Abstract
The chemokines are a large family of mainly secreted molecules involved in the regulation of numerous physiological and pathophysiological processes. Despite many years of investigation, the precise cellular sources of most chemokines have remained incompletely defined as a consequence of the limited availability of suitable reagents to visualize the expression of chemokine proteins at the single-cell level. Here, we developed a simple flow cytometry–based assay using commercially available chemokine-specific antibodies for efficient cell-associated detection of 37 of 39 murine chemokines. To demonstrate the utility of this methodology, we used it to reevaluate the nature of homeostatic chemokines in the hematopoietic compartment, to delineate the complete chemokine profiles of NK cells and B cells in response to major polyclonal stimuli, and to assess the chemokine response of DCs to bacterial infection. The versatility of this analytical methodology was further demonstrated by its application to selected human chemokines and should greatly facilitate any future investigation into chemokine biology at large.
Introduction
The term chemokine, a linguistic contraction derived from chemotactic cytokine, describes a large family of mostly secreted small molecules involved in numerous physiological and pathophysiological processes. Although their nominal function, the control of directed cell migration, constitutes a defining attribute, chemokines exert a host of additional activities that modulate many fundamental properties of cellular function (1–13). With the completion of the human and murine genomes, the rapid discovery of novel chemokines has come to an apparent conclusion, and the chemokines may in fact be one of the first mammalian superfamilies known in its entirety (14). Based on a defining tetracysteine motif, chemokines can be divided into 4 distinct subfamilies (15): CC chemokines (CCL1–CCL28) contain 2 adjacent cysteine residues near their amino terminus that are separated by a single nonconserved amino acid in the CXC chemokines (CXCL1–CXCL17) and by 3 amino acids in the sole CX3C member, CX3CL1; the C chemokines (XCL1/2 in humans, only XCL1 in mouse) lack 1 of the first 2 cysteine residues found in the other subfamilies. In general, this subdivision limits chemokine binding to members of certain chemokine receptor subgroups. However, among the CC and CXC subfamilies, a degree of redundancy and promiscuity exists, with some receptors able to bind multiple chemokines and some chemokines capable of engaging multiple receptors. It should be noted that the structure-based taxonomy of chemokines does not supersede, but rather complements, an older classification according to functional properties (homeostatic, inflammatory, and dual-use chemokines) or the alternative clustering based on genomic organization (14).
The extraordinary complexity of the chemokine system emerges from the confluence of several factors. Excluding pseudogenes and multiple copy numbers for some chemokine genes, the murine genome contains 40 distinct genes that give rise to 39 unique chemokine proteins (14), as the Ccl21b and Ccl21c gene products are identical (Table 1). In addition to the large number of chemokine family members (at least 46 in humans), the presence of splice variants and extensive posttranslational modifications, existence of promiscuous receptor binding and receptor-independent binding, formation of hetero-oligomeric chemokine complexes, dynamic expression patterns and functional diversity combine to generate an exceedingly broad spectrum of possible chemokine activities (1, 16, 17). Although the transcriptional expression patterns of many chemokines have been detailed in various experimental and clinical settings, analytical access to specific chemokine-secreting cell types has remained somewhat limited, given methodological approaches preferentially reliant on immunoblots, ELISA assays, and/or immunohistochemistry (IHC).
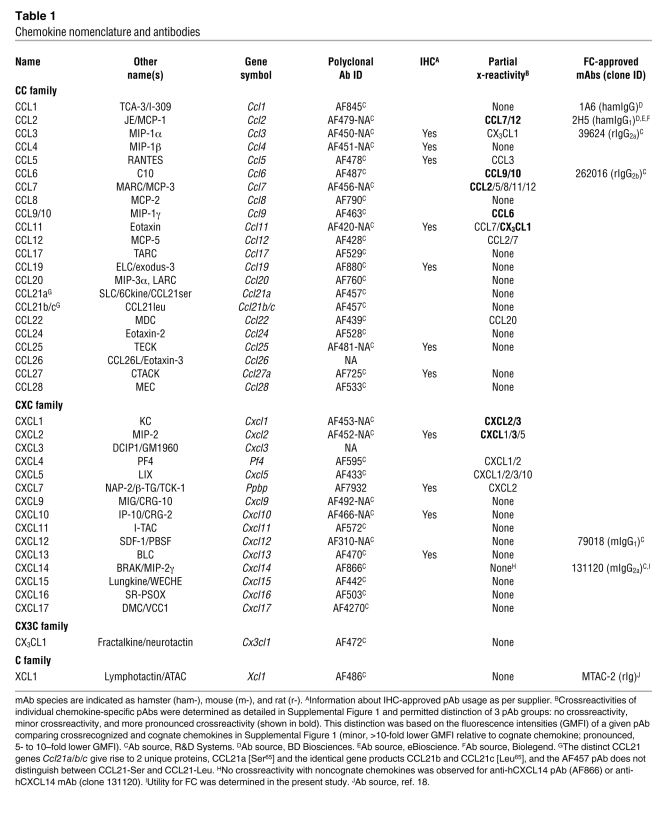
Table 1 .
Chemokine nomenclature and antibodies
The analytical method of choice for the detection of chemokine proteins in defined cellular subsets is flow cytometry (FC), which allows for multiparametric analysis of individual chemokine-producing cells within larger cell populations of interest. Here, the preferred tools are chemokine-specific mAbs conjugated to fluorochromes; however, although the list of FC-approved mAbs is growing, no such reagents are available for the majority of murine chemokines (Table 1). Polyclonal Abs (pAbs) constitute an appropriate alternative, and indeed have been used for the flow cytometric detection of selected murine chemokines in a variety of immune cell subsets, such as T cells, NK cells, NKT cells, DCs, monocyte/macrophages (Mo/Mϕ), granulocytes, and others (18–27). However, not all studies have rigorously excluded potential crossreactivities of these reagents, and, to our knowledge, direct visualization by means of FC has not been reported for most murine chemokines.
The use of pAbs rather than mAbs for detection of intracellular antigens offers a number of challenges and some advantages that have to be addressed in order to assure their reliable usage for FC (see Methods). With the aim to develop comprehensive analytical access to all known murine chemokines, we have selected, tested, and validated a panel of commercially available affinity-purified pAbs specific for 37 of 39 murine chemokines for use in FC (Table 1). To demonstrate the principal utility of our approach to chemokine FC, we applied this methodology to an identification of homeostatic chemokines and the principal hematopoietic cell subsets in the spleen involved in their expression (Table 2). In addition, we have delineated the complete chemokine profiles of NK and B cells in response to major stimuli and defined the DC chemokine response to infection (Table 2).
Table 2 .
Summary of chemokine expression patterns and cellular subsets
Results
Development of a FC-based assay for detection of murine chemokines
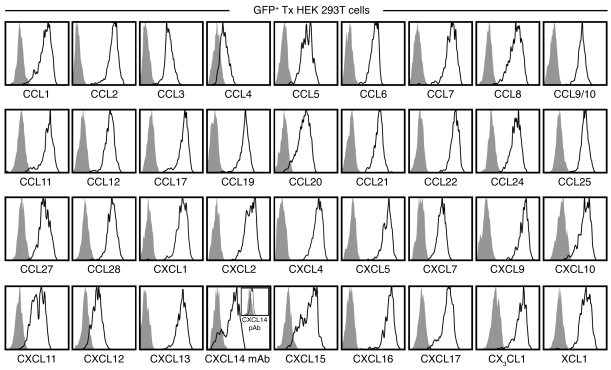
To develop a comprehensive tool set for the detection of murine chemokines by FC, we evaluated a large panel of commercially available chemokine-specific Abs. Given the scarcity of mAbs suitable for this application (Table 1), we focused our attention on pAbs and defined several criteria for their effective and reliable use in FC (see Methods). Here, HEK 293T cells were transfected with bicistronic GFP vectors containing individual chemokine genes and subsequently stained with the respective chemokine-specific pAbs for concurrent visualization of the reporter gene and chemokine protein by FC (see Methods). Our results, displayed in Figure 1 and summarized in Table 1, identified 36 chemokine-specific pAbs suitable for the flow cytometric detection of 37 of 39 cell-associated chemokines (the anti-CCL21 pAb does not discriminate between CCL21-Ser and CCL21-Leu; Table 1). Detection of CXCL14 posed a particular challenge, as a result of several pAbs being found unsuitable for FC and a failure of intracellular CXCL14 protein accumulation (data not shown). The latter observation likely resulted from proteasomal degradation, as previously reported for human CXCL14 (hCXCL14) transfected into HEK and other cancer cell lines (28), as well as the fact that the unique “destruction sequence” identified by Peterson et al. (28) is largely preserved in murine CXCL14 (mCXCL14). We therefore cultured Cxcl14-transfected HEK cells in the presence of a protease inhibitor, permitting specific detection of CXCL14 at low levels with the anti-hCXCL14 pAb AF866, but not the anti-mCXCL14 pAb AF730 (Figure 1 and data not shown; mouse and human CXCL14 are 95% identical). In contrast, robust CXCL14 expression was revealed with an anti-hCXCL14 mAb, which consequently became our preferred reagent for analysis of mCXCL14 expression by FC. Finally, no suitable reagents could be identified for the specific detection of the 2 remaining chemokines, CCL26 and CXCL3 (data not shown).
Figure 1. Validation of chemokine-specific pAb use for FC.
HEK 293T cells were transfected (Tx) with individual pIRES2-AcGFP1 vectors containing Il15 (negative control) or 36 distinct murine chemokines and cultured for 18 hours. BFA was added for the final 14 hours to limit chemokine secretion, and cells were analyzed for expression of corresponding chemokine proteins by FC as detailed in Methods. All histograms are gated on GFP+ HEK cells comparing Il15 transfectants (gray solid) and respective chemokine transfectants (black tracing) stained with the same chemokine-specific pAb. To reduce proteasomal degradation, Cxcl14-transfected HEK cells were cultured in the presence of 10 μM of the protease inhibitor MG-132 and stained with anti-hCXCL14 mAb clone 131120 or with an anti-hCXCL14 pAb (inset).
Perhaps the main concern regarding the use of pAbs pertains to their enhanced potential for crossreactivity. The specificities of many pAbs used in the present study have been verified by immunoblot and/or ELISA using large panels of recombinant chemokines (see data sheets for pAbs listed in Table 1), yet none of the pAbs have been evaluated in a systematic fashion for potential crossreactivities when used in FC. To this end, we determined the reactivity of all 36 chemokine pAbs with each of the 36 HEK chemokine transfectants, as shown in Supplemental Figure 1 (supplemental material available online with this article; doi: 10.1172/JCI40645DS1) and summarized in Table 1. Although the majority of chemokine pAbs exclusively recognized their cognate antigen, 14 pAbs exhibited partial reactivity with noncognate chemokines. The majority of crossreactivities were focused on chemokines that belong to the CC-MCP (CCL2/7/8/11/12) or CXC-GRO (CXCL1–CXCL7) clusters (ref. 14 and Supplemental Figure 1), emphasizing the role of structural homologies in shaping the crossreactive pAb repertoire. Importantly, however, the precise degree of crossreactivity was for the most part minor (>10-fold lower staining intensities for crossrecognized versus cognate chemokines; Table 1); moreover, as detailed below, the respective pAbs retained their principal utility for analyses of primary cells, especially when used in combination and complemented with transcriptome analyses.
