Abstract
The southern and northern Japanese populations of the medaka fish provide useful tools to gain insights into the comparative genomics and speciation of vertebrates, because they can breed to produce healthy and fertile offspring despite their highly divergent genetic backgrounds compared with those of human–chimpanzee. Comparative genomics analysis has suggested that such large genetic differences between the two populations are caused by higher molecular evolutionary rates among the medakas than those of the hominids. The argument, however, was based on the assumption that the two Japanese populations diverged approximately at the same time (4.0–4.7 Myr ago) as the human–chimpanzee lineage (5.0–6.0 Myr ago). This can be misleading, because the divergence time of the two populations was calculated based on estimated, extremely higher molecular evolutionary rates of other fishes with an implicit assumption of a global molecular clock. Here we show that our estimate, based on a Bayesian relaxed molecular-clock analysis of whole mitogenome sequences from 72 ray-finned fishes (including 14 medakas), is about four times older than that of the previous study (18 Myr). This remarkably older estimate can be reconciled with the vicariant events of the Japanese archipelago, and the resulting rates of molecular evolution are almost identical between the medaka and hominid lineages. Our results further highlight the fact that reproductive isolation may not evolve despite a long period of geographical isolation.
Keywords: divergence time, vicariance, plate tectonics, single nucleotide polymorphism
1. Introduction
The medaka Oryzias latipes is a small (ca 3–4 cm) oviparous freshwater teleost, placed in the family Adrianichthyidae of the order Beloniformes (Nelson 2006). The medaka has long been used as an excellent model organism for various fields of biology, including ecotoxicology, carcinogenesis, sex determination and developmental genetics (Wittbrodt et al. 2002). Recently, the medaka has attracted considerable attention as a model system for evolutionary biology and comparative genomics, because of the availability of inbred strains derived from two highly diverged, regional populations in Japan that can mate and produce healthy and fertile offspring (Ishikawa 2000).
A high-quality draft whole-genome sequence from one of the inbred strains (Hd-rR) was published recently (Kasahara et al. 2007), enabling the use of the medaka as a model organism for comparative genomics. The comparative genomics of model teleosts have provided novel insights into the structure and evolution of whole genomes in vertebrates. For example, comparative studies of whole genome sequences from two puffer fishes (Aparicio et al. 2002; Jaillon et al. 2004) with that of the human have illuminated the evolution of vertebrate genomes, such as the unequivocal confirmation of a whole-genome duplication event in the common ancestor of teleosts and the unprecedented reconstruction of the proto-karyotype of bony vertebrates (Jaillon et al. 2004).
Comparative genomics analysis of the medaka, on the other hand, revealed that the average pairwise divergence per nucleotide site between the two Japanese populations was 3.42 per cent, the highest rate seen in any vertebrate species (referred to as single nucleotide polymorphisms (SNPs) rate in Kasahara et al. (2007) and Takeda (2008) and hereafter following their usage). Further, Takeda (2008) stated that SNP rates of the two medaka populations were significantly higher than those of the human–chimpanzee (3.42% versus 1.23%) based on the assumption that the two Japanese populations diverged approximately at the same time (4.0–4.7 Myr ago) as the human–chimpanzee lineage (5.0–6.0 Myr ago). The statement, however, can be misleading because a divergence time of the medaka (4.0–4.7 Myr ago) was calculated using estimated, extremely higher molecular evolutionary rates from other fishes (2.5–2.8% Myr−1; Takehana et al. 2003), which implicitly assumes a global molecular clock.
Recent developments in the molecular estimation of divergence times have provided promising tools to introduce time scales for phylogenetic trees (Yang 2006). One of the most significant advances common to these new methods is a departure from the molecular clock assumption and another one is the use of time constraints at multiple nodes for rate calibration, usually based on the fossil record. In higher teleosts, however, the fossil record is scarce and fragmentary, and alternative calibration points based on biogeographical events are expected to be useful for the divergence time estimation. Azuma et al. (2008) recently reported that estimated divergence times of cichlid fishes showed excellent agreement with the history of Gondwanan continental fragmentations, arguing that such vicariant events can be used as effective time constraints in dating teleostean divergences. Biogeographical information from cichlids should be useful for divergence time estimation of the medaka and its congeneric relatives (hereafter called ‘medaka fishes’) as multiple calibration points for both minimum and maximum time constraints, because these two fish groups are demonstrably more closely related to each other than previously thought (e.g. Setiamarga et al. 2008).
