Abstract
In flowering plants, pollen formation depends on the differentiation and interaction of two cell types in the anther: the reproductive cells, called microsporocytes, and somatic cells that form the tapetum. The microsporocytes generate microspores, whereas the tapetal cells support the development of microspores into mature pollen grains. Despite their importance to plant reproduction, little is known about the underlying genetic mechanisms that regulate the differentiation and interaction of these highly specialized cells in the anther. Here, we report the identification and characterization of a novel TAPETUM DETERMINANT1 (TPD1) gene that is required for the specialization of tapetal cells in the Arabidopsis anther. Analysis of the male-sterile mutant, tpd1, showed that functional interruption of TPD1 caused the precursors of tapetal cells to differentiate and develop into microsporocytes instead of tapetum. As a results, extra microsporocytes were formed and tapetum was absent in developing tpd1 anthers. Molecular cloning of TPD1 revealed that it encodes a small protein of 176 amino acids. In addition, tpd1 was phenotypically similar to excess microsporocytes1/extra sporogenous cells (ems1/exs) single and tpd1 ems1/exs double mutants. These data suggest that the TPD1 product plays an important role in the differentiation of tapetal cells, possibly in coordination with the EMS1/EXS gene product, a Leu-rich repeat receptor protein kinase.
INTRODUCTION
In higher plants, the formation of the haploid gametophytes from the diploid sporophytes results from two sequential processes: sporogenesis and gametogenesis. Sporogenesis starts with the differentiation of hypodermal cells of ovules and anthers to form archesporial cells, which further generate the reproductive sporocytes and supportive somatic cells, whereas gametogenesis is characterized by the development of haploid spores into mature gametophytes. Genetic studies have revealed that the interaction between the sporocytes and sporophytic tissues is important to spore formation (Yang and Sundaresan, 2000). In male organs of flowering plants, called anthers, the formation of microspores relies on the interaction of reproductive cells called microsporocytes with several types of somatic anther wall cells, including the tapetum. Therefore, the development of tapetal cells also is important to microspore formation. Although several sporophytic mutants that affect the development of tapetal cells have been reported (Dawson et al., 1993; Taylor et al., 1998; Sanders et al., 1999; Wilson et al., 2001; Ito and Shinozaki, 2002), the molecular and genetic mechanisms that control the interaction of tapetal cells with microsporocytes are poorly understood.
Our previous study of the Arabidopsis SPOROCYTELESS/NOZZLE (SPL/NZZ) gene revealed that its product plays an important role at early stages of anther development (Schiefthaler et al., 1999; Yang et al., 1999). SPL/NZZ encoded a putative MADS-related transcription factor, and its expression was restricted in the developing microsporocytes in the anther. The fact that the spl mutation blocked not only microsporocyte formation but also somatic anther wall development suggested that the development of the somatic anther wall cells, including the tapetum, is dependent on signaling from developing microsporocytes. However, it is still not clear which genes are involved in the signal transduction from developing microsporocytes to developing tapetal cells. Recently, a gene called EXCESS MICROSPOROCYTES1/EXTRA SPOROGENOUS CELLS (EMS1/EXS) was reported to be involved in the development of tapetal cells in Arabidopsis (Canales et al., 2002; Zhao et al., 2002).
Characterization of ems1/exs mutants showed that the EMS1/EXS product was required for tapetal cell fate determination. Disruption of EMS1/EXS caused the precursors of tapetal cells to develop into microsporocytes instead of tapetum. EMS1/EXS encoded a putative plasma membrane–bound Leu-rich repeat receptor protein kinase, which possibly defines a signaling pathway that controls tapetal cell fate. The predominant expression of EMS1/EXS in developing tapetal cells suggested that its product may perceive a signal received by the developing tapetal cells. However, the source and nature of the signal remains unknown. Here, we report the isolation and characterization of a novel TAPETUM DETERMINANT1 (TPD1) gene that plays a role in the specialization of tapetal cells in the Arabidopsis anther. Our results indicate that TPD1 and EMS1/EXS have similar genetic functions in tapetum development, because tpd1 was phenotypically similar to ems1/exs single and tpd1 ems1/exs1-2 double mutants. TPD1 encodes a small protein of 176 amino acids and is expressed mostly in developing microsporocytes in the anther. Our data suggest that the TPD1 product may play a key role in the specialization of tapetal cell fate, possibly in coordination with EMS1/EXS.