Reevaluation of homeostatic chemokine expression by means of FC
The distinction between homeostatic and inflammatory chemokines emphasizes the biological context in which individual chemokines exert their function and reflects a consensus that has been established in a multitude of different experimental systems (14). The more recent introduction of a third category, dual-use chemokines, testifies to the evolving nature of this conceptual distinction, as it incorporates novel observations that resist a precise assignment to either of the 2 prevailing categories (14). Because the exact criteria guiding attribution — to the category of homeostatic chemokine in particular — are thus in flux, we introduce a definition for the purpose of the present study based on chemokine FC as the principal analytical readout: Homeostatic chemokines are those chemokines detectable in defined cellular subsets in the absence of exogenous stimuli. More precisely, we distinguish constitutive chemokine protein expression (detectable directly ex vivo) from spontaneous chemokine production (after a 5-hour culture in the presence of the protein transport inhibitor brefeldin A [BFA], but in the absence of specific stimuli) that most likely results from the translation of preexisting chemokine transcripts. An investigation into the extent to which the homeostatic chemokines identified here contribute to immune homeostasis, however, is beyond the scope of the present study.
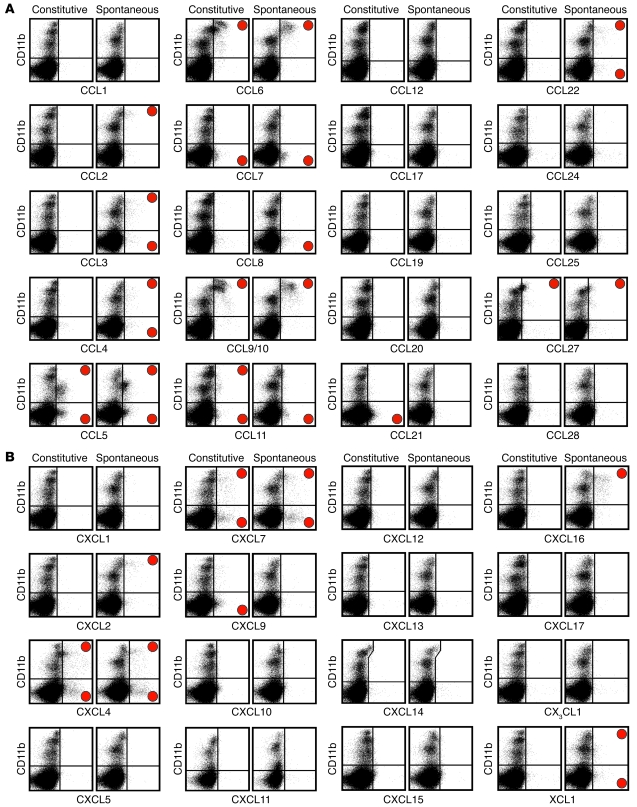
Remarkably, nearly half the murine chemokines evaluated (18 of 37) were produced in a constitutive and/or spontaneous fashion in cells recovered from the spleens of unmanipulated B6 mice (Figure 2). Pronounced constitutive chemokine expression was observed for CCL5, CCL6, CCL9/10, CCL21, CCL27, CXCL4, and CXCL7. In contrast, only low-level expression of CCL7, CCL8, CCL11, CXCL9, and XCL1 was detected directly ex vivo in smaller splenic hematopoietic cell subsets, and several additional chemokines (CCL2, CCL3, CCL4, CCL22, CXCL2, and CXCL16) were synthesized after a brief in vitro culture in a spontaneous manner. Two-thirds of the above chemokines, namely CCL2/3/4/5/6/7/8/11 and CXCL2/7/9/16, are traditionally regarded as inflammatory (14), and their constitutive and/or spontaneous expression observed here suggests that these chemokines may also exert previously unrecognized homeostatic functions. Whether the presence of the latter chemokines under conditions of immune homeostasis warrants their reclassification as dual-use members of the chemokine superfamily may remain a matter of debate. However, it is important to emphasize that the simple presence of many inflammatory chemokines, as shown here, does not necessarily reflect activation of immune cells in response to pathological processes.
Figure 2. Visualization and identification of homeostatic chemokines in splenic hematopoietic cells.
(A and B) Spleen cells obtained from naive B6 mice (or, for CXCL11 analyses, from Balb/c mice) were analyzed for chemokine expression directly ex vivo (constitutive) or after a 5-hour culture in the presence of BFA but absent any exogenous stimuli (spontaneous) as detailed in Methods. All plots are gated on “live” cells, as determined by forward/side scatter properties. Red circles denote chemokine-positive staining in the respective quadrants. (A) CC family. (B) CXC, CX3C, and C families. Note that our analyses of CXCL14 expression with a preconjugated anti-hCXCL14 mAb (clone 131120) were associated with nonspecific staining of a CD11b++ cell subset (identical staining pattern observed with a preconjugated mIgG2a isotype control; not shown). Data are compiled from multiple independent experiments (n = 2–3 mice), with individual chemokine stains repeated at least twice.
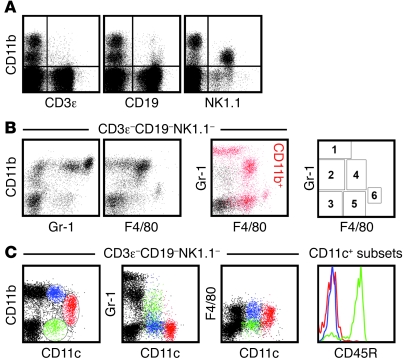
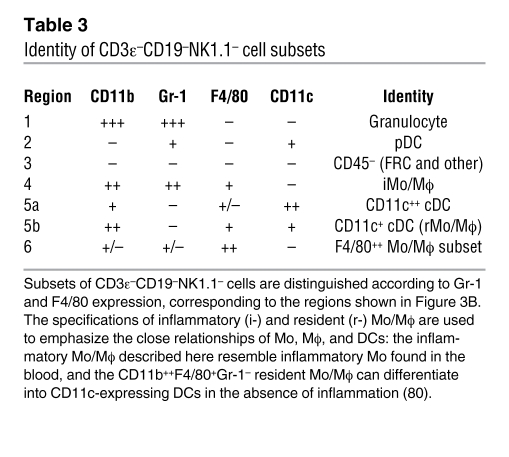
To identify cellular subsets capable of homeostatic chemokine production, we initially focused on cells expressing CD11b (Figure 2), an integrin prominently expressed by granulocyte, Mo/Mϕ, and DC subsets. However, CD11b is also expressed by activated T cells, small B cell subsets, and mature NK cells (refs. 29–32 and Figure 3A), and, a more detailed demarcation of innate immune cell populations can therefore be achieved by analytical exclusion of B cells (CD19+), T cells (CD3ε+), NK cells (NK1.1+), and NKT cells (NK1.1+CD3ε+) and by expression analysis of 2 additional markers: Gr-1, composed of Ly6C and Ly6G antigens, and the glycoprotein F4/80 (Figure 3B). The resulting delineation of 6 cellular subsets, combined with a determination of CD11c expression levels (Figure 3C), permits the identification of granulocytes, Mo/Mϕ, conventional DCs (cDCs, with CD11c+ and CD11c++ subsets), CD45R/B220+ plasmacytoid DCs (pDCs), a small population of F4/80++ Mo/Mϕ, and a group of CD11b–Gr-1–F4/80–CD11c– cells, many of which lack expression of CD45 (Table 3). These distinctions correspond well to a contemporary phenotypic differentiation of Mo/Mϕ, DC, and granulocyte subsets (33, 34).
Figure 3. Phenotypic distinction of CD3ε–CD19–NK1.1– cell populations.
Single-cell suspensions prepared from spleens of naive B6 mice were analyzed directly ex vivo for expression of various cell surface markers. (A) CD11b expression by small subsets of CD3ε+ T and CD19+ B cells as well as mature NK cells. (B) Analytical exclusion of T cells (CD3ε+), B cells (CD19+), NK cells (NK1.1+), and NKT cells (NK1.1+CD3ε+) permitted delineation of 6 cellular subsets according to expression of CD11b, Gr-1 (Ly6C/G), and F4/80 antigens. In the 2-color dot plot, CD11b-expressing cells are identified as red events. The plot at right demarcates regions corresponding to the 6 distinct cell subsets in the 2-color dot plot, whose identities are summarized in Table 3. (C) Major DC subsets were distinguished based on CD11b and CD11c expression patterns (left), with respective Gr-1, F4/80, and CD45R (B220) expression profiles shown at right, distinguished by color.
Table 3 .
Identity of CD3ε–CD19–NK1.1– cell subsets
Delineation of cellular subsets exhibiting homeostatic or induced chemokine expression
The combined analysis of complex cellular phenotypes and chemokine expression by FC allows for the precise characterization of chemokine-producing cell subsets under varied experimental conditions. Taking into account considerations and experimental strategies that address the partial crossrecognition of noncognate chemokines by several of the pAbs (Table 1), we first focused on homeostatic chemokine expression (Figure 4) and complemented these analyses with a general evaluation of induced chemokine production (Supplemental Figures 2 and 3) before assessing the complete chemokine response of specific immune cell subsets (NK cells, B cells, and DCs) in more detail. All data on cellular chemokine expression profiles are summarized in Table 2.
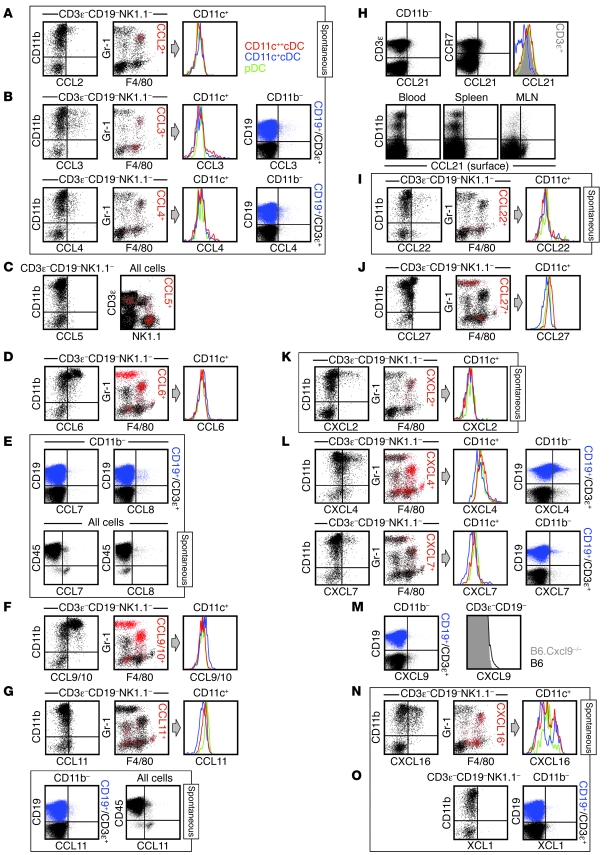
Figure 4. Delineation of cellular subsets expressing homeostatic chemokines.