In this paper, we estimated the divergence time of medaka fishes within the framework of the entire evolutionary history of the ray-finned fishes using a Bayesian relaxed molecular-clock method. In accordance with Azuma et al. (2008), we used a suggested 21 fossil-based and six biogeography-based time constraints with slight modifications. The resulting estimated divergence times are remarkably older than the previous ones, but can be reconciled with the vicariant events of the Japanese archipelago, providing a more likely time scale for evolutionary history of the medaka fishes and thereby offering alternative interpretations for evolution of the vertebrate genomes.
2. Material and methods
Taxonomic sampling other than the atherinomorphs (medaka fishes and their relatives) follows that of Azuma et al. (2008) who used all major actinopterygian lineages represented by 50 species and two sarcopterygians (coelacanth and lungfish) plus two sharks as outgroups for rooting trees. Twenty-two atherinomorphs (including 14 medaka fishes) were newly added and the total number of species analysed in this study was 76 (electronic supplementary material, table S1). Oryzias latipes designated with Hd-rR (southern Japanese population) is the same strain as used in Kasahara et al. (2007) for whole genome sequencing.
We newly determined whole mitogenome sequences from eight medaka fishes using a combination of long and short polymerase chain reactions and direct cycle sequencing techniques following the methods developed by Miya & Nishida (1999).
Mitogenome sequences from the 22 atherinomorphs were concatenated with the pre-aligned sequences used in Azuma et al. (2008) with MAFFT v. 6 (Katoh & Toh 2008). The dataset comprises 6966 positions from first and second codon positions of the 12 protein-coding genes (excluding the ND6 gene), 1673 positions from the two rRNA genes and 1407 positions from the 22 tRNA genes (a total of 10 046 positions).
Unambiguously aligned sequences were divided into four partitions (first, second codon positions, rRNA and tRNA genes) and subjected to the partitioned maximum-likelihood (ML) analysis using RAxML v. 7.0.4 (Stamatakis 2006). The best-scoring ML tree was estimated using a general time reversible + gamma model of sequence evolution with 1000 bootstrap replicates (–f an option in RAxML).
Divergence time was estimated using a multidistribute program package (Thorne & Kishino 2002) by assuming a topology obtained thereby but without assuming the molecular clock (i.e. by allowing heterogeneity in molecular evolutionary rates along branches). Upper and/or lower time constraints at selected nodes were set for the Bayesian Markov chain Monte Carlo processes to estimate divergence times (posterior means and 95% credible intervals). Twenty-one fossil-based and six biogeography-based time constraints from Azuma et al. (2008) were used in this study (electronic supplementary material, table S2).
More details of the methods can be found in the electronic supplementary material.
3. Results and discussion
Partitioned ML analysis recovered a tree topology that is identical to those obtained in Azuma et al. (2008); Setiamarga et al. (2008) and Takehana et al. (2005) overall for ray-finned fishes, atherinomorphs and medaka fishes, respectively (electronic supplementary material, figure S1). Relationships between three species groups of medaka fishes ((javanicus, celebensis), latipes) were reproduced with higher bootstrap probabilities than those obtained in Takehana et al. (2005) (89% versus 68%).
Divergence time estimates in the present study show excellent agreement with the previous studies (electronic supplementary material, table S3; figure 1), indicating that the analysis is not sensitive to differences in taxon sampling (addition of 22 atherinomorphs) and settings of the priors (updated time constraints). The medaka fishes (family Adrianichthyidae) are estimated to have diverged from an ancestral lineage of the Belonidae 118 Myr ago (106–130 Myr ago; 95% credible interval) (figure 1). Although a common ancestor of the medaka fishes has failed to leave extant lineages for about 50 Myr, it has subsequently diversified into the three major species groups in a relatively short time period, around a boundary between the Cretaceous and Paleogene (K/P boundary) 65 Myr ago (figure 1).
Figure 1.
Divergence times of ray-finned fishes estimated from the partitioned Bayesian analysis using a multidistribute program package (Thorne & Kishino 2002). A total of 27 nodes (A–U) were used for time constraints (for details, see table S2 in the electronic supplementary material). Horizontal bars indicate 95% credible intervals of the divergence time estimates.