RESULTS
Isolation of tpd1
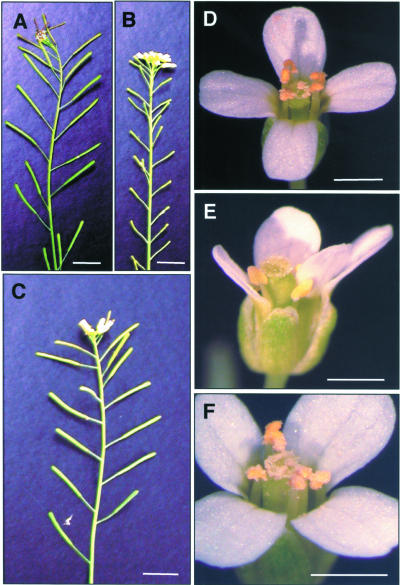
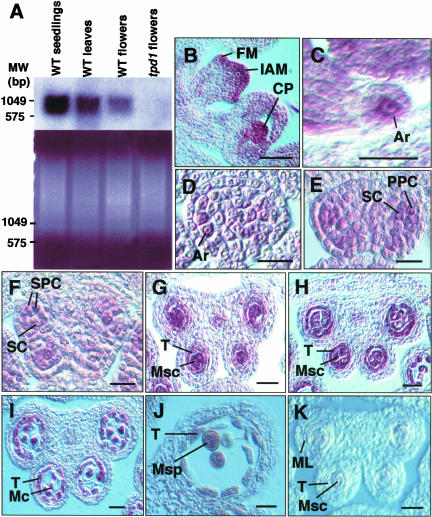
The tpd1 mutant was identified by its complete male sterility (Figure 1B) in a phenotypic screen of the enhancer-trap and gene-trap Dissociation (Ds) insertion lines in Arabidopsis ecotype Landsberg erecta (Sundaresan et al., 1995). The tpd1 plant produced no pollen grain (Figure 1E) and thus was male sterile. Analysis of anther sections showed that tpd1 was devoid of tapetal cells (Figures 2E and 2J). The second male-sterile mutant was identified in the same screen (see Figure 5C). When it was used as a female parent in a cross with tpd1 heterozygous plants, the F1 plants from the crosses segregated as 1:1 (fertile:male sterile), indicating that the second male-sterile mutant was a new allele of tpd1. Thus, we renamed the first allele tpd1-1 and the second allele tpd1-2. Both alleles produced morphologically normal flowers with a normal number of floral organs (Figure 1E). No morphological abnormality in vegetative parts was observed.
Figure 1.
Phenotype of tpd1-1 and a Complemented Mutant.
(A) A wild-type plant with fertility indicated by the siliques with normal seed set.
(B) A tpd1-1 plant with sterility indicated by the small siliques without seeds.
(C) Plant from a transgenic tpd1-1 background line with normal fertility complemented by a wild-type 5.281-kb TPD1 genomic DNA fragment.
(D) A wild-type flower, showing the anthers with pollen grains.
(E) A tpd1-1 flower, showing the anthers without pollen grains.
(F) A complemented tpd1-1 flower, showing the anthers with pollen grains.
Bars = 1 cm in (A) to (C) and 1 mm in (D) to (F).
