University). Anti-actin and anti-FLAG antibodies were purchased from Sigma-Aldrich. Anti-HA antibody was purchase from Stressgen, and anti-ATF6 and anti-GFP antibodies were purchased from Santa Cruz Biotechnology. Antigen retrieval was used for the anti-ATF6 antibody. Anti-ubiquitin and anti-IRE1 antibodies were purchased from Cell Signaling. Anti–alpha 5 20s proteasome antibody was purchased from Biomol, and anti-PERK antibody was purchased from Rockland Inc. Anti–c-myc antibody was purchased from Roche. The anti–alpha-1-antitrypsin antibody was purchased from DakoCytomation. Anti-Hrd1 antibody was generated in rabbits using a KLH-conjugated synthetic peptide, TCRMDVLRASLPAQS. The antibody specificity was tested by peptide/antigen competition.
DTT chase.
INS-1 832/13 cells were treated with 1 mM DTT for 2 hours. The DTT was washed out with normal media (RPMI 1640 supplemented with 10% FBS) for 0, 1, or 2 hours. Cells were lysed and subjected to IP with anti-WFS1 antibodies.
Fractionation.
The ER was isolated from INS-1 832/13 cells using an Endoplasmic Reticulum Isolation kit (Sigma-Aldrich). The ER pellet was then lysed in ice-cold TNE buffer containing 1% NP-40 and protease inhibitors, and the lysates were cleared and normalized. The ER lysates (1.0 ml) were loaded on top of a glycerol gradient (10%–40%) prepared in PBS containing 1 mM DTT and 2 mM ATP and centrifuged at 4°C and 80,000 g for 20 hours. We collected 32 fractions from the top of the tubes. Of each fraction, 200 μl was precipitated with acetone, and the remaining pellet was lysed with 50 μl sample buffer. Precipitated proteins were then separated using a 4%–20% linear gradient SDS-PAGE and electroblotted.
IP.
Cells were lysed in ice-cold TNE buffer with protease inhibitors for 15 minutes on ice; the lysates were then cleared by centrifuging the cells at 12,000 g for 20 minutes at 4°C. For IP of endogenous WFS1, 500 μg whole cell extract from each sample was incubated with Protein G Sepharose 4 Fast Flow beads (GE Healthcare) and 4 μg anti-WFS1 antibody overnight at 4°C with rotation. After incubation, the beads were washed 3 times with TNE buffer followed by a final wash in 1× PBS. The IPs were resolved by SDS-PAGE and then subjected to IB. For IP of ATF6, 6 μg anti-ATF6 antibody was used; for HA, 2 μg anti-HA antibody was used; and for Hrd1, 4 μg anti-Hrd1 antibody was used. As a control, lysates were subjected to IP as described above using rabbit IgG.
Real-time PCR.
Total RNA was isolated from the cells using the RNeasy Mini Kit (Qiagen) and reverse transcribed using 1 μg total RNA from cells with Oligo-dT primer. For the thermal cycle reaction, the iQ5 system (BioRad) was used at 95°C for 10 minutes, then 40 cycles at 95°C for 10 seconds, and 55°C for 30 seconds. The relative amount for each transcript was calculated by a standard curve of cycle thresholds for serial dilutions of cDNA sample and normalized to the amount of actin. PCR was performed in triplicate for each sample; all experiments were repeated 3 times. The following sets of primers and Power SYBR Green PCR Master Mix (Applied Biosystems) were used for real-time PCR: rat actin, GCAAATGCTTCTAGGCGGAC and AAGAAAGGGTGTAAAACGCAGC; rat BiP, TGGGTACATTTGATCTGACTGGA and CTCAAAGGTGACTTCAATCTGGG; rat Chop, AGAGTGGTCAGTGCGCAGC and CTCATTCTCCTGCTCCTTCTCC; rat total XBP-1, TGGCCGGGTCTGCTGAGTCCG and ATCCATGGGAAGATGTTCTGG; rat ERO1-α, GAGAAGCTGTAATAGCCACGAGG and GAGCCTTTCAATAAGCGGACTG; rat GLUT2, GTGTGAGGATGAGCTGCCTAAA and TTCGAGTTAAGAGGGAGCGC; rat INS2, ATCCTCTGGGAGCCCCGC and AGAGAGCTTCCACCAAG.
Luciferase assay.
COS7 cells were mock transfected, transfected with full-length ATF6 or ΔATF6 with pcDNA3.0, or transfected with ATF6 with WFS1 expression plasmids along with rat GRP78 (ERSE) promoter luciferase reporter gene, WT ATF6 binding site luciferase reporter gene (ATF6GL3), or mutant ATF6 binding site luciferase reporter gene (ATF6m1GL3) using Lipofectamine 2000 (Invitrogen). At 48 hours after transfection, lysates were prepared using a Luciferase Assay System kit (Promega). The light produced from the samples was read by a standard plate reading luminometer. Each sample was read in triplicate and normalized against the signal produced from mock wells. All experiments were repeated 3 times.
Wfs1–/– animals.