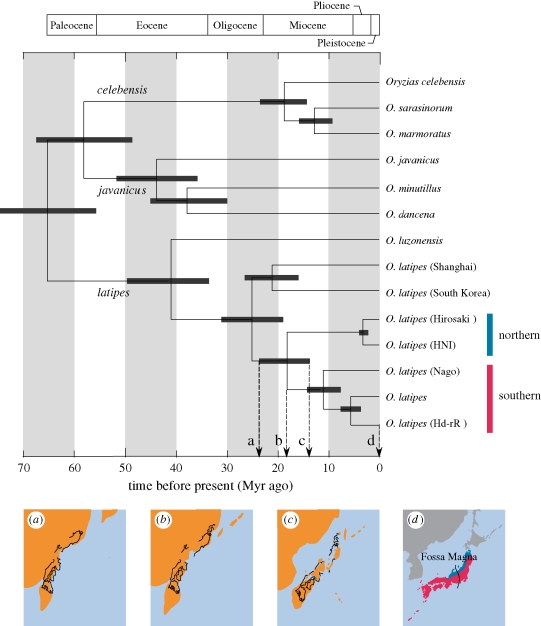
A common ancestor of the medaka (O. latipes) was estimated to have diverged from a sister lineage (Oryzias luzonensis) 41 Myr ago (33–50 Myr ago), which is followed by the divergence between continental (Shanghai and South Korea) and Japanese populations (southern and northern populations) 25 Myr ago (20–33 Myr ago) (figure 2). Subsequently, the two regional Japanese populations diverged 18 Myr ago (13–24 Myr ago), which is about four times older than the previous estimate (4.0–4.7 Myr ago; Takehana et al. 2003). The latter younger estimate, however, was calculated uncritically based on an estimated nucleotide substitution rate of freshwater gobies of the genus Gymonogobius (2.5–2.8% Myr−1; Harada et al. 2002), which is one of the fastest rates assumed for freshwater fishes (Watanabe et al. 2006), and application of such a rate to the divergence time of distantly related medaka fishes requires a global molecular-clock assumption.
Figure 2.
Divergence times of the medaka and medaka fishes. (a)–(c) Plate tectonic process of the Japanese archipelago ((a) 24 Myr ago, (b) 18 Myr ago and (c) 13 Myr ago; reconstructed from Ogasawara (1993)) schematically shown under the time tree with the present distribution ranges (d). Oryzias latipes (Hd-rR) is the same strain used in Kasahara et al. (2007) for whole-genome sequencing.
With the absence of a reliable fossil record for the medaka (but see Parenti 2008), our older divergence time estimate of 18 Myr ago (13–24 Myr ago in 95% credible interval) can be reconciled with the vicariant events of the Japanese archipelago (figure 2a–c). Recent palaeomagnetic studies demonstrated that the Japanese archipelago started to separate from the Asian continent during the early Miocene about 20 Myr ago, with the initial eastward drift of the northeastern landmasses followed by that of the southwestern landmasses 15 Myr ago, while undergoing clockwise and counterclockwise tectonic rotations, respectively (Watanabe et al. 2006). This putative plate tectonic process of the Japanese archipelago is consistent with both divergence patterns and time of the Japanese medaka, strongly suggesting the ‘out-of-Asia’ vicariant origins of the southern and northern Japanese populations. Of course, we acknowledge that formation of the present-day distribution patterns of the two populations require dispersal scenarios across the boundary between the two landmasses called ‘Fossa Magna’ (figure 2d).
If the estimated divergence time (18 Myr ago) is used for scaling the genomics comparisons as has been done in the previous studies (e.g. Takeda 2008), then nucleotide substitution rates for the SNPs of the medaka and human–chimpanzee are almost identical, at 0.19% Myr−1 and 0.21% Myr−1, respectively. This good agreement contradicts the previous argument for significantly faster molecular evolutionary rates in the medaka, providing an alternative view that accumulation of SNPs has occurred at a similar rate in two distantly related lineages. Patterns of the accumulation of SNPs, however, may differ from each other, because it was suggested that differential selective pressures act on specific gene categories in the two lineages (Kasahara et al. 2007). Nevertheless, our older estimated divergence time between the two regional populations of the medaka, while providing a new and more reliable time scale for comparative genomics of model organisms, highlights the fact that reproductive isolation may not evolve despite a long period of geographical isolation, and a resulting morphological and ecological diversification (Naruse et al. 2004).
Acknowledgements
Sincere thanks to K. Naruse, Y. Ozawa, A. Shimada, H. Takeda and K. Yamahira for providing materials, to H. Katoh and S. Isaji for geological information and to D. Koop and two anonymous reviewers for their critical comments on the manuscript. This study has been supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan (12NP0201, 15380131, 17207007 and 19207007).