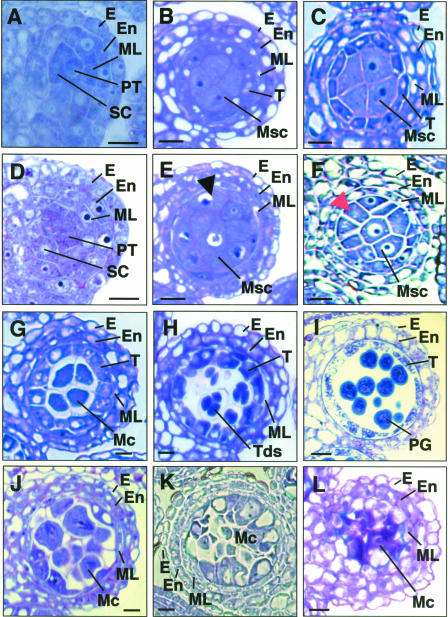
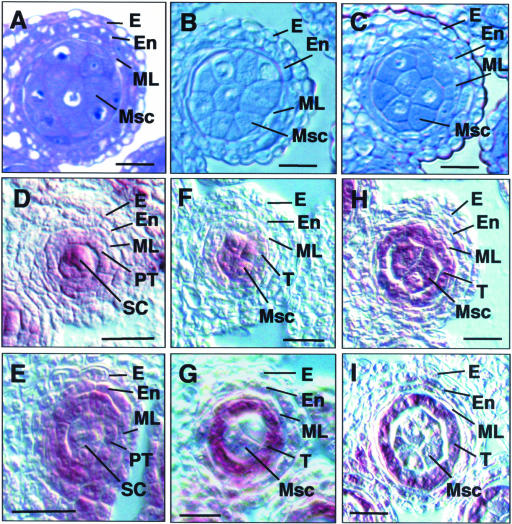
Figure 2.
Anther Development in tpd1-1 Compared with the Wild Type.
The micrographs show one of the four lobes in transverse anther sections.
(A) A wild-type anther at stage 4, showing the normal endothecium, middle layer, primary tapetum, and sporogenous cells.
(B) and (C) Wild-type anthers at early (B) and late (C) stage 5, showing the well-organized four anther wall cell layers and larger microsporocytes at locule center.
(D) A tpd1-1 anther at stage 4, showing the morphologically normal endothecium, middle layer, primary tapetum, and sporogenous cells.
(E) and (F) tpd1-1 anthers at early (E) and late (F) stage 5, showing the extra microsporocytes at layer 4, indicated by arrows.
(G) A wild-type anther at stage 6, showing the meiocytes detaching from tapetal cells and from each other by depositing callose on the cell walls and the collapsing middle layer.
(H) A wild-type anther at stage 7, showing the tetrads.
(I) A wild-type anther at stage 11, showing the pollen grains and collapsing tapetal cells.
(J) A tpd1-1 anther at stage 6, showing the extra meiocytes and the abnormal middle layer.
(K) A tpd1-1 anther at stage 7, showing that no tetrad was formed and that the middle layer was abnormal.
(L) A tpd1-1 anther at stage 14, showing the cell debris from the degenerated meiocytes and the highly vacuolated middle layer.
E, epidermis; En, endothecium; Mc, meiocyte; ML, middle layer; Msc, microsporocyte; PG, pollen grain; PT, primary tapetum; SC, sporogenous cell; T, tapetum; Tds, tetrads. Bars = 10 μm in (A) to (H) and (J) to (K) and 20 μm in (I) and (L).
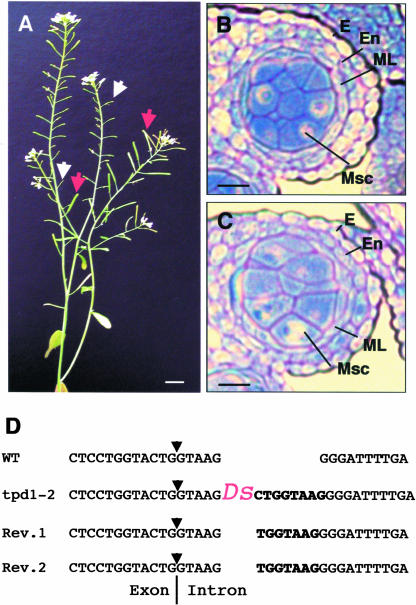
Figure 5.