Spleen cells obtained from naive mice were analyzed directly ex vivo (constitutive) or after 5 hours of in vitro culture in the presence of BFA (spontaneous; identified by labeled boxes). For chemokines preferentially expressed by CD11b+ cell subsets (A, B, D, F, G, I–L, and N), dot plots are gated on CD3ε–CD19–NK1.1– as indicated. In the 2-color black/red dot plots, chemokine-expressing cells are identified as red events; histograms are gated on CD11c-expressing cells and color-coded as in A. For chemokines expressed by CD11b– cells (B, E, G, and L–N), 2-color dot plots are gated on B cells (CD19+CD3ε–CD11b–, blue) and T cells (CD19–CD3ε+CD11b–, black). (C, E, G, H, and M) Note the partially different gating strategies used for analysis of CCL5/7/8/11/21 and CXCL9 expression. (C) Constitutive CCL5 was not detectable in CD3ε–CD19–NK1.1– cells (left), but was detected in NK cells, subpopulations of T cells, and some NKT cells (right). (E and G) Spontaneous CCL7/11, but not CCL8, production in cell subsets lacking CD45 expression. (H) Top: Ex vivo detectable CCL21 expression by T cells (left) and CCR7+ cells (middle). Histogram at right is gated on the 3 DC subsets (colored traces) and CD3ε+T cells (gray solid). Bottom: CCL21 surface stains of cells recovered from blood, spleen, and mesenteric lymph node (MLN). (M) Constitutive CXCL9 expression by T cells (histograms gated on CD3ε+T cells from Cxcl9–/– and B6 mice). Experiments were conducted in multiple independent experiments with groups of 3 mice and performed at least twice.
CCL1.
Although we did not detect homeostatic expression of CCL1 in splenic hematopoietic cells (Figure 2A), this chemokine was readily produced by subpopulations of activated effector and memory T cells (TE and TM, respectively). In fact, induced CCL1 expression identifies a subset of highly polyfunctional CD8+ and CD4+ TM cells (J. Eberlein and D. Homann, unpublished observation).
CCL2.
In the absence of ex vivo detectable CCL2, spontaneous production of this chemokine was restricted to a subset of Mo/Mϕ (Figure 4A). LPS stimulation enhanced the fraction of CCL2+ Mo/Mϕ only slightly, but increased CCL2 expression levels and also induced production by a minor CD11c+cDC subset (Supplemental Figure 2A). Although these findings were confirmed by the absence of detectable CCL2 in tissues from Ccl2–/– mice (Supplemental Figure 2B), the anti-CCL2 pAb was also crossreactive with CCL7/12 (Supplemental Figure 1). We therefore conducted corresponding analyses with the anti-CCL2 mAb 2H5 and observed similar, but notably weaker, CCL2 expression patterns in B6 mice (Supplemental Figure 2C). Unexpectedly, the anti-CCL2 mAb, which did not react with CCL7, also recognized CCL12, a chemokine that was not spontaneously synthesized by spleen cells (Figure 2A and Supplemental Figure 2C). That the specificity of a widely used mAb was not limited to its cognate chemokine provides an example that the phenomenon of crossreactivty is not restricted to pAbs and illustrates the need to extend comprehensive specificity analyses to mAbs.
CCL3 and CCL4.
Spontaneous expression of CCL3 and CCL4 occurred in both CD11b+ and CD11b– populations (Figure 4B). In the former group, CCL3/4 production was observed in subsets of Mo/Mϕ and cDCs (Figure 4B), cell types that also responded with significantly increased CCL3/4 synthesis upon LPS stimulation (in particular, Mo/Mϕ and CD11c+cDCs, but also some granulocytes; Supplemental Figure 2A). Inducible CCL3/4 production is also a major component of the pathogen-specific T cell response, but constitutive or spontaneous expression is only observed at a very low level in TE but not TM populations (J. Eberlein and D. Homann, unpublished observation). In agreement with this finding, only marginal spontaneous CCL3/4 expression was found in T cells of undefined specificity. Rather, B cell subsets constituted the largest CD11b– cell type capable of spontaneous CCL3/4 synthesis (Figure 4B). Finally, the minor crossreactivity of the anti-CCL3 pAb with CX3CL1 (Figure 2B), a chemokine not produced by splenic hematopoietic cells, did not result in false positive signals in CCL3-deficient cells stained with this pAb (data not shown).
CCL5.
We identified CCL5 as one of the most prominent homeostatic chemokines (Figure 2A) with a constitutive expression pattern largely restricted to NK cells and several T cell subsets (Figure 4C; note the absence of constitutive CCL5 in the CD3ε–CD19–NK1.1– population). Notwithstanding the fact that CCL5 can be induced in many other cell types, including Mo/Mϕ and DCs (Supplemental Figure 2A), the lack of constitutive CCL5 expression by the latter cells was systemic, applying to lymphatic and nonlymphatic tissues alike (blood, spleen, LNs, BM, peritoneal cavity, liver, and lung; data not shown). CD44hi memory-phenotype CD8+ T cells represent the major population of T cells featuring constitutive CCL5 expression, whereas subsets of γδTCR+ T cells and, to a much lesser extent, memory-phenotype CD4+ and NKT cells, are also CCL5+ (J. Eberlein and D. Homann, unpublished observation). Interestingly, pathogen-specific CD8+ TM within the memory-phenotype CD8+ population express particularly high levels of constitutive CCL5 and account for the largest source for ex vivo detectable CCL5 in pathogen-immune animals (J. Eberlein and D. Homann, unpublished observation). Finally, the weak crossreactivity of the anti-CCL5 pAb with CCL3 appears to be negligible, as we observed no residual staining in various CCL5-deficient cell subsets evaluated under homeostatic or inflammatory conditions (data not shown).
CCL6 and CCL9/10.
The murine chemokines CCL6 and CCL9/10, together with their closest human homologs, CCL23 and CCL15, belong to the N6 subfamily of proteins that have an N-terminal extension relative to other β-chemokines (14). The constitutive expression of both chemokines was rather pronounced and confined to the same cell subsets (granulocytes, Mo/Mϕ, and some CD11c+cDCs; Figure 4, D and F), but stood in apparent contrast to their homeostatic regulation: CCL9/10 can be found at extraordinarily high levels in the circulation of normal mice (~1 μg/ml; ref. 35), yet serum levels of the inflammatory chemokine CCL6 (36) are estimated to be more than 10,000-fold lower (A. Coelho, unpublished observation). The above considerations, however, are tempered by our observation that the complementary crossreactivities of anti-CCL6 and anti-CCL9/10 pAbs were the most pronounced of all chemokine pAbs evaluated (Supplemental Figure 1, A and B). Nevertheless, the expression pattern described above for CCL6 was confirmed with a noncrossreactive anti-CCL6 mAb (Supplemental Figure 2D), and the ostensible phenotype of CCL9/10+ cells as well as the intensity of the anti-CCL9/10 pAb staining signal are consistent with the notion that granulocytes and Mo/Mϕ are in fact coproducers of CCL9/10 and CCL6. In other cell types, attribution of the anti-CCL9/10 pAb signal to CCL9/10 production can be more definitive, for example, in activated T cell subsets that bind the anti-CCL9/10 pAb, but not the anti-CCL6 mAb (data not shown).
CCL7, CCL8, and CCL11.
Anti-CCL7 and anti-CCL11 are part of a group of 4 pAbs that feature noncognate interactions with CC-MCP chemokines. The former pAb in particular exhibited an unusually broad spectrum of mostly low-level crossreactivities, including recognition of CCL2/11/12; conversely, the pAbs specific for CCL2/11/12 also partially recognize CCL7 (Table 1). In spite of these caveats, the results of a complementary evaluation of CCL7/8/11 and other crossrecognized chemokine expression led us to conclude the following. Production of CCL7/8/11, detectable ex vivo at low levels preferentially in the CD11b– compartment, remained largely unchanged after short-term in vitro culture (Figure 2A), and some of the CCL8+ and CCL11+ cells appeared to be B cells (Figure 4, E and G). Although borderline expression can be found in T cells, pathogen-specific T cells neither contain Ccl7/8/11 mRNA transcripts nor produce the corresponding proteins upon TCR engagement (J. Eberlein and D. Homann, unpublished observation). In addition, constitutive CCL11 was found in some CD11b+ cells, but its expression — mostly excluded from granulocytes and slightly more pronounced in pDCs — was not preferentially associated with a specific immune cell subset (Figure 4G). Upon stimulation with LPS, induced chemokine expression was observed for CCL7 (mostly in Mo/Mϕ and some DCs), but not for CCL8/11 (Supplemental Figure 2A and data not shown). Finally, about half of the CCL7+ cells and approximately one-third of CCL11+ cells, but no CCL8+ cells, analyzed for constitutive and spontaneous chemokine production lacked expression of CD45. This population remains to be defined in detail, but did include fibroblastic reticular cells (FRCs), as determined by CD31 and/or gp38/podoplanin expression (Figure 4, E and G, and data not shown). However, the apparent CCL7/11 production by CD45– cells must be interpreted with caution, as CCL7-deficient mice, which demonstrated the expected lack of LPS-induced CCL7 expression in CD11b+ populations (Supplemental Figure 2E), nevertheless exhibited higher background staining in nonhematopoietic cells (data not shown); further analyses of FRCs will also require optimized cell dissociation protocols (37).
CCL12, CCL17, CCL19, and CCL20.
None of these chemokines were expressed by splenic hematopoietic cells under homeostatic conditions (Figure 2A). The lack of CCL12 is noteworthy because the chemokines weakly crossrecognized by the anti-CCL12 pAb (CCL2/7) were expressed under homeostatic conditions. We also evaluated CCL17 expression in LN DCs, which are reported to constitutively express Ccl17 mRNA (38), but did not detect any protein (data not shown). The apparent absence of homeostatic CCL19 in spleen and LNs (Figure 2A and data not shown) is interesting because a recent publication proposed that CCL19 is surface-bound to T cells expressing its receptor CCR7 (39). This conclusion was based on differential staining intensities using a CCR7-specific Ab versus CCL19 fusion protein, and, although the CCL19-FP may well constitute a more sensitive reagent for the indirect determination of CCR7 occupancy by CCL19, direct Ab staining of surface-bound CCL19 was not performed (39).
CCL21.
CCL21 exhibited a peculiar expression pattern: ex vivo detectable CCL21 in CD11b– cells disappeared after 5 hours of in vitro culture (Figure 2A). This observation suggests that CCL21 may be acquired rather than produced by CD11b– cells and is supported by a recent report that documents the capture and surface immobilization of CCL21 by a variety of immune cells in the LN (26). Our subset analyses demonstrated that T cells constituted the major population of CCL21+ cells in the spleen and likely bind CCL21 via its receptor, CCR7 (Figure 4H). The notion that T cell CCL21 is preferentially captured is also supported by the observation that pathogen-specific T cells neither contain Ccl21 message nor upregulate mRNA or protein expression after specific in vitro stimulation (J. Eberlein and D. Homann, unpublished observation). However, our conclusions are in seeming contradiction to the finding by Friedman et al. that CD11c+ DCs bind CCL21 at higher levels than any other immune cell subset (26). We therefore evaluated surface CCL21 expression in LNs, spleen, and blood, tissues previously shown to contain a wide range of distinct CCL21 levels (about 2,000 ng/g, about 300 ng/g, and <10 ng/ml serum, respectively; refs. 40, 41). Indeed, the level of detectable surface CCL21 declined in the expected order (LN, then spleen, then blood), and although few CD11c+ cells were CCL21+ in the spleen, the overall mean fluorescence intensity of CCL21 stains was higher in the CD11c++ cDC subset than in other cells (Figure 4H). Because CCL21 capture and presentation by DCs facilitates the specific priming of T cells by establishing initial antigen nonspecific interactions (26), it is possible that the reciprocal interaction, namely the binding of CCL21-bearing T cells to CCR7+ DCs, may also contribute to improved T cell activation.
CCL22.
In the absence of constitutive expression, CCL22 was spontaneously synthesized by cDC subsets, and to a lesser degree by CD45+CD11b–Gr-1–F4/80– cells (Figure 4I and data not shown). The possibility of B cells being a source for induced CCL22 synthesis is discussed below. None of these or other splenic hematopoietic cells produced CCL20, the chemokine with which the anti-CCL22 pAb crossreacted weakly.