Wfs1–/– mice were provided by M.A. Permutt (Washington University, St. Louis, Missouri). All procedures were reviewed and approved by the University of Massachusetts Medical School IACUC (assurance no. A-3306-01).
Statistics.
To determine whether the treatment was significantly different from the control, 2-tailed paired Student’s t test was used. A P value less than 0.01 was considered statistically significant. In the figures, numbers below the lanes of blots denote relative protein amounts, as quantified by ImageJ software, normalized to the respective control. All graphical data are shown as mean ± SD.
Supplementary Material
Acknowledgments
We thank Keiji Tanaka, Hideki Yashiroda, Reid Gilmore, Sara Evans, and Michael R. Green for comments on the manuscript; Julie Zhu for statistical analysis; Karen Sargent, Jessica Crisci, Yuan Lee, Linda Leehy, and Cris Welling for technical assistance; and Carlos Fonseca for support and advice. This work was supported by grants from NIDDK, NIH (R01 DK067493), the Diabetes and Endocrinology Research Center at the University of Massachusetts Medical School (5 P30 DK32520), the Juvenile Diabetes Research Foundation International, the Worcester Foundation for Biomedical Research, and the Iacocca Foundation to F. Urano.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(3):744–755. doi:10.1172/JCI39678.
References
- 1.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 2.Rutkowski DT, Kaufman RJ. That which does not kill me makes me stronger: adapting to chronic ER stress. Trends Biochem Sci. 2007;32(10):469–476. doi: 10.1016/j.tibs.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10(11):3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshida H, et al. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20(18):6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye J, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6(6):1355–1364. doi: 10.1016/S1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 6.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3(1):99–111. doi: 10.1016/S1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 7.Wu J, et al. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13(3):351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto K, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13(3):365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 9.Hong M, Li M, Mao C, Lee AS. Endoplasmic reticulum stress triggers an acute proteasome-dependent degradation of ATF6. J Cell Biochem. 2004;92(4):723–732. doi: 10.1002/jcb.20118. [DOI] [PubMed] [Google Scholar]
- 10.Takeda K, et al. WFS1 (Wolfram syndrome 1) gene product: predominant subcellular localization to endoplasmic reticulum in cultured cells and neuronal expression in rat brain. Hum Mol Genet. 2001;10(5):477–484. doi: 10.1093/hmg/10.5.477. [DOI] [PubMed] [Google Scholar]
- 11.Fonseca SG, et al. WFS1 is a novel component of the unfolded protein response and maintains homeostasis of the endoplasmic reticulum in pancreatic {beta}-cells. J Biol Chem. 2005;280(47):39609–39615. doi: 10.1074/jbc.M507426200. [DOI] [PubMed] [Google Scholar]
- 12.Strom TM, et al. Diabetes insipidus, diabetes mellitus, optic atrophy and deafness (DIDMOAD) caused by mutations in a novel gene (wolframin) coding for a predicted transmembrane protein. Hum Mol Genet. 1998;7(13):2021–2028. doi: 10.1093/hmg/7.13.2021. [DOI] [PubMed] [Google Scholar]
- 13.Inoue H, et al. A gene encoding a transmembrane protein is mutated in patients with diabetes mellitus and optic atrophy (Wolfram syndrome). Nat Genet. 1998;20(2):143–148. doi: 10.1038/2441. [DOI] [PubMed] [Google Scholar]
- 14.Lyssenko V, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. . N Engl J Med. 2008;359(21):2220–2232. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 15.Franks PW, et al. Replication of the association between variants in WFS1 and risk of type 2 diabetes in European populations. Diabetologia. 2008;51(3):458–463. doi: 10.1007/s00125-007-0887-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandhu MS, et al. Common variants in WFS1 confer risk of type 2 diabetes. Nat Genet. 2007;39(8):951–953. doi: 10.1038/ng2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada T, et al. WFS1-deficiency increases endoplasmic reticulum stress, impairs cell cycle progression and triggers the apoptotic pathway specifically in pancreatic beta-cells. Hum Mol Genet. 2006;15(10):1600–1609. doi: 10.1093/hmg/ddl081. [DOI] [PubMed] [Google Scholar]
- 18.Riggs AC, et al. Mice conditionally lacking the Wolfram gene in pancreatic islet beta cells exhibit diabetes as a result of enhanced endoplasmic reticulum stress and apoptosis. Diabetologia. 2005;48(11):2313–2321. doi: 10.1007/s00125-005-1947-4. [DOI] [PubMed] [Google Scholar]
- 19.Kakiuchi C, Ishiwata M, Hayashi A, Kato T. XBP1 induces WFS1 through an endoplasmic reticulum stress response element-like motif in SH-SY5Y cells. J Neurochem. 2006;97(2):545–555. doi: 10.1111/j.1471-4159.2006.03772.x. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275(35):27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- 21.Shen J, Snapp EL, Lippincott-Schwartz J, Prywes R. Stable binding of ATF6 to BiP in the endoplasmic reticulum stress response. Mol Cell Biol. 2005;25(3):921–932. doi: 10.1128/MCB.25.3.921-932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu H, Kaung G, Kobayashi S, Kopito RR. Cytosolic degradation of T-cell receptor alpha chains by the proteasome. J Biol Chem. 1997;272(33):20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