Phenotype and Characterization of tpd1-2.
(A) A tpd1-2 plant with the reversion sectors, showing the revertant siliques (red arrows) adjacent to the small infertile mutant siliques (white arrows).
(B) and (C) tpd1-1 and tpd1-2 exhibited similar phenotypes, as demonstrated by the formation of the extra microsporocytes in both tpd1-1 (B) and tpd1-2 (C) anthers at stage 5.
(D) The Ds insertion site in the tpd1-2 genome. Boldface letters indicate a typical 8-bp repeat sequence generated by the Ds insertion in the tpd1-2 genome and 7-bp footprint sequences in the revertant genomes. The arrows indicate the splicing site of the third intron.
Bars = 1 cm in (A) and 20 μm in (B) and (C).
In both tpd1 alleles, male sterility segregated as a single recessive mutation. When tpd1-1 was used as the female parent in crosses with the wild-type plant, nearly all ovules (563 of 569) produced viable seeds similar to the wild-type siliques, indicating that the tpd1-1 mutation did not affect female fertility. A similar result was obtained when tpd1-2 was used as the female parent and the wild type was used as the male parent. All F1 seedlings resulting from these crosses were resistant to kanamycin upon germination and were fertile when mature. The F2 progeny of heterozygous tpd1 plants segregated 3:1 (resistant:sensitive) for kanamycin. One-third of the kanamycin-resistant F2 seedlings gave rise to male-sterile plants. This result indicated linkage between the male sterility mutation and a kanamycin selection marker carried by a single Ds element in both alleles. We further confirmed a single Ds insertion in each tpd1 mutant genome by DNA gel blot hybridization using a 755-bp 5′ end fragment of the Ds element as a probe (data not shown). In addition, when tpd1-1 heterozygous plants were used in reciprocal crosses with wild-type plants, the F1 seedlings from both crosses showed the same segregation ratio of 1:1 kanamycin resistant to kanamycin sensitive (209:212 and 252:243, respectively). Therefore, the tpd1 mutations do not affect either male or female gametophytic development.
Phenotypic Characterization of tpd1
To elucidate the biological function of TPD1, we compared the development of anthers in the wild type and in tpd1 mutants. In Arabidopsis, anther development and microspore formation follow the pattern typical of dicotyledonous plants (Sanders et al., 1999; Yang et al., 1999). The entire process has been divided into 14 stages (Sanders et al., 1999). The anther at stage 1 is a three-layer structure. At stage 2, hypodermal cells at four corners expand radially and develop into archesporial cells. Then, archesporial cells undergo a periclinal division to give rise to outer primary parietal cells and inner primary sporogenous cells at stage 3. The primary sporogenous cells develop further into microsporocytes at stages 4 to 5. The primary parietal cells subsequently divide periclinally and anticlinally to form two sets of secondary parietal cells at early stage 4. During stage 4, the inner set of secondary parietal cells adjacent to sporogenous cells divides and develops into primary tapetum. The outer set of secondary parietal cells undergoes one more periclinal division to generate the outer endothecium and the inner middle layer (Figure 2A). In the tpd1 anther, all cell types were formed normally during stages 1 to 3. No differences in morphological structures between wild-type and tpd1 anthers were observed at stage 4 (Figure 2D).
A clear defect in the tpd1 anther was first observed at early stage 5. During stage 5, the wild-type anther develops into a four-lobe structure (Goldberg et al., 1993; Sanders et al., 1999). In each lobe, five cell layers, from the exterior to the interior—the epidermis, endothecium, middle layer, tapetum, and microsporocyte—are well organized. The microsporocytes typically are larger than other cells and packed at the center of each locule. The tapetal cells also are recognizable, with their rectangular shape and position between the larger microsporocytes and the thinner middle layer (Figures 2B and 2C). In the tpd1 anther at stage 5, the endothecium, middle layer, and microsporocytes were morphologically normal. However, the cells at layer 4 were not well organized. They became larger and were morphologically similar to microsporocytes instead of tapetal cells (Figures 2E and 2F). Thus, the tpd1 locule appeared to contain excess microsporocytes and to lack a tapetum. In the wild-type anther at stage 6, the tapetal cells become binucleate and vacuolated and the microsporocytes are isolated from tapetum and one from another by the deposition of callose on the cell walls. Meanwhile, the middle layer degenerates and collapses into a thin line (Figure 2G). In the tpd1 anther at stage 6, the microsporocytes also became isolated as in the wild type. However, the middle layer was relatively thicker than that in the wild type (Figure 2J).