CCL24, CCL25, and CCL28.
No homeostatic expression of these chemokines was observed in splenic hematopoietic cells (Figure 2A). However, in agreement with the previously reported spontaneous secretion of CCL25 by in vitro cultured small intestinal epithelial cells (42), constitutive CCL25 expression by these cells was readily visualized by FC (Supplemental Figure 2F).
CCL27.
Based on its homeostatic expression primarily in keratinocytes, CCL27 appears to be a tissue-specific chemokine (43). Therefore, the unique expression pattern of CCL27 described here — ex vivo detectable CCL27 in splenic granulocytes accompanied by weaker expression in DC subsets (Figure 2A and Figure 4J) — was surprising. This observation is of particular interest in light of a recent publication on the differential expression of CCL27 protein and mRNA in skin-draining LNs: even under homeostatic conditions, CCL27 protein is transported by as-yet undetermined mechanisms from the epidermis to the LNs, where it may assist in the recruitment of CCR10-bearing T cells (44). Whether granulocytes and/or DCs indeed participate in CCL27 transport in the context of immune homeostasis or inflammation is currently under investigation.
CXCL1, CXCL2, and CXCL3.
The determination of CXCL1/2 expression patterns is complicated by the partial crossreactivity of anti-CXCL1/2 pAbs (Table 1), a likely consequence of the pronounced homology among these chemokines (14). However, although no gene products were detected in splenic hematopoietic cells analyzed directly ex vivo with anti-CXCL1/2 pAbs, distinct expression patterns were discerned for spontaneous chemokine production. Specifically, CXCL2, but not CXCL1, was detected in Mo/Mϕ subsets (Figure 2B and Figure 4K), even though the anti-CXCL1 pAb exhibited the highest degree of CXCL2 crossreactivity (Supplemental Figure 1C). As expected, LPS stimulation caused the rapid and robust upregulation of both CXCL1/2 by Mo/Mϕ, some CD11c+cDC subsets, and, in the case of CXCL2, granulocytes (Supplemental Figure 3A).
CXCL4 and CXCL7.
The constitutive expression of CXCL4 and CXCL7 by a wide variety of immune cells (Mo/Mϕ, granulocytes, DCs, B cells, and some T cells, but <2% of CD45– cells; Figure 2B and Figure 4L) was unexpected. In spite of the weak crossreactivity of anti-CXCL4/7 pAbs with CXCL1/2, the latter chemokines were not expressed in a constitutive fashion, as discussed above (Figure 2B and Table 2), and the specificity of the anti-CXCL4 pAb was confirmed by stains using CXCL4-deficient cells (Supplemental Figure 3B). Although recent work identified monocytes as an auxiliary CXCL4/7 source, and blood-borne DC subsets constitutively transcribe Cxcl4/7 mRNA (45–47), CXCL4/7 expression is usually regarded as an exclusive feature of the megakaryocyte lineage. In fact, CXCL4/7 are the 2 most abundantly expressed chemokines in platelets and are detectable in normal sera at micromolar concentrations (48). It is thus conceivable that CXCL4 and CXCL7 are in part captured rather than synthesized by the different immune cell subsets: the CXCL7 receptors CXCR1/2 are preferentially expressed by Mo/Mϕ and granulocytes, and these cells constituted the majority of CXCL7+ cells in the CD11b+ myeloid compartment (Figure 4L). The identity of the murine CXCL4 receptor remains unknown, but CXCR3 appears to be a promising candidate. In humans, CXCL4 was initially shown to only bind an alternative CXCR3 splice variant, termed CXCR3B (49), but more recent work indicates that it also binds to the native CXCR3 receptor (50). Among murine CXCR3-expressing cells, there appeared to be direct correlation between CXCR3 and CXCL4 expression levels (data not shown), but high levels of CXCL4 were also detected in CXCR3– cells, most notably B cells (Figure 4L). The notion that CXCL4 is predominantly captured by various cellular subsets is indeed supported by our preliminary findings (data not shown), and the surprising expression patterns of both CXCL4 and CXCL7 will clearly require further investigation.
CXCL9, CXCL10, and CXCL11.
In accordance with the published literature (51), our FC analyses of primary murine spleen cells stimulated with IFN-γ identified Mo/Mϕ as a principal source for induced CXCL9/10 production. Furthermore, DCs constituted an additional source for these inflammatory chemokines, and the CD11c+ cDC subset consistently demonstrated the highest level of inducible CXCL10 expression (Supplemental Figure 3C). Interestingly, we also found that a small population of T cells was characterized by weak constitutive CXCL9 expression (Figure 4M). Similar to CCL21, detectable CXCL9 disappeared after in vitro culture (Figure 2B); moreover, pathogen-specific T cells do not synthesize Cxcl9 mRNA or protein (J. Eberlein and D. Homann, unpublished observation). Thus, it is possible that T cell CXCL9 is also preferentially surface-bound, perhaps through its receptor CXCR3, one of the most prominently TM-expressed chemokine receptors (data not shown). In contrast to the robust CXCL9/10 production following IFN-γ stimulation, induced CXCL11 expression, as analyzed in spleen cells of Balb/c origin, remained comparatively weak (Supplemental Figure 3D). Finally, using mice deficient for Cxcl9 or Cxcl10 and regular B6 mice that lack a functional Cxcl11 gene (52), we confirmed the specificity of our pAb stains (Figure 4M and Supplemental Figure 3D).
CXCL5, CXCL12, CXCL13, CXCL14, CXCL15, and CXCL17.
Splenic hematopoietic cells evaluated under homeostatic conditions expressed none of these chemokines (Figure 2B).
CXCL16 and CX3CL1.
CXCL16 and CX3CL1 share the distinction of being the only cell membrane–anchored chemokines (14, 23). However, whereas CX3CL1 was not expressed by splenic hematopoietic cells under homeostatic conditions, robust spontaneous CXCL16 production was observed for Mo/Mϕ, cDCs, and, to a lesser extent, pDC subsets (Figure 4N), and the anti-CXCL16 pAb specificity was validated in analyses of Cxcl16–/– tissues (Supplemental Figure 3E). In agreement with previous findings (23), LPS stimulation enhanced CXCL16 expression by DCs and Mo/Mϕ (data not shown).
XCL1.
Some constitutive and spontaneous XCL1 expression was found in both CD11b– and CD11b+ cells (Figure 2B). Because the analytical exclusion of NK, T, and B cells also removed XCL1+ populations, these cells contain the relevant sources of homeostatic XCL1 production. Indeed, NK cells exhibited low levels of constitutive and spontaneous XCL1 expression (see below), as did small subsets of T and B cells (Figure 4O). In addition, robust XCL1 production is readily induced in large subpopulations of pathogen-specific CD8+TE and TM, and inducible XCL1 expression, similar to IL-2, progressively increases in aging CD8+ TM populations (J. Eberlein and D. Homann, unpublished observation).
In summary, our application of chemokine FC has confirmed, extended, and identified the homeostatic expression patterns of 18 chemokines in defined murine hematopoietic cells (Table 2). The remarkable diversity of homeostatic chemokines expressed by these cells likely generates a complex network for cellular interactions that is further modulated by the contribution of nonhematopoietic cells and the overall composition of specific tissue microenvironments (e.g., constitutive CCL25+ intestinal epithelial cells in the small intestine; Supplemental Figure 2F).
Chemokine profiles of murine and human NK cells
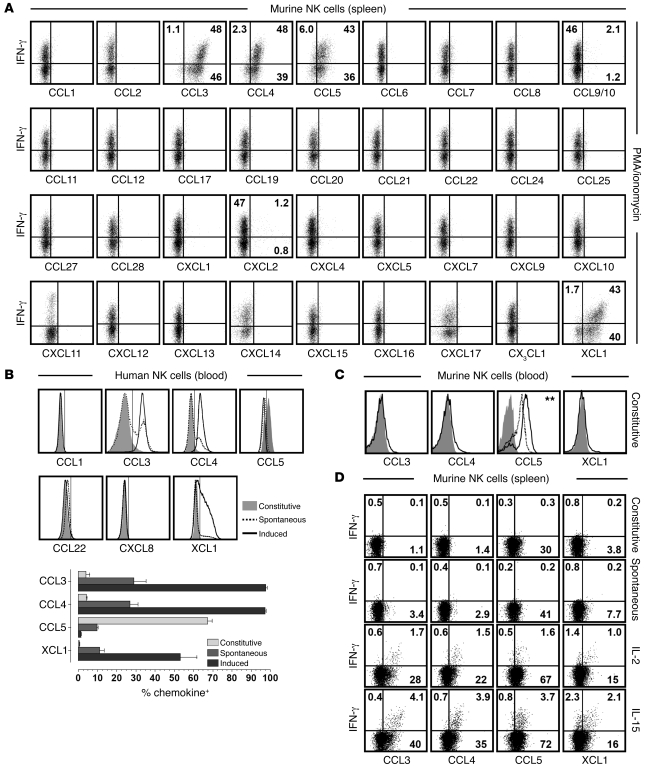
Among the immune cell subsets evaluated for their capacity to produce chemokines, NK cells constitute one of the best-characterized populations (53), as evidenced by the NK cell production of CCL3/4/5 and XCL1 in vitro or in vivo in the course of murine cytomegalovirus infection (18, 21). Our results confirmed the robust induction of these chemokines by PMA/ionomycin-stimulated NK cells, demonstrated the presence of polyfunctional NK cell subsets (i.e., most pronounced coproduction of CCL3/4/5 and XCL1 in conjunction with IFN-γ), and revealed the synthesis of CCL9/10 and CXCL2 by minor NK cell subsets (Figure 5A and Supplemental Figure 4, A and B). The absence of any other NK cell–produced chemokine was particularly noteworthy in regard to 2 chemokines, CCL1 and CCL22: whereas prolonged activation of human NK cells elicits their secretion (54, 55), short-term stimulation of murine NK cells did not (Figure 5A).
Figure 5. Chemokine profiles of murine and human NK cells.
(A) Splenocytes obtained from B6 mice (or Balb/c mice for CXCL11 analyses) were cultured for 5 hours with PMA/ionomycin and BFA and processed for chemokine FC. Plots are gated on NK1.1+CD3ε– (B6) or DX5+CD3ε– (Balb/c) cells; numbers within plots indicate mean percent IFN-γ+ and/or chemokine-positive cells in the respective quadrants. (B) Chemokine expression by human NK cells (CD56+CD3–) was determined directly ex vivo (constitutive), after 5 hours of culture in the presence of BFA (spontaneous) or stimulation with PMA/ionomycin plus BFA (induced). Vertical markers were set according to goat IgG or isotype control stains. The fraction of chemokine+ NK cells obtained from 4 healthy volunteers is shown below (representative data from 3–5 independent experiments). (C) Blood-borne murine NK cells (NK1.1+CD3ε–) were analyzed for constitutive chemokine expression (black tracings). Different controls (gray solid) are featured in the individual plots (CCL3 control, CCL3 stains of Ccl3–/– NK cells; CCL4 and XCL1 controls, goat IgG stains; CCL5 controls, CCL5 stains of Ccl5–/– and Ccl5+/– NK cells, the latter shown by dotted tracing). The CCL5 expression level (GMFI) of Ccl5+/– NK cells were slightly, but significantly, lower than those of wild-type NK cells (P = 0.01). (D) Constitutive, spontaneous, and cytokine-induced (100 ng/ml IL-2 or IL-15) CCL3/4/5 and XCL1 expression by splenic NK cells was determined in conjunction with IFN-γ; numbers denote average percentages of IFN-γ+ and/or chemokine-positive cells in corresponding quadrants. All data obtained for murine NK cells are representative for multiple independent experiments in 2–3 mice each.