- 23.Yu H, Kopito RR. The role of multiubiquitination in dislocation and degradation of the alpha subunit of the T cell antigen receptor. J Biol Chem. 1999;274(52):36852–36858. doi: 10.1074/jbc.274.52.36852. [DOI] [PubMed] [Google Scholar]
- 24.Hosokawa N, et al. A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2001;2(5):415–422. doi: 10.1093/embo-reports/kve084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaneko M, et al. A different pathway in the endoplasmic reticulum stress-induced expression of human HRD1 and SEL1 genes. FEBS Lett. 2007;581(28):5355–5360. doi: 10.1016/j.febslet.2007.10.033. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto K, et al. Human HRD1 promoter carries a functional unfolded protein response element to which XBP1 but not ATF6 directly binds. J Biochem. 2008;144(4):477–486. doi: 10.1093/jb/mvn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hampton RY, Gardner RG, Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1996;7(12):2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardner RG, et al. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000;151(1):69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishihara H, et al. Disruption of the WFS1 gene in mice causes progressive beta-cell loss and impaired stimulus-secretion coupling in insulin secretion. Hum Mol Genet. 2004;13(11):1159–1170. doi: 10.1093/hmg/ddh125. [DOI] [PubMed] [Google Scholar]
- 30.Kudo T, et al. A molecular chaperone inducer protects neurons from ER stress. Cell Death Differ. 2008;15(2):364–375. doi: 10.1038/sj.cdd.4402276. [DOI] [PubMed] [Google Scholar]
- 31.Martindale JJ, et al. Endoplasmic reticulum stress gene induction and protection from ischemia/reperfusion injury in the hearts of transgenic mice with a tamoxifen-regulated form of ATF6. Circ Res. 2006;98(9):1186–1193. doi: 10.1161/01.RES.0000220643.65941.8d. [DOI] [PubMed] [Google Scholar]
- 32.Ye R, et al. Grp78 heterozygosity promotes adaptive unfolded protein response and attenuates diet-induced obesity and insulin resistance. Diabetes. 2010;59(1):6–16. doi: 10.2337/db09-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han D, et al. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138(3):562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipson KL, et al. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 2006;4(3):245–254. doi: 10.1016/j.cmet.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Cnop M, et al. Selective inhibition of eukaryotic translation initiation factor 2 alpha dephosphorylation potentiates fatty acid-induced endoplasmic reticulum stress and causes pancreatic beta-cell dysfunction and apoptosis. J Biol Chem. 2007;282(6):3989–3997. doi: 10.1074/jbc.M607627200. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell. 2003;4(2):265–271. doi: 10.1016/S1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 37.Fonseca SG, Lipson KL, Urano F. Endoplasmic reticulum stress signaling in pancreatic beta-cells. Antioxid Redox Signal. 2007;9(12):2335–2344. doi: 10.1089/ars.2007.1790. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura A, et al. A novel mutation of WFS1 gene in a Japanese man of Wolfram syndrome with positive diabetes-related antibodies. Diabetes Res Clin Pract. 2006;73(2):215–217. doi: 10.1016/j.diabres.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 39.Awata T, et al. Missense variations of the gene responsible for Wolfram syndrome (WFS1/wolframin) in Japanese: possible contribution of the Arg456His mutation to type 1 diabetes as a nonautoimmune genetic basis. Biochem Biophys Res Commun. 2000;268(2):612–616. doi: 10.1006/bbrc.2000.2169. [DOI] [PubMed] [Google Scholar]
- 40.Meex SJ, et al. Activating transcription factor 6 polymorphisms and haplotypes are associated with impaired glucose homeostasis and type 2 diabetes in Dutch Caucasians. J Clin Endocrinol Metab. 2007;92(7):2720–2725. doi: 10.1210/jc.2006-2280. [DOI] [PubMed] [Google Scholar]
- 41.Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52(1):102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 42.Swift RG, Sadler DB, Swift M. Psychiatric findings in Wolfram syndrome homozygotes. Lancet. 1990;336(8716):667–669. doi: 10.1016/0140-6736(90)92157-D. [DOI] [PubMed] [Google Scholar]
- 43.Swift RG, Polymeropoulos MH, Torres R, Swift M. Predisposition of Wolfram syndrome heterozygotes to psychiatric illness. Mol Psychiatry. 1998;3(1):86–91. doi: 10.1038/sj.mp.4000344. [DOI] [PubMed] [Google Scholar]
- 44.Swift M, Swift RG. Wolframin mutations and hospitalization for psychiatric illness. Mol Psychiatry. 2005;10(8):799–803. doi: 10.1038/sj.mp.4001681. [DOI] [PubMed] [Google Scholar]
- 45.Campeau E, et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS ONE. 2009;4(8):e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.