During stage 7, wild-type microsporocytes undergo meiosis and form tetrads (Figure 2H). Then, individual microspores are released from tetrads by degeneration of the callose cell walls and develop further into pollen grains during stages 8 to 12 (Figure 2I). By contrast, no tetrad was formed in the tpd1 anther at stage 7 (Figure 2K). Meiotic analysis showed that the meiotic nuclear division was normal (Figure 2H). Obviously, meiotic cytokinesis did not occur or failed to complete, resulting in the degeneration of microsporocytes. As a result, the mature tpd1 anther contained no pollen grains and only cell debris were observed (Figure 2L). The middle layer in the tpd1 anther remained abnormal until the final stage of development and eventually became highly vacuolated (Figure 2L). To summarize, we conclude that the tpd1 mutation affects the formation of tapetum and functional microspores and the normal degeneration of the middle layer.
The tpd1 Mutation Alters the Identity of Tapetal Cells
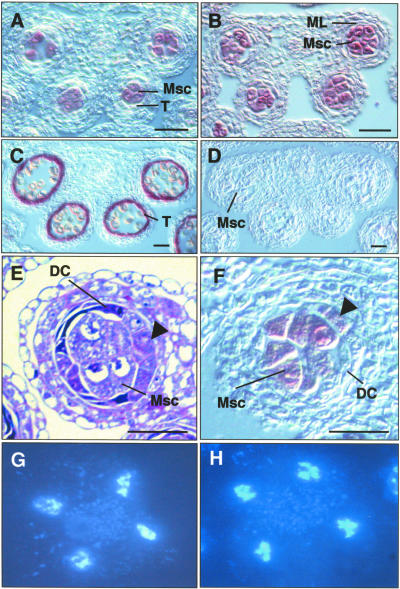
To confirm the identity of the extra microsporocytes in the tpd1 anther, we performed RNA in situ hybridization on sections using antisense SOLO DANCERS (SDS) RNA as a microsporocyte-specific probe (Figure 3A) (Azumi et al., 2002) and antisense ARABIDOPSIS THALIANA ANTHER7 (ATA7) RNA as a tapetum-specific probe (Figure 3C) (Rubinelli et al., 1998). The antisense SDS RNA probe strongly hybridized to the microsporocytes in the tpd1 anthers at stage 5 (Figure 3B), but the antisense ATA7 RNA probe did not (Figure 3D). These results confirm the finding that the tpd1 anther produced extra microsporocytes and lacked a tapetum.
Figure 3.
tpd1-1 Anthers with Extra Microsporocytes and Normal Meiotic Nuclear Divisions but without Tapetum.
(A) to (D) and (F) RNA in situ hybridization on transverse sections of wild-type and tpd1 anthers.
(A) A wild-type anther at stage 5, showing the specific expression of SDS in microsporocytes.
(B) A tpd1 anther at stage 5, showing the expression of SDS in the extra microsporocytes.
(C) A wild-type anther at stage 8, showing the specific expression of ATA7 in the tapetal cells.
(D) A tpd1 anther at stage 8, showing the absence of the tapetum-specific ATA7 RNA signal.
(E) A tpd1 anther at stage 5. The arrow indicates the partially formed microsporocyte cell layer, which is occupied by the tapetum in the wild-type anther.
(F) RNA in situ hybridization on a tpd1 anther at stage 5. The arrow indicates the expression signal of SDS in the extra microsporocytes at layer 4.
