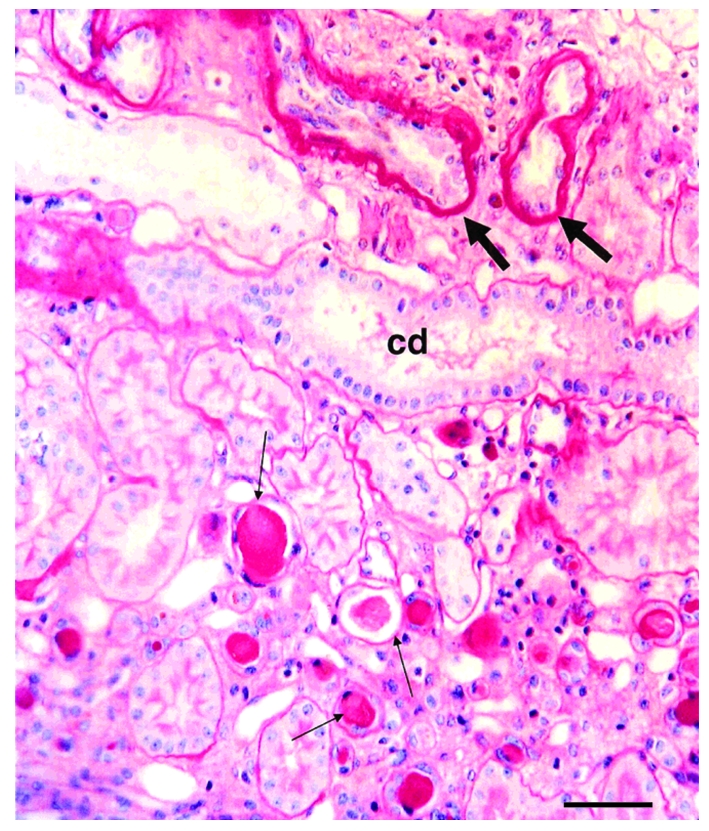
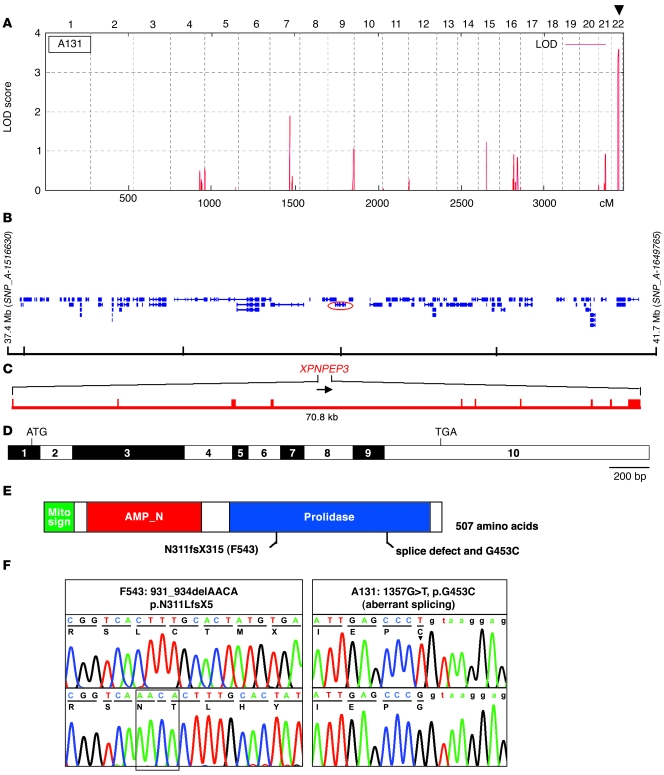
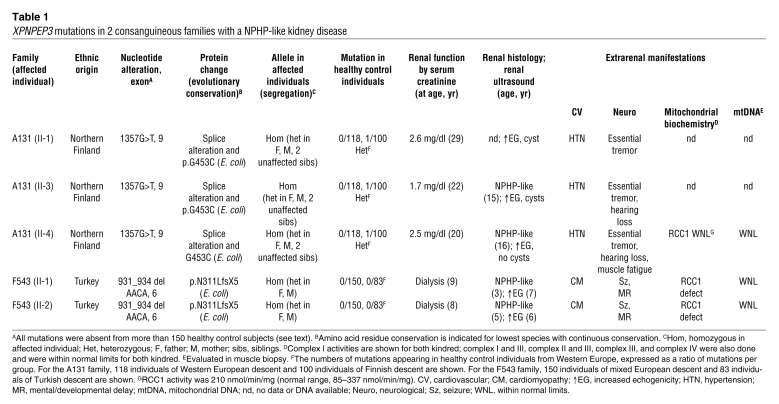
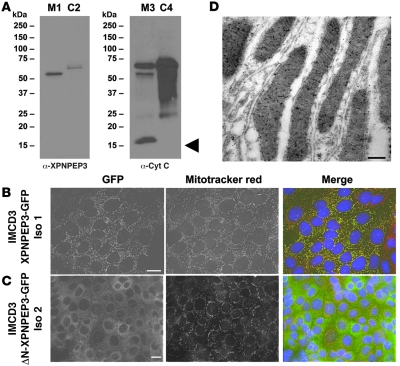
John F O’Toole
John F O’Toole
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
1,
Yangjian Liu
Yangjian Liu
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
2,
Erica E Davis
Erica E Davis
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
2,3,
Christopher J Westlake
Christopher J Westlake
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
4,
Massimo Attanasio
Massimo Attanasio
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
1,
Edgar A Otto
Edgar A Otto
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
1,
Dominik Seelow
Dominik Seelow
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
5,6,
Gudrun Nurnberg
Gudrun Nurnberg
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
5,
Christian Becker
Christian Becker
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
5,
Matti Nuutinen
Matti Nuutinen
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
7,
Mikko Kärppä
Mikko Kärppä
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
7,
Jaakko Ignatius
Jaakko Ignatius
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
7,
Johanna Uusimaa
Johanna Uusimaa
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
7,
Salla Pakanen
Salla Pakanen
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
7,
Elisa Jaakkola
Elisa Jaakkola
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
7,
Lambertus P van den Heuvel
Lambertus P van den Heuvel
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
8,
Henry Fehrenbach
Henry Fehrenbach
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
9,
Roger Wiggins
Roger Wiggins
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
10,
Meera Goyal
Meera Goyal
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
10,
Weibin Zhou
Weibin Zhou
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
1,
Matthias TF Wolf
Matthias TF Wolf
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
1,
Eric Wise
Eric Wise
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
1,
Juliana Helou
Juliana Helou
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
1,
Susan J Allen
Susan J Allen
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
1,
Carlos A Murga-Zamalloa
Carlos A Murga-Zamalloa
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
11,
Shazia Ashraf
Shazia Ashraf
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
1,
Moumita Chaki
Moumita Chaki
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
1,
Saskia Heeringa
Saskia Heeringa
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
1,
Gil Chernin
Gil Chernin
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
1,
Bethan E Hoskins
Bethan E Hoskins
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
1,
Hassan Chaib
Hassan Chaib
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
1,
Joseph Gleeson
Joseph Gleeson
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
12,
Takehiro Kusakabe
Takehiro Kusakabe
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
13,14,
Takako Suzuki
Takako Suzuki
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
13,14,
R Elwyn Isaac
R Elwyn Isaac
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
15,
Lynne M Quarmby
Lynne M Quarmby
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
16,
Bryan Tennant
Bryan Tennant
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
16,
Hisashi Fujioka
Hisashi Fujioka
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
17,
Hannu Tuominen
Hannu Tuominen
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
18,
Ilmo Hassinen
Ilmo Hassinen
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
19,
Hellevi Lohi
Hellevi Lohi
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
20,
Judith L van Houten