References
- Aparicio S., et al. 2002Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297, 1301–1310 (doi:10.1126/science.1072104) [DOI] [PubMed] [Google Scholar]
- Azuma Y., Kumazawa Y., Miya M., Mabuchi K., Nishida M.2008Mitogenomic evaluation of the historical biogeography of cichlids toward reliable dating of teleostean divergences. BMC Evol. Biol. 8, 215 (doi:10.1186/1471-2148-8-215) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S., Jeon S.-R., Kinoshita I., Tanaka M., Nishida M.2002Phylogenetic relationships of four species of floating gobies (Gymnogobius) as inferred from partial mitochondrial cytochrome b gene sequences. Ichthyol. Res. 49, 324–332 (doi:10.1007/s102280200048) [Google Scholar]
- Ishikawa Y.2000Medaka fish as a model system for vertebrate developmental genetics. BioEssays 22, 487–495 (doi:10.1002/(SICI)1521-1878(200005)22:5<487::AID-BIES11>3.0.CO;2-8) [DOI] [PubMed] [Google Scholar]
- Jaillon O., et al. 2004Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431, 946–957 (doi:10.1038/nature03025) [DOI] [PubMed] [Google Scholar]
- Kasahara M., et al. 2007The medaka draft genome and insights into vertebrate genome evolution. Nature 447, 714–719 (doi:10.1038/nature05846) [DOI] [PubMed] [Google Scholar]
- Katoh K., Toh H.2008Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinformatics 9, 286–298 (doi:10.1093/bib/bbn013) [DOI] [PubMed] [Google Scholar]
- Miya M., Nishida M.1999Organization of the mitochondrial genome of a deep-sea fish Gonostoma gracile (Teleostei: Stomiiformes): first example of transfer RNA gene rearrangements in bony fishes. Mar. Biotechnol. 1, 416–426 (doi:10.1007/PL00011798) [DOI] [PubMed] [Google Scholar]
- Naruse K., Hori H., Shimizu N., Kohara Y., Takeda H.2004Medaka genomics: a bridge between mutant phenotype and gene function. Mech. Dev. 121, 619–628 (doi:10.1016/j.mod.2004.04.014) [DOI] [PubMed] [Google Scholar]
- Nelson J. S.2006Fishes of the world, 4th edn Hoboken, NJ: John Wiley & Sons [Google Scholar]
- Ogasawara K.1993Neogene paleogeography and marine climate of the Japanese Islands based on shallow-marine molluscs. Palaeogeogr. Palaeoclimatol. Palaeoecol. 108, 335–351 (doi:10.1016/0031-0182(94)90241-0) [Google Scholar]
- Parenti L. R.2008A phylogenetic analysis and taxonomic revision of ricefishes, Oryzias and relatives (Beloniformes: Adrianichthyidae). Zool. J. Linn. Soc. 154, 494–610 [Google Scholar]
- Setiamarga D. H. E., Miya M., Yamanoue Y., Mabuchi K., Satoh T. P., Inoue J. G., Nishida M.2008Interrelationships of Atherinomorpha (medakas, flyingfishes, killifishes, silversides, and their relatives): the first evidence based on whole mitogenome sequences. Mol. Phylogenet. Evol. 49, 598–605 (doi:10.1016/j.ympev.2008.08.008) [DOI] [PubMed] [Google Scholar]
- Stamatakis A.2006RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (doi:10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- Takeda H.2008Draft genome of the medaka fish: a comprehensive resource for medaka developmental genetics and vertebrate evolutionary biology. Dev. Growth Differ. 50, S157–S166 (doi:10.1111/j.1440-169X.2008.00992.x) [DOI] [PubMed] [Google Scholar]
- Takehana Y., Naga N., Matsuda M., Tsuchiya K., Sakaizumi M.2003Geographic variation and diversity of the cytochrome b gene in Japanese wild populations of medaka Oryzias latipes. Zool. Sci. 20, 1279–1291 (doi:10.2108/zsj.20.1279) [DOI] [PubMed] [Google Scholar]
- Takehana M., Naruse K., Sakaizumi M.2005Molecular phylogeny of the medaka fishes genus Oryzias (Beloniformes: Adrianichthyidae) based on nuclear and mitochondrial DNA sequences. Mol. Phylogenet. Evol. 36, 417–428 (doi:10.1016/j.ympev.2005.01.016) [DOI] [PubMed] [Google Scholar]
- Thorne J. L., Kishino H.2002Divergence time and evolutionary rate estimation with multilocus data. Syst. Biol. 51, 689–702 (doi:10.1080/10635150290102456) [DOI] [PubMed] [Google Scholar]
- Watanabe K., et al. 2006Biogeographical history of Japanese freshwater fishes: phylogeographic approaches and perspectives. Jpn J. Ichthyol. 53, 1–38 [Google Scholar]
- Wittbrodt J., Shima A., Schartl M.2002Medaka: a model organism from the Far East. Nat. Genet. 3, 53–64 (doi:10.1038/nrg704) [DOI] [PubMed] [Google Scholar]
- Yang Z.2006Computational molecular evolution New York, NY: Oxford University Press [Google Scholar]