To determine whether human NK cells produce an expanded range of chemokines compared with their murine counterparts, we evaluated the expression of all chemokines reported to be secreted by human NK cells (53), including CCL1, CCL22, and CXCL8 (IL-8), a chemokine/IL not found in the mouse. For detection of CCL1 and XCL1, to our knowledge not previously analyzed by FC, we used affinity-purified pAbs in analogy to our murine chemokine assays; we also found that a CCL5-specific pAb provided more consistent results than did fluorochrome-conjugated CCL5 mAbs (Supplemental Figure 4C and data not shown). Interestingly, small subsets of human NK cells constitutively expressed CCL3/4, and larger NK populations spontaneously synthesized CCL3/4 and XCL1 in the absence of added stimuli. Upon activation with PMA/ionomycin, practically all NK cells rapidly produced CCL3/4, and about half also expressed XCL1 (Figure 5B). At the same time, the induced IFN-γ response was far more limited, usually with less than 10% of NK cells producing this cytokine (data not shown). In agreement with a recent report (56), we also found substantial constitutive CCL5 expression in human NK cells, but, contrary to their murine counterpart, stimulation resulted in depletion of CCL5 stores rather than intracellular CCL5 accumulation. A similar difference was also noted in constitutive and induced CCL5 expression by murine versus human T cells and remains unexplained — although it may be related to different sensitivities to protein transport inhibition by BFA and monensin (57, 58). Finally, human NK cells did not exhibit constitutive, spontaneous, or induced expression of CCL1, CCL22, or CXCL8 (Supplemental Figure 4D, control stains for hCCL1). The lack of induced CCL1 synthesis in particular defines a functional difference between NK and T cells: after activation, both populations produce copious amounts of CCL3/4/5 and XCL1 as well as some CCL9/10 in the mouse, but only T cells are induced to express CCL1 (J. Eberlein and D. Homann, unpublished observation). Interestingly, CXCL2 — the only other T cell–produced chemokine detected in a small subset of stimulated CD4+T cells (data not shown) — also constituted a minor component of the murine NK cell response (Figure 5A). Thus, at the level of induced chemokine production, an otherwise remarkable similarity between NK and T cells (18) contrasted the unique capacity for CCL1 secretion by CD8+ and CD4+ T cells (Table 2).
Our further observations emphasized quantitative differences among constitutively NK cell–expressed murine chemokines in different anatomic compartments and illustrated the exquisite sensitivity of NK cells to cytokine-induced chemokine production. In comparison to the spleen, blood-borne NK cells contained high amounts of constitutive CCL5 and exhibited somewhat more pronounced constitutive CCL3/4 and XCL1 expression (Figure 5, C and D), a finding that complements the constitutive and spontaneous production of the same chemokines by human NK cells. The functional status of resting murine NK cells was recently detailed by Fehniger, Ley, and colleagues, who documented the presence of mRNA transcripts for several cytolytic effector molecules (granzyme A [GzmA], granzyme B [GzmB], and perforin; ref. 59). However, whereas GzmA protein is expressed in a constitutive fashion, GzmB and perforin are translated from preexisting mRNA species only upon stimulation with cytokines such as IL-2 and IL-15 (59). Our analysis of the microarray data generated by Fehniger et al. (59) revealed that among chemokine mRNA species, only Ccl3/4/5 and Xcl1 transcripts were expressed at high levels in resting NK cells (data not shown), and their low-level (CCL3/4 and XCL1) to high-level (CCL5) constitutive translation was rapidly increased by treatment with IL-2 or IL-15 (Figure 5D). Because IL-15–induced protein production was not accompanied by a corresponding increase of chemokine mRNA (data not shown), we conclude that IL-15, and possibly also IL-2, promote the early NK chemokine response, apparently through release from a partial translational blockade. In summary, our data provide evidence that the earliest stage of the murine and human NK cell response is dominated by the synthesis and secretion of chemokines that precede IFN-γ production and the eventual acquisition of cytolytic effector functions mediated by GzmB and perforin (59).
Chemokine profiles of activated murine B cells
B cells are recognized as a biologically relevant source for selected cytokines and chemokines (60–62), but the spectrum of chemokines synthesized by stimulated B cells remains incompletely defined. We first determined the range of chemokines produced by splenic B cells obtained from unmanipulated mice in response to LPS, a widely used stimulus that mediates B cell activation through TLR4. Compared with spontaneous B cell chemokine expression, LPS treatment significantly increased CCL3/4 and CCL22 synthesis (Supplemental Figure 5A). However, overall LPS-induced chemokine expression levels are weak (CCL3/4) or restricted to small B cell subsets (CCL22) and not associated with enhanced transcription of Ccl3, Ccl4, or Ccl22 mRNA species (61, 63). No other chemokines were synthesized by LPS-activated B cells (data not shown), and the absence of induced CCL6 and CXCL1/2 production should be emphasized here, given the reported upregulation of corresponding chemokine mRNA transcripts (63).
Inspection of the chemokine mRNA species expressed after stimulation of the B cell receptor (anti-IgM or antigen) revealed somewhat more robust and diversified induction of chemokine transcripts compared with LPS stimulation (Ccl2/3/4/5/6/9/22, Cxcl16, Cx3cl1, and/or Xcl1; refs. 61, 63). However, when evaluated for the production of chemokine proteins after IgM/CD40 engagement, B cells failed to synthesize most of these chemokines (Supplemental Figure 5B). The low-level production of CCL3/4, similar to the results obtained after LPS stimulation, was the only exception, and the synthesis of CCL22 after IgM/CD40 stimulation was at best marginal. Of further interest was the absence of spontaneous or induced CCL5 protein expression under any experimental conditions, particularly in light of the fact that Ccl5 is the most abundantly expressed chemokine mRNA transcript in purified B cells (61, 63). In comparison to the rapid production of multiple chemokines by innate immune cells (NK cells, Mo/Mϕ, and DCs) and pathogen-specific T cells (ref. 18 and data not shown), the chemokine response by B cells obtained from naive mice was therefore limited in regard to both the spectrum (CCL3/4/22) and the quantity of chemokines produced.
The DC chemokine response to infection
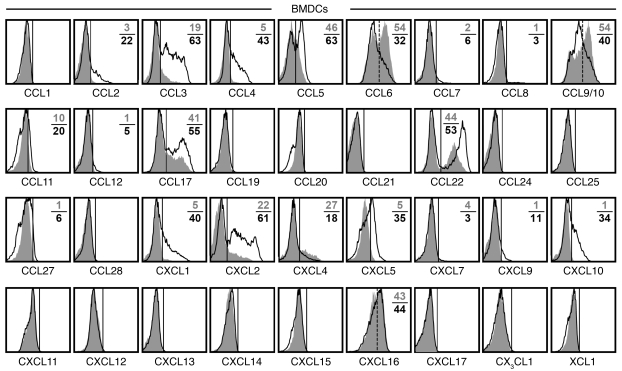
Given the diverse array of constitutive, spontaneous, and inducible chemokine expression by DCs (Figure 4, Supplemental Figure 2A, and Supplemental Figure 3, A and C) and their importance for the effective coordination of adaptive immune responses (64, 65), we surveyed the DC chemokine response to in vitro infection with the bacterium L. monocytogenes. In the absence of infection, BM- derived DCs (BMDCs) synthesize an array of chemokines at variable levels, similar to those of primary DCs recovered from the spleen. However, in contrast to splenic DCs, cultured BMDCs also expressed CCL5/17 and CXCL1/2 (Figure 6), possibly resulting from partial activation under the culture conditions. Upon L. monocytogenes infection, BMDCs displayed a chemokine response similar to that of LPS/IFN-γ–stimulated splenic DCs. Specifically, robust induction of CCL2/3/4/5 and CXCL1/2/5/10 was accompanied by more modest synthesis of CCL11/CXCL9 as well as marginal upregulation of CCL7/8/12/27 (Figure 6). Moreover, 6 additional chemokines were highly expressed in uninfected BMDCs but differentially regulated upon infection: increased CCL17/22 production contrasted with unaltered CXCL16 levels and downmodulated CCL6, CCL9/10, and CXCL4 (and perhaps CXCL7; Figure 6). Thus, the DC response to infection was distributed over approximately 20 distinct chemokines and composed of 2 major components: (a) the modulation of homeostatic chemokine expression (potentiation, CCL3/4/5/11/17/22 and CXCL1/2; no change, CXCL16; inhibition, CCL6, CCL9/10, and CXCL4/7); and (b) the de novo induction of inflammatory chemokines generally found at somewhat lower levels (CCL2/7/8/12/27 and CXCL5/9/10). Interestingly, several of the above chemokines (CCL2/5/17 and CXCL1/2/10) were previously reported to be spontaneously produced by CD11bhi cDCs isolated from the lungs of naive mice (24). Because these chemokines were not produced by primary splenic DCs in the absence of stimulation (Figures 2 and 4), peripheral DCs residing in an organ exposed to the exterior environment apparently exhibit a chemokine signature consistent with heightened activation and a predisposition to readily process inflammatory insults.
Figure 6. DC chemokine response to bacterial infection.
BMDCs were obtained from B6 mice (or Balb/c mice for CXCL11 analyses), propagated by culture in GM-CSF, and infected with rLM-OVA at MOI 1 as described in Methods. At 4 hours after L. monocytogenes or mock infection, BFA was added to cultures for an additional 5 hours, followed by quantitation of chemokine expression by uninfected versus infected DCs. Histograms are gated on CD11c+ BMDCs and display chemokine expression by uninfected (gray solid) and infected (black tracing) cells. Solid vertical markers demarcate chemokine-expressing from nonexpressing cells, dashed markers for CCL6, CCL9/10, and CXCL16 distinguish chemokine++ and chemokine+/– cells. Numbers indicate the percentage of chemokine-positive DCs in the absence (gray) or presence (black) of infection; values are the average of duplicate experimental samples. Data are representative for 1 of 4 similar experiments. Similar results were obtained after 24 hours of rLM-OVA infection and prevention of cell death by addition of antibiotics after initial establishment of infection (not shown).
Discussion
The considerable complexity of the chemokine system and its involvement in numerous physiological and pathophysiological processes are the subjects of many ongoing investigations. Here, our direct visualization of practically all murine chemokines by FC should greatly facilitate the precise delineation of chemokine-expressing cells in varied experimental settings, permit detailed evaluation of chemokine coexpression, and assist in efficient analyses pertaining to multiple aspects of chemokine biology in general. The use of pAbs for chemokine FC will eventually be superseded by the increased use of fluorochrome-conjugated mAbs. However, given the multiplicity of applications for individual pAbs, these reagents will continue to provide greater methodological flexibility. For example, our demonstration that all 25 pAbs currently not approved for IHC (Table 1) retain specific reactivity under conditions of PFA fixation indicates that they will also be suitable for chemokine detection in paraffin-embedded tissues, a contention that we have already verified for selected pAbs (data not shown). Furthermore, our general approach to pAb selection and validation can readily be adopted for visualization of chemokine production by cells from other species, including humans (Figure 5B and Supplemental Figure 4, C and D).
A perennial concern pertaining to pAbs is their potential for recognition of noncognate antigens, and we have addressed this issue, which is certainly not limited to pAbs (Supplemental Figure 2C), in an extended series of experiments determining pAb crossreactivities at the level of murine chemokines (Supplemental Figure 1). Although our results may serve as a general template for the utility and limitations of individual chemokine-specific pAbs in FC and other applications, it remains imperative for any experimental design to assure the validity of experimental readouts by inclusion of appropriate controls (see Methods).
Our exemplary implementation of chemokine FC to the characterization of hematopoietic cells capable of homeostatic and induced chemokine expression underscores the relative ease with which this methodology can identify both the cellular sources of individual chemokines and the entire spectrum of chemokines produced by defined cellular subsets. Indeed, several of the expression patterns described in Results may serve as a foundation for future investigations into specific functions exerted by particular combinations of chemokines and the cell populations involved in their synthesis (a detailed investigation of the chemokine signatures of pathogen-specific T cells is in progress, although pertinent findings have been reported herein as unpublished observations). Because of its comprehensive nature, chemokine FC can also, within the limits of defined experimental settings and the analytical sensitivity afforded by FC, demonstrate the absence of multiple chemokine proteins. This insight is particularly useful because the correlation of mRNA and protein expression is in general unpredictable, and chemokine production detected by other means may be confounded by the generation of artifacts when assessed after prolonged in vitro culture. Finally, we emphasize that a delineation of functional profiles, of which the chemokine response is a major component, must complement any phenotypic differentiation of cellular subsets in order to develop an integrative understanding of their roles in health and disease.
Methods
Cell lines, vectors, and transfection.
Murine chemokine cDNA clones were purchased from ATCC (Ccl17) or Open Biosystems (all other clones), sequenced, and subcloned into the pIRES2-AcGFP1 vector (Clontech). For further details, see Supplemental Table 1. A frameshift error in the cds of the Cxcl12 clone (IMAGE:6406409) was corrected using PCR-driven overlap extension. HEK 293T cells and/or Cos7 cells were transfected with chemokine vectors or Il15 vector (negative control; production of murine IL-15 verified by immunoblot and FC, data not shown; mIl15 vector provided by T. Sosinowski, University of Colorado Denver and National Jewish Health) using FuGENE (Roche) according to the manufacturer’s protocol, or CaPO4 as described previously (66), and cultured for 18–36 hours. For the final 14 hours, 1 μg/ml BFA (Sigma-Aldrich) was added, and the cells were processed and stained with chemokine-specific Abs as detailed below. In the case of Cxcl14/15, an ER retention sequence (KDEL) was introduced to enhance intracellular chemokine accumulation, and Cxcl14-transfected HEK cells were additionally cultured in the presence of 10 μM of the protease inhibitor MG-132 (Fisher) to reduce proteasomal degradation.
Animals.
C57BL/6J (B6) and Balb/c as well as Ccl2–/– (B6.129S4-Ccl2tm1Rol/J), Ccl3–/– (B6.129P2-Ccl3tm1Unc/J), and Cxcl10–/– (B6.129S4-Cxcl10tm1Adl/J) mice on a B6 background were purchased from The Jackson Laboratory. B6.Ccl5–/– mice (identical to the commercially available B6.129P2-Ccl5tm1Hso/J strain; ref. 67) were obtained from M. von Herrath (LIAI, La Jolla, California) and backcrossed to B6 mice, and F1 mice were intercrossed to yield B6.Ccl5–/–, B6.Ccl5+/–, and B6.Ccl5+/+ littermates. Additional chemokine-deficient mice or tissues were provided by I. Charo (University of California, San Francisco; B6.Ccl7–/– and B6.Cxcl16–/–; refs. 68, 69), A. Kowalska (Children’s Hospital of Philadelphia, Pennsylvania; B6.Cxcl4–/–; ref. 70), and J. Farber (NIH/NIAID, Bethesda, Maryland; B6.Cxcl9–/–; ref. 71). All animals were evaluated at 8–12 weeks of age, with the exception of B6.Ccl7–/– and B6.Cxcl16–/– mice (approximately 28 weeks), and all procedures were performed in accordance with regulations as set forth by the University of Colorado IACUC.
Primary cell preparation and purification.
Single-cell suspensions were prepared from murine spleen, LNs, blood, peritoneal cavity, and BM according to standard procedures (72, 73). For isolation of cells from liver and lung, mice were terminally anesthetized with Avertin and sacrificed by total body perfusion through the left ventricle with PBS prior to tissue processing, as described previously (73). Suspensions of small intestinal epithelial cells were prepared using PBS-flushed and inverted small intestines incubated with 30 mM EDTA/PBS buffer at 37°C for 30 minutes on a rotating platform with supernatant harvest/buffer replacement every 5 minutes (74). Human PBMCs were obtained from healthy subjects and isolated from whole blood using cellular preparation tubes (BD). Informed consent was obtained in all cases, and the study protocol was approved by the University of Colorado Institutional Review Board.
Primary cell culture.
To determine spontaneous chemokine production by defined immune cell subsets, primary cells obtained from spleen or blood of B6 mice were cultured for 5 hours in RPMI plus 10% FCS in the presence of 1 μg/ml BFA but absence of any exogenous stimuli. Stimulation of murine NK cells was performed by a 5-hour culture of spleen cells in the presence of BFA as well as 20 ng/ml PMA and 1 μg/ml ionomycin (Sigma-Aldrich), recombinant human IL-2 (100 ng/ml, i.e., 1,000 U/ml; eBioscience), or recombinant murine IL-15 (100 ng/ml; Peprotech). Mo/Mϕ stimulation was achieved by 5 hours of spleen cell incubation with 500 ng/ml LPS (Sigma-Aldrich) or 1,000 U/ml recombinant murine IFN-γ (Peprotech) and BFA; B cell stimulation was provided by 5 hours of treatment with 500 ng/ml LPS, or 10 μg/ml donkey anti-IgM F(ab′)2 (Jackson Immunoresearch) and 15 μg/ml anti-CD40 (FGK45.5; gift from R. Torres, University of Colorado Denver and National Jewish Health), plus BFA. Human PBMCs were cultured for 5 hours in the presence or absence of PMA and ionomycin using protocols identical to the stimulation of murine NK cells.
BMDC propagation and infection.
BMDCs were generated as described previously (75). In brief, BM cells obtained from femurs and tibias of B6 mice were cultured for 6 days in the presence of 10 ng/ml recombinant GM-CSF (BD Biosciences), yielding a population of greater than 90% CD11c+ cells that for the most part expressed low levels of CD40 and CD86 characteristic of immature DCs (data not shown). Recombinant L. monocytogenes expressing full-length ovalbumin (rLM-OVA; provided by L. Lenz, University of Colorado Denver and National Jewish Health; ref. 76) was grown and titered as described previously (77) and used at MOI 1 for in vitro BMDC infection. Infection was allowed to proceed for 9 hours, with BFA added for the final 5 hours. As assessed by FC, infected DCs exhibited slightly increased CD11c expression and elevated SSC properties, likely resulting from the presence of intracellular bacteria (data not shown).
General FC.
mAbs used in FC were purchased as purified, biotinylated and/or fluorophore-conjugated reagents from BD Biosciences, eBioscience, Biolegend, R&D Systems, or Invitrogen (Supplemental Table 2); our protocols for cell surface and intracellular FACS staining using saponin-based permeabilization buffers were as previously described (73, 78). Alternatively, transfected cells were fixed with 2% PFA (10 min at room temperature), permeabilized with 90% methanol, and stored at –20°C in 90% methanol prior to chemokine staining and analysis. All samples were acquired on FACSCalibur, FACSCanto, or LSR II flow cytometers (BD Biosciences) and analyzed with CellQuest, DIVA (BD Biosciences), and/or FlowJo (TreeStar) software.
Selection of chemokine-specific pAbs for use in FC.
Based on initial observations, we defined several criteria for the choice of chemokine-specific pAbs potentially suitable for use in FC. (a) Immunizing antigen: Use of full-length chemokines rather than peptide epitopes generates a spectrum of pAbs likely containing subsets that retain reactivity even under conditions of cell fixation. However, because the relative preponderance of these subsets is unpredictable, the intensities of pAb-specific stains over background (signal/noise ratio) is somewhat variable. (b) Affinity purification: This limits the spectrum of pAb species to those specific for the target antigen. (c) Host species: In our hands, pAbs generated in goats and sheep demonstrate less background staining than those generated in rabbits or hamsters. (d) Detection Abs: Fluorochrome-conjugated F(ab′)2 fragments with minimal species crossreactivity are the preferred reagents. For reasons that remain unclear, we found that the same donkey anti-goat F(ab′)2 conjugated to FITC or Cy5 consistently provided better results than did the PE conjugate.
Evaluation of pAbs for chemokine FC.
To assure pAb specificity and appropriate sensitivity, it is essential to combine multiple complementary controls. (a) We used HEK or Cos7 cells transfected with bicistronic vectors (GFP/chemokine) to identify suitable pAbs (Figure 1), evaluate staining protocols (all pAbs described here work under conditions of both saponin and methanol permeabilization), and determine the extent of pAb crossreactivities (Supplemental Figure 1). (b) For those chemokines known to be produced in response to defined stimuli, we cultured cells in the absence or presence of specific stimuli prior to intracellular chemokine staining. (c) As a general negative control, preimmune serum [IgG] from the same host species in which the pAb of interest was raised was used at a concentration commensurate to that of chemokine-specific Abs. However, the extent of background staining obtained with different pAbs, even if generated by standardized experimental protocols, is somewhat variable, so the data must be interpreted with great care. (d) The preincubation of pAbs with corresponding recombinant chemokines and use of the resulting pAb/chemokine complex as a staining reagent (cold-block control) is widely used (e.g., ref. 24); however, it cannot rule out specific crossreactivity: if the Ab specifically recognizes antigens A and B, blockade with antigen A will also abolish recognition of antigen B. Nevertheless, cold-block controls are well-suited, and preferable to IgG controls, for establishing the level of background staining for individual pAbs. (e) Chemokine-deficient tissues are perhaps the best negative control, but are obviously limited to the availability of respective mouse strains. Nevertheless, it is possible, depending on the nature of the gene-targeting construct, that expression of truncated chemokine proteins does not completely abolish chemokine detection even if an effective functional loss is achieved. (f) The utility of chemokine FC is limited by its relative sensitivity, i.e., the number of specific molecules expressed by individual cells and the specific signal/noise ratios of the staining reagents. However, the overall robust signal/noise ratios of chemokine-specific Abs selected for our study (Figure 1 and Supplemental Figure 1), provide a practical orientation for their effective use as reagents for FC-based distinction of chemokine-expressing and nonexpressing cell subsets.
Chemokine FC.
Intracellular chemokine stains were performed using affinity-purified pAbs (CXCL17 pAb, sheep; all other pAbs, goat) as well as goat and sheep IgG controls, all provided by F. Mortari (R&D Systems, Minneapolis, Minnesota; Table 1). Chemokine expression was visualized by staining of cells with no more than 0.1 μg pAb for 45 minutes at room temperature in a volume of 50 μl, followed by detection of primary pAb with FITC- or Cy5-conjugated donkey anti-goat or -sheep F(ab′)2 fragments (Jackson Immunoresearch) for 30 minutes at room temperature. Where available, chemokine-specific mAbs were also used (PE-conjugated anti-CCL2, clone 2H5, Biolegend; and PE-conjugated anti-CCL3, clone 39624, allophycocyanin-conjugated anti-CCL6, clone 262016, anti-CXCL12, clone 79018, and anti-hCXCL14, clone 131120, R&D Systems). Preconjugation of mAbs, pAbs, and corresponding control Abs to Alexa Fluor 488 or Alexa Fluor 647 was performed using Zenon technology (Invitrogen). For detection of human chemokines, we used fluorochrome-conjugated mAbs (allophycocyanin-conjugated anti-CCL3, clone 93342, FITC-conjugated anti-CCL4, clone 24006, FITC-conjugated anti-CCL5, clone 21445, PE-conjugated anti-CCL22, clone 57203, and FITC-conjugated anti-CXCL8, clone 6217) and affinity-purified goat pAbs specific for CCL1 (AF272), CCL5 (AF278-NA), and XCL1 (AF695; all from R&D Systems). Although not formally tested, the XCL1-specific AF695 pAb likely recognizes both XCL1 and XCL2, as these chemokines differ by only 2 amino acids in positions 7 and 8 (79).
Microarray data and analysis.
We used deposited data from gene array analyses of resting and IL-15–stimulated murine NK cells, performed by Fehniger et al. (59), from the NCBI GEO Web site (accession GSE7764). For the purpose of our investigations, we downloaded the raw data, performed MAS5 normalizations, and determined mRNA expression levels for all murine chemokines. The only statistically significant difference between resting and IL-15–stimulated NK cells at the level of chemokine transcripts pertained to the approximately 3.5-fold downregulation of Ccl5 mRNA after 24 hours of IL-15 treatment (data not shown). In addition, the public Reference Database of Immune Cells (63) was used to assess global chemokine mRNA profiles of various murine and human immune cell subsets.
Statistics.
Data handling, analysis, and graphic representation (all shown as mean ± SEM) was performed using Prism 4.0a (GraphPad Software). Statistical significance was calculated by 2-tailed Student’s t test, with P < 0.05 considered significant (*P < 0.05; **P < 0.01; ***P < 0.001).
Supplementary Material
Acknowledgments
This work was supported by Juvenile Diabetes Research Foundation Career Development Award 2-2007-240, NIH grant AG026518-01, a Barbara Davis Center Pilot and Feasibility grant, and Diabetes and Endocrinology Research Center grant P30-DK057516 (to D. Homann) as well as by NIH grants RO1 DK060590 and U19 A 1066328-01 and a VA Merit Review Grant (to H.R. Rosen). We thank R. Torres for the FGK45.5 mAb and expert advice; L. Lenz for rLM-OVA; T. Sosinowski for the mIl15 construct; C. Henry for advice about BMDC cultures; the University of Colorado Denver Diabetes and Endocrinology Research Center Molecular Biology Core, R. Wong, and C. Patel for assistance with vector design, PCR, and sequencing; N. Castelblanco for technical assistance; and F. Mortari for the gift of most of the chemokine pAbs used in this study. Due to the large number of chemokines discussed, we have not been able to provide a truly comprehensive overview of the relevant literature, and we apologize to the authors whose work we have not cited here.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(3):907–923. doi:10.1172/JCI40645.
References
- 1.Rot A, von Andrian UH. Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annu Rev Immunol. 2004;22:891–928. doi: 10.1146/annurev.immunol.22.012703.104543. [DOI] [PubMed] [Google Scholar]
- 2.Rossi D, Zlotnik A. The biology of chemokines and their receptors. Annu Rev Immunol. 2000;18:217–242. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- 3.Kunkel EJ, Butcher EC. Chemokines and the tissue-specific migration of lymphocytes. Immunity. 2002;16(1):1–4. doi: 10.1016/S1074-7613(01)00261-8. [DOI] [PubMed] [Google Scholar]
- 4.Cyster JG. Chemokines, sphingosine-1-phosphate, and cell migration in secondary lymphoid organs. Annu Rev Immunol. 2005;23:127–159. doi: 10.1146/annurev.immunol.23.021704.115628. [DOI] [PubMed] [Google Scholar]
- 5.Cyster JG. Chemokines and cell migration in secondary lymphoid organs. Science. 1999;286(5447):2098–2102. doi: 10.1126/science.286.5447.2098. [DOI] [PubMed] [Google Scholar]
- 6.Ansel KM, Cyster JG. Chemokines in lymphopoiesis and lymphoid organ development. Curr Opin Immunol. 2001;13(2):172–179. doi: 10.1016/S0952-7915(00)00201-6. [DOI] [PubMed] [Google Scholar]
- 7.Luther SA, Cyster JG. Chemokines as regulators of T cell differentiation. Nat Immunol. 2001;2(2):102–107. doi: 10.1038/84205. [DOI] [PubMed] [Google Scholar]
- 8.Campbell DJ, Kim CH, Butcher EC. Chemokines in the systemic organization of immunity. Immunol Rev. 2003;195:58–71. doi: 10.1034/j.1600-065X.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- 9.Gerard C, Rollins BJ. Chemokines and disease. Nat Immunol. 2001;2(2):108–115. doi: 10.1038/84209. [DOI] [PubMed] [Google Scholar]
- 10.Zlotnik A. Chemokines and cancer. Ernst Schering Res Found Workshop. 2004;(45):53–58. doi: 10.1007/978-3-662-05403-1_4. [DOI] [PubMed] [Google Scholar]
- 11.Locati M, Murphy PM. Chemokines and chemokine receptors: biology and clinical relevance in inflammation and AIDS. Annu Rev Med. 1999;50:425–440. doi: 10.1146/annurev.med.50.1.425. [DOI] [PubMed] [Google Scholar]
- 12.Kunkel SL, Godessart N. Chemokines in autoimmunity: from pathology to therapeutics. Autoimmun Rev. 2002;1(6):313–320. doi: 10.1016/S1568-9972(02)00085-X. [DOI] [PubMed] [Google Scholar]
- 13.Yopp AC, Krieger NR, Ochando JC, Bromberg JS. Therapeutic manipulation of T cell chemotaxis in transplantation. Curr Opin Immunol. 2004;16(5):571–577. doi: 10.1016/j.coi.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Zlotnik A, Yoshie O, Nomiyama H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol. 2006;7(12):243. doi: 10.1186/gb-2006-7-12-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12(2):121–127. doi: 10.1016/S1074-7613(00)80165-X. [DOI] [PubMed] [Google Scholar]
- 16.Allen SJ, Crown SE, Handel TM. Chemokine: receptor structure, interactions, and antagonism. Annu Rev Immunol. 2007;25:787–820. doi: 10.1146/annurev.immunol.24.021605.090529. [DOI] [PubMed] [Google Scholar]
- 17.Weber C, Koenen RR. Fine-tuning leukocyte responses: towards a chemokine ‘interactome’. Trends Immunol. 2006;27(6):268–273. doi: 10.1016/j.it.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Dorner BG, et al. MIP-1alpha, MIP-1beta, RANTES, and ATAC/lymphotactin function together with IFN-gamma as type 1 cytokines. Proc Natl Acad Sci U S A. 2002;99(9):6181–6186. doi: 10.1073/pnas.092141999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber SE, et al. Adaptive islet-specific regulatory CD4 T cells control autoimmune diabetes and mediate the disappearance of pathogenic Th1 cells in vivo. J Immunol. 2006;176(8):4730–4739. doi: 10.4049/jimmunol.176.8.4730. [DOI] [PubMed] [Google Scholar]
- 20.Cantor J, Haskins K. Recruitment and activation of macrophages by pathogenic CD4 T cells in type 1 diabetes: evidence for involvement of CCR8 and CCL1. J Immunol. 2007;179(9):5760–5767. doi: 10.4049/jimmunol.179.9.5760. [DOI] [PubMed] [Google Scholar]
- 21.Dorner BG, et al. Coordinate expression of cytokines and chemokines by NK cells during murine cytomegalovirus infection. J Immunol. 2004;172(5):3119–3131. doi: 10.4049/jimmunol.172.5.3119. [DOI] [PubMed] [Google Scholar]
- 22.Faunce DE, Stein-Streilein J. NKT cell-derived RANTES recruits APCs and CD8+ T cells to the spleen during the generation of regulatory T cells in tolerance. J Immunol. 2002;169(1):31–38. doi: 10.4049/jimmunol.169.1.31. [DOI] [PubMed] [Google Scholar]
- 23.Matloubian M, David A, Engel S, Ryan JE, Cyster JG. A transmembrane CXC chemokine is a ligand for HIV-coreceptor Bonzo. Nat Immunol. 2000;1(4):298–304. doi: 10.1038/79738. [DOI] [PubMed] [Google Scholar]
- 24.Beaty SR, Rose CE, Jr, Sung SS. Diverse and potent chemokine production by lung CD11bhigh dendritic cells in homeostasis and in allergic lung inflammation. J Immunol. 2007;178(3):1882–1895. doi: 10.4049/jimmunol.178.3.1882. [DOI] [PubMed] [Google Scholar]
- 25.Matzer SP, Rodel F, Strieter RM, Rollinghoff M, Beuscher HU. Constitutive expression of CXCL2/MIP-2 is restricted to a Gr-1high, CD11b+, CD62Lhigh subset of bone marrow derived granulocytes. Int Immunol. 2004;16(11):1675–1683. doi: 10.1093/intimm/dxh169. [DOI] [PubMed] [Google Scholar]
- 26.Friedman RS, Jacobelli J, Krummel MF. Surface-bound chemokines capture and prime T cells for synapse formation. Nat Immunol. 2006;7(10):1101–1108. doi: 10.1038/ni1384. [DOI] [PubMed] [Google Scholar]
- 27.Lore K, Sonnerborg A, Spetz AL, Andersson U, Andersson J. Immunocytochemical detection of cytokines and chemokines in Langerhans cells and in vitro derived dendritic cells. J Immunol Methods. 1998;214(1–2):97–111. doi: 10.1016/S0022-1759(98)00040-4. [DOI] [PubMed] [Google Scholar]
- 28.Peterson FC, Thorpe JA, Harder AG, Volkman BF, Schwarze SR. Structural determinants involved in the regulation of CXCL14/BRAK expression by the 26 S proteasome. J Mol Biol. 2006;363(4):813–822. doi: 10.1016/j.jmb.2006.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christensen JE, Andreasen SO, Christensen JP, Thomsen AR. CD11b expression as a marker to distinguish between recently activated effector CD8(+) T cells and memory cells. Int Immunol. 2001;13(4):593–600. doi: 10.1093/intimm/13.4.593. [DOI] [PubMed] [Google Scholar]
- 30.Lin Y, Roberts TJ, Sriram V, Cho S, Brutkiewicz RR. Myeloid marker expression on antiviral CD8+ T cells following an acute virus infection. Eur J Immunol. 2003;33(10):2736–2743. doi: 10.1002/eji.200324087. [DOI] [PubMed] [Google Scholar]
- 31.Ghosn EE, Yang Y, Tung J, Herzenberg LA. CD11b expression distinguishes sequential stages of peritoneal B-1 development. Proc Natl Acad Sci U S A. 2008;105(13):5195–5200. doi: 10.1073/pnas.0712350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lanier LL, Phillips JH, Hackett J, Jr, Tutt M, Kumar V. Natural killer cells: definition of a cell type rather than a function. J Immunol. 1986;137(9):2735–2739. [PubMed] [Google Scholar]
- 33.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5(12):953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 34.Vremec D, et al. Production of interferons by dendritic cells, plasmacytoid cells, natural killer cells, and interferon-producing killer dendritic cells. Blood. 2007;109(3):1165–1173. doi: 10.1182/blood-2006-05-015354. [DOI] [PubMed] [Google Scholar]
- 35.Poltorak AN, et al. MIP-1 gamma: molecular cloning, expression, and biological activities of a novel CC chemokine that is constitutively secreted in vivo. J Inflamm. 1995;45(3):207–219. [PubMed] [Google Scholar]
- 36.Coelho AL, et al. The chemokine CCL6 promotes innate immunity via immune cell activation and recruitment. J Immunol. 2007;179(8):5474–5482. doi: 10.4049/jimmunol.179.8.5474. [DOI] [PubMed] [Google Scholar]
- 37.Link A, et al. Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nat Immunol. 2007;8(11):1255–1265. doi: 10.1038/ni1513. [DOI] [PubMed] [Google Scholar]