(G) Meiotic nuclear division of a wild-type microsporocyte.
(H) Meiotic nuclear division of a tpd1-1 microsporocyte.
DC, degenerated cell; ML, middle layer; Msc, microsporocyte; T, tapetum. Bars = 20 μm.
In the wild-type anther, tapetal cells originate from the inner secondary parietal cells, which are formed at early stage 4. These cells divide and differentiate into primary tapetal cells at late stage 4 and develop further into tapetum at stage 5. In the tpd1 anther, the inner secondary parietal cells were formed normally (Figure 2D). To determine whether the additional microsporocytes that were formed at stage 5 originated from the inner secondary parietal cells or from the primary tapetal cells, we examined 63 and 61 independent locules of transverse middle sections of wild-type and tpd1 anthers, respectively. The average numbers of microsporocytes and tapetal cells per wild-type locule were 5.0 ± 1.0 and 12.9 ± 1.4, respectively. By contrast, the average number of microsporocytes per tpd1 locule was 16.2 ± 2.0, indicating that this number is the sum of microsporocytes and tapetal cells per wild-type locule. These results suggested that the extra microsporocytes in tpd1 anthers originated from the primary tapetal cells. Additional support was provided by the fact that the extra microsporocytes were formed at the position of layer 4 at early stage 5 (Figure 2E), where tapetal cells are formed in wild-type anthers. Furthermore, we occasionally found that some of tpd1 anther lobes at stage 5 contained a partially formed cell layer that was clearly located at the position of layer 4 (Figure 3E), which normally is occupied by tapetal cells in wild-type anthers. In situ hybridization using SDS antisense RNA probe showed signals in some of the cells at the partially formed layer 4 (Figure 3F), indicating that they were microsporocytes. These results strongly suggest that TPD1 is required for the specialization of tapetal cells and prevents the differentiation of primary tapetal cells into microsporocytes.
Molecular Cloning of TPD1
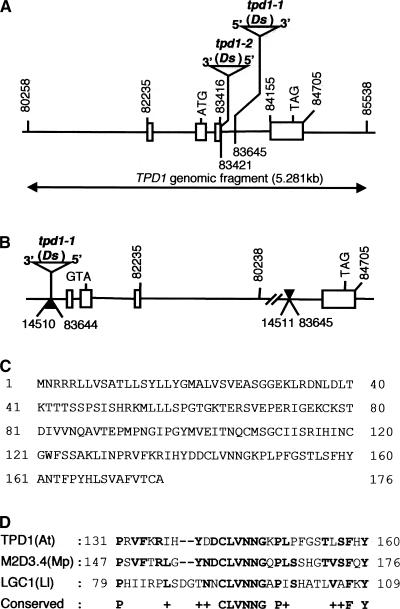
To identify the TPD1 gene, we performed thermal asymmetric interlaced PCR (TAIL-PCR) (Liu et al., 1995; Grossniklaus et al., 1998) to obtain the genomic flanking sequences adjacent to both ends of the Ds element in tpd1-1 and tpd1-2. Sequencing of the TAIL-PCR products revealed that the insertion of the Ds element caused an inversion translocation of a 69.113-kb genomic DNA fragment on chromosome IV (BAC F6I7) (Figure 4B) in the tpd1-1 genome. In particular, the flanking sequence adjacent to the 3′ end of the Ds element was located in an intergene region, whereas that adjacent to the 5′ end of the Ds element was located inside the third intron of a predicted gene related to At4g24973 (Figures 4A and 4B). In tpd1-2, Ds also inserted into the third intron of the same gene (Figure 4A).
Figure 4.
Characterization of the TPD1 Gene and tpd1 Mutations.
(A) Organization of the TPD1 gene and Ds insertion sites. The rectangular boxes show the exons, and the lines between the exons indicate the positions of the introns. The arrow line indicates the TPD1 genomic fragment used for the complementation of tpd1-1. The numbers indicate the positions of the start and stop nucleotides of the fragments located on BAC clone F6I7.