Judith L van Houten
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
21,
Agnes Rotig
Agnes Rotig
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
22,
John A Sayer
John A Sayer
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
1,23,
Boris Rolinski
Boris Rolinski
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
24,
Peter Freisinger
Peter Freisinger
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
24,
Sethu M Madhavan
Sethu M Madhavan
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
25,
Martina Herzer
Martina Herzer
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
26,
Florence Madignier
Florence Madignier
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
26,
Holger Prokisch
Holger Prokisch
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
26,27,
Peter Nurnberg
Peter Nurnberg
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
5,28,
Peter Jackson
Peter Jackson
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
4,
Hemant Khanna
Hemant Khanna
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
11,
Nicholas Katsanis
Nicholas Katsanis
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
2,3,29,30,
Friedhelm Hildebrandt
Friedhelm Hildebrandt
1Department of Pediatrics, University of Michigan, Ann Arbor.
2McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
3Center for Human Disease Modeling, Department of Cell Biology, Duke University Medical Center, Durham, North Carolina.
4Genentech Inc., South San Francisco, California.
5Cologne Center for Genomics and Institute for Genetics, University of Cologne, Germany.
6Department of Neuropediatrics, Charite – Universitaetsmedizin Berlin, Germany.
7Department of Pediatrics and Adolescents, Department of Clinical Genetics, and Department of Neurology, Oulu University Hospital, Finland.
8Department of Pediatrics, Nijmegen Centre for Mitochondrial Disorders, Radboud University Nijmegen Medical Centre, Netherlands.
9Kinderklinik Memmingen, Germany.
10Department of Internal Medicine,
11Department of Ophthalmology and Visual Sciences, W.K. Kellogg Eye Center, University of Michigan.
12Howard Hughes Medical Institute, Department of Neurosciences, University of California at San Diego, La Jolla.
13Department of Life Science, Graduate School of Life Science, University of Hyogo, Japan.
14Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan.
15Institute of Integrative and Comparative Biology, Faculty of Biological Sciences, University of Leeds, United Kingdom.
16Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, Canada.
17Department of Pharmacology, Case Western Reserve University, Cleveland, Ohio.
18Department of Pathology and
19Department of Medical Biochemistry, University of Oulu, Finland.
20Department of Internal Medicine, Lappland Central Hospital, Rovaniemi, Finland.
21Department of Biology and Vermont Chemosensory Group, University of Vermont, Burlington.
22Département de Génétique, Université Paris Descartes, Unité INSERM U781, Hôpital Necker-Enfants Malades, France.
23Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, United Kingdom.
24Academic Hospital Munchen-Schwabing, Institute for Clinical Chemistry and Metabolic Disease Center, and Department of Pediatrics, Technical University Munich, Germany.
25Department of Medicine, MetroHealth Medical System, Cleveland, Ohio.
26Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Center for Environmental Health, Neuherberg, Germany.
27Institute of Human Genetics, Klinikum rechts der Isar, Technical University Munich, Germany.
28Center for Molecular Medicine and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, University of Cologne.
29Wilmer Eye Institute and
30Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine.
31Human Genetics and
32Howard Hughes Medical Institute, University of Michigan.
1,31,32