- 38.Alferink J, et al. Compartmentalized production of CCL17 in vivo: strong inducibility in peripheral dendritic cells contrasts selective absence from the spleen. J Exp Med. 2003;197(5):585–599. doi: 10.1084/jem.20021859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Britschgi MR, Link A, Lissandrin TK, Luther SA. Dynamic modulation of CCR7 expression and function on naive T lymphocytes in vivo. J Immunol. 2008;181(11):7681–7688. doi: 10.4049/jimmunol.181.11.7681. [DOI] [PubMed] [Google Scholar]
- 40.Mueller SN, et al. Regulation of homeostatic chemokine expression and cell trafficking during immune responses. Science. 2007;317(5838):670–674. doi: 10.1126/science.1144830. [DOI] [PubMed] [Google Scholar]
- 41.Unsoeld H, et al. Abrogation of CCL21 chemokine function by transgenic over-expression impairs T cell immunity to local infections. Int Immunol. 2007;19(11):1281–1289. doi: 10.1093/intimm/dxm098. [DOI] [PubMed] [Google Scholar]
- 42.Stenstad H, Svensson M, Cucak H, Kotarsky K, Agace WW. Differential homing mechanisms regulate regionalized effector CD8alphabeta+ T cell accumulation within the small intestine. Proc Natl Acad Sci U S A. 2007;104(24):10122–10127. doi: 10.1073/pnas.0700269104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Homey B, et al. CCL27-CCR10 interactions regulate T cell-mediated skin inflammation. Nat Med. 2002;8(2):157–165. doi: 10.1038/nm0202-157. [DOI] [PubMed] [Google Scholar]
- 44.Huang V, et al. Cutting edge: rapid accumulation of epidermal CCL27 in skin-draining lymph nodes following topical application of a contact sensitizer recruits CCR10-expressing T cells. J Immunol. 2008;180(10):6462–6466. doi: 10.4049/jimmunol.180.10.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaffner A, Rhyn P, Schoedon G, Schaer DJ. Regulated expression of platelet factor 4 in human monocytes--role of PARs as a quantitatively important monocyte activation pathway. J Leukoc Biol. 2005;78(1):202–209. doi: 10.1189/jlb.0105024. [DOI] [PubMed] [Google Scholar]
- 46.El-Gedaily A, Schoedon G, Schneemann M, Schaffner A. Constitutive and regulated expression of platelet basic protein in human monocytes. J Leukoc Biol. 2004;75(3):495–503. doi: 10.1189/jlb.0603288. [DOI] [PubMed] [Google Scholar]
- 47.Piqueras B, Connolly J, Freitas H, Palucka AK, Banchereau J. Upon viral exposure, myeloid and plasmacytoid dendritic cells produce 3 waves of distinct chemokines to recruit immune effectors. Blood. 2006;107(7):2613–2618. doi: 10.1182/blood-2005-07-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Hundelshausen P, Petersen F, Brandt E. Platelet-derived chemokines in vascular biology. Thromb Haemost. 2007;97(5):704–713. doi: 10.1160/th07-01-0066. [DOI] [PubMed] [Google Scholar]
- 49.Lasagni L, et al. An alternatively spliced variant of CXCR3 mediates the inhibition of endothelial cell growth induced by IP-10, Mig, and I-TAC, and acts as functional receptor for platelet factor 4. J Exp Med. 2003;197(11):1537–1549. doi: 10.1084/jem.20021897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mueller A, et al. CXCL4-induced migration of activated T lymphocytes is mediated by the chemokine receptor CXCR3. J Leukoc Biol. 2008;83(4):875–882. doi: 10.1189/jlb.1006645. [DOI] [PubMed] [Google Scholar]
- 51.Farber JM. Mig and IP-10: CXC chemokines that target lymphocytes. J Leukoc Biol. 1997;61(3):246–257. [PubMed] [Google Scholar]
- 52.Sierro F, et al. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc Natl Acad Sci U S A. 2007;104(37):14759–14764. doi: 10.1073/pnas.0702229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robertson MJ. Role of chemokines in the biology of natural killer cells. J Leukoc Biol. 2002;71(2):173–183. [PubMed] [Google Scholar]
- 54.Cosman D, et al. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14(2):123–133. doi: 10.1016/S1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 55.Andrew DP, et al. STCP-1 (MDC) CC chemokine acts specifically on chronically activated Th2 lymphocytes and is produced by monocytes on stimulation with Th2 cytokines IL-4 and IL-13. J Immunol. 1998;161(9):5027–5038. [PubMed] [Google Scholar]
- 56.Kumar D, Hosse J, von Toerne C, Noessner E, Nelson PJ. JNK MAPK pathway regulates constitutive transcription of CCL5 by human NK cells through SP1. J Immunol. 2009;182(2):1011–1020. doi: 10.4049/jimmunol.182.2.1011. [DOI] [PubMed] [Google Scholar]
- 57.Catalfamo M, et al. Human CD8+ T cells store RANTES in a unique secretory compartment and release it rapidly after TcR stimulation. Immunity. 2004;20(2):219–230. doi: 10.1016/S1074-7613(04)00027-5. [DOI] [PubMed] [Google Scholar]
- 58.Stegelmann F, et al. Coordinate expression of CC chemokine ligand 5, granulysin, and perforin in CD8+ T cells provides a host defense mechanism against Mycobacterium tuberculosis. J Immunol. 2005;175(11):7474–7483. doi: 10.4049/jimmunol.175.11.7474. [DOI] [PubMed] [Google Scholar]
- 59.Fehniger TA, et al. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 2007;26(6):798–811. doi: 10.1016/j.immuni.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 60.Harris DP, et al. Reciprocal regulation of polarized cytokine production by effector B and T cells. Nat Immunol. 2000;1(6):475–482. doi: 10.1038/82717. [DOI] [PubMed] [Google Scholar]
- 61.Bystry RS, Aluvihare V, Welch KA, Kallikourdis M, Betz AG. B cells and professional APCs recruit regulatory T cells via CCL4. Nat Immunol. 2001;2(12):1126–1132. doi: 10.1038/ni735. [DOI] [PubMed] [Google Scholar]
- 62.Schaniel C, et al. Activated murine B lymphocytes and dendritic cells produce a novel CC chemokine which acts selectively on activated T cells. J Exp Med. 1998;188(3):451–463. doi: 10.1084/jem.188.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hijikata A, et al. Construction of an open-access database that integrates cross-reference information from the transcriptome and proteome of immune cells. Bioinformatics. 2007;23(21):2934–2941. doi: 10.1093/bioinformatics/btm430. [DOI] [PubMed] [Google Scholar]
- 64.Castellino F, Huang AY, Altan-Bonnet G, Stoll S, Scheinecker C, Germain RN. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature. 2006;440(7086):890–895. doi: 10.1038/nature04651. [DOI] [PubMed] [Google Scholar]
- 65.Lukacs-Kornek V, Engel D, Tacke F, Kurts C. The role of chemokines and their receptors in dendritic cell biology. Front Biosci. 2008;13:2238–2252. doi: 10.2741/2838. [DOI] [PubMed] [Google Scholar]
- 66.Swift S, Lorens J, Achacoso P, Nolan GP. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. Curr Protoc Immunol. 2001;Chapter 10:Unit 10.17C. doi: 10.1002/0471142735.im1017cs31. May . [DOI] [PubMed] [Google Scholar]
- 67.Makino Y, et al. Impaired T cell function in RANTES-deficient mice. Clin Immunol. 2002;102(3):302–309. doi: 10.1006/clim.2001.5178. [DOI] [PubMed] [Google Scholar]
- 68.Tsou CL, et al. Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J Clin Invest. 2007;117(4):902–909. doi: 10.1172/JCI29919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aslanian AM, Charo IF. Targeted disruption of the scavenger receptor and chemokine CXCL16 accelerates atherosclerosis. Circulation. 2006;114(6):583–590. doi: 10.1161/CIRCULATIONAHA.105.540583. [DOI] [PubMed] [Google Scholar]
- 70.Eslin DE, et al. Transgenic mice studies demonstrate a role for platelet factor 4 in thrombosis: dissociation between anticoagulant and antithrombotic effect of heparin. Blood. 2004;104(10):3173–3180. doi: 10.1182/blood-2003-11-3994. [DOI] [PubMed] [Google Scholar]
- 71.Park MK, et al. The CXC chemokine murine monokine induced by IFN-gamma (CXC chemokine ligand 9) is made by APCs, targets lymphocytes including activated B cells, and supports antibody responses to a bacterial pathogen in vivo. J Immunol. 2002;169(3):1433–1443. doi: 10.4049/jimmunol.169.3.1433. [DOI] [PubMed] [Google Scholar]
- 72.Homann D, McGavern DB, Oldstone MB. Visualizing the viral burden: phenotypic and functional alterations of T cells and APCs during persistent infection. J Immunol. 2004;172(10):6239–6250. doi: 10.4049/jimmunol.172.10.6239. [DOI] [PubMed] [Google Scholar]
- 73.Lenz DC, et al. IL-7 regulates basal homeostatic proliferation of antiviral CD4+T cell memory. Proc Natl Acad Sci U S A. 2004;101(25):9357–9362. doi: 10.1073/pnas.0400640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ericsson A, Kotarsky K, Svensson M, Sigvardsson M, Agace W. Functional characterization of the CCL25 promoter in small intestinal epithelial cells suggests a regulatory role for caudal-related homeobox (Cdx) transcription factors. J Immunol. 2006;176(6):3642–3651. doi: 10.4049/jimmunol.176.6.3642. [DOI] [PubMed] [Google Scholar]
- 75.Henry CJ, Ornelles DA, Mitchell LM, Brzoza-Lewis KL, Hiltbold EM. IL-12 produced by dendritic cells augments CD8+ T cell activation through the production of the chemokines CCL1 and CCL17. J Immunol. 2008;181(12):8576–8584. doi: 10.4049/jimmunol.181.12.8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pope C, et al. Organ-specific regulation of the CD8 T cell response to Listeria monocytogenes infection. J Immunol. 2001;166(5):3402–3409. doi: 10.4049/jimmunol.166.5.3402. [DOI] [PubMed] [Google Scholar]
- 77.Humann J, Bjordahl R, Andreasen K, Lenz LL. Expression of the p60 autolysin enhances NK cell activation and is required for listeria monocytogenes expansion in IFN-gamma-responsive mice. J Immunol. 2007;178(4):2407–2414. doi: 10.4049/jimmunol.178.4.2407. [DOI] [PubMed] [Google Scholar]
- 78.Homann D, Teyton L, Oldstone MB. Differential regulation of antiviral T-cell immunity results in stable CD8+ but declining CD4+ T-cell memory. Nat Med. 2001;7(8):913–919. doi: 10.1038/90950. [DOI] [PubMed] [Google Scholar]
- 79.Yoshida T, et al. Structure and expression of two highly related genes encoding SCM-1/human lymphotactin. FEBS Lett. 1996;395(1):82–88. doi: 10.1016/0014-5793(96)01004-6. [DOI] [PubMed] [Google Scholar]
- 80.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19(1):71–82. doi: 10.1016/S1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.