(B) The mutation caused by inversion translocation in the tpd1-1 genome. The solid arrows indicate the recombination sites. The site adjacent to the 3′ end of the Ds element is located in an intergene area, which does not interrupt any predicted gene; the other end is located in the third intron of TPD1 and breaks down TPD1.
(C) Sequence of the TPD1 protein with 176 amino acids.
(D) Comparison of the TPD1 region at amino acids 131 to 160 with M2D3.4 from liverwort and LGC1 from lily. Boldface letters indicate amino acids that are identical in any two sequences.
To determine whether the Ds insertions were responsible for male sterility in tpd1 mutants, an Activator (Ac) element was reintroduced to both tpd1 allele plants to remobilize the Ds element (Sundaresan et al., 1995; Yang et al., 1999). If the male sterility was caused by the Ds insertions, tpd1 homozygous plants in the presence of Ac should exhibit sectors of reversion, resulting in wild-type flowers with normal siliques adjacent to mutant flowers with infertile siliques. In the case of tpd1-1, no revertant was obtained. Characterization of the Ds border sequences by TAIL-PCR revealed a 3-bp deletion within the 3′ end terminus of Ds (data not shown). Therefore, Ds had lost its capability for mobilization. However, revertants were obtained in tpd1-2 homozygous plants in the presence of Ac (Figure 5A). Further investigation of the insertion site by PCR showed that the reversions resulted from remobilization of the Ds element from the third intron of TPD1 (Figure 5D). We conclude that the male sterility in tpd1 mutants was caused by independent Ds insertions into the same gene.
To further confirm that knocking out TPD1 is responsible for the tpd1 phenotype, a 5.281-kb genomic DNA fragment including the predicted promotor, transcription region, and 3′ end nontranscription region (Figure 4A) was amplified by high-fidelity PCR and introduced into tpd1-1 heterozygous plants by infiltration. Sixty-three independent transformants were obtained in the screen. Sixty-two of 63 T1 seedlings gave rise to fully fertile plants. In the T2 generation, 100% of the seedlings from 15 of 62 T2 families were resistant to kanamycin, indicating that they were tpd1-1 homozygous plants. All hygromycin-selected seedlings from the 15 T2 families gave rise to normal fully fertile plants (Figures 1C and 1F), whereas nonselected T2 plants segregated in male sterility, in concordance with hygromycin-resistant segregation. These results indicate that the 5.281-kb genomic fragment from chromosome IV contains all of the genetic information required for the normal functioning of TPD1.
A Basic Local Alignment Search Tool (BLASTN) search of EST databases was performed with the 5.281-kb genomic sequence and showed that a 580-bp EST sequence was a match to the 5′ end sequence of the predicted transcription region of TPD1. However, RNA gel blot hybridization indicated that the full-length mRNA is likely to be ∼1 kb in length (Figure 6A). We used reverse transcriptase–mediated PCR to clone the full-length cDNA. The cloned cDNA has 935 nucleotides. Phenotypic complementation of tpd1-1 by the introduction of the 935-bp cDNA fragment driven by the TPD1 promoter and terminator showed that the 935-bp mRNA was sufficient to encode the full functional product of TPD1 (data not shown).
Figure 6.
Expression of TPD1.
(A) Expression of TPD1 in wild-type seedlings, leaves, and flower buds, as revealed by RNA gel blot hybridization. The bottom gel shows mRNA loading controls. Five micrograms of mRNA was loaded in each lane.
(B) to (K) Expression patterns in wild-type flowers, as revealed by RNA in situ hybridization.
(B) TPD1 expression in inflorescence apical meristem, floral meristem, and carpel primordia.
(C) TPD1 expression in ovule primordia, showing the stronger TPD1 signal in the female archesporial cell.
(D) TPD1 expression in archesporial cells in an anther at stage 2.
(E) TPD1 expression in primary parietal cells and sporogenous cells in an anther at stage 3.
(F) TPD1 expression in secondary parietal cells and sporogenous cells in an anther at stage 4.
(G) TPD1 expression in an anther at early stage 5, showing the predominant signal in microsporocytes.
(H) TPD1 expression in an anther at late stage 5, showing that the TPD1 RNA signal was predominant in both tapetal cells and microsporocytes.
(I) TPD1 expression was reduced greatly in an anther at stage 6.
(J) TPD1 expression was not detected in an anther at stage 10.
(K) Control section of an anther at stage 5, which was hybridized with the sense TPD1 RNA probe, showing only the background signal compared with the stronger signals on the other sections hybridized with the antisense TPD1 RNA probe.
Ar, archesporial cell; CP, carpel primordia; FM, floral meristem; IAM, inflorescence apical meristem; Mc, meiocyte; ML, middle layer; Msc, microsporocyte; Msp, microspore; MW, molecular weight markers; PPC, primary parietal cell; SC, sporogenous cell; SPC, secondary parietal cell; T, tapetum; WT, wild type. Bars = 5 μm in (C) and 20 μm in all other panels.
TPD1 Encodes an Unknown Protein
The TPD1 mRNA encodes an unknown protein of 176 amino acids (Figure 4C). A BLASTP search performed with the full-length sequence of TPD1 showed no significant similarity to other known proteins. However, it shared low identity in different regions with a group of unknown small proteins (Table 1). A putative rice protein shared the greatest identity (44%) with TPD1. More detailed analysis showed that TPD1 contained a six–amino acid region (CLVNNG) that was conserved in two other proteins that were less similar to TPD1 (Figure 4D). One is M2D3.4 from male tissues of the liverwort Marchantia polymorpha (Ishizaki et al., 2002); the other is LGC1 from Lilium longiflorum (Xu et al., 1999). However, the most similar rice protein mentioned above does not contain this conserved region.
Table 1.
Small Proteins That Share Identity with TPD1
| Protein Name | Size (Amino Acids) | Regions TPD1/– | Percent Identical (Amino Acids) |
Species and Characterization | Accession No. |
|---|---|---|---|---|---|
| Unknown | 169 | 75-175/69-168 | 44 (45 of 101) | Rice (Oryza sativa); unknown | AC092387 |
| M2D3.4 | 195 | 51-174/66-190 | 34 (43 of 126) | Liverwort (Marchantia polymorpha) male organ-specific expression |
AAN74746 |
| LGC1 | 128 | 77-170/27-120 | 27 (26 of 96) | Lily (Lilium longiflorum); pollen-specific expression |
AAD19962 |
| Unknown | 124 | 95-166/42-109 | 33 (24 of 72) | Arabidopsis thaliana; unknown | NP194937 |
| Unknown | 134 | 99-168/55-124 | 33 (24 of 72) | Arabidopsis thaliana; unknown | AAL87299 |
| orf121 | 121 | 99-171/17-89 | 24 (18 of 74) | Staphylococcus aureus; unknown | NP075473 |
Expression of TPD1
RNA gel blot hybridization showed that TPD1 was expressed in leaves, young seedlings, and flower buds (Figure 6A). To further investigate the expression of TPD1 in anther development, we performed RNA in situ hybridization with sections of wild-type flower buds at different stages. TPD1 RNA was detectable in archesporial cells at stage 2 (Figure 6D), in primary sporogenous cells and primary parietal cells at stage 3 (Figure 6E), and in secondary parietal cells at early stage 4 (Figure 6F). TPD1 RNA signals in microsporocytes became stronger at stage 5 (Figure 6G). At late stage 5, TPD1 RNA was expressed predominantly in both tapetum and microsporocytes (Figure 6H). Expression of TPD1 in tapetum and microsporocytes was reduced greatly at stage 6 (Figure 6I) and then declined gradually until it was not detectable at stage 10 (Figure 6J). Compared with the control, in which sense TPD1 RNA was used as a probe for hybridization, only a slight background signal was observed (Figure 6K). Therefore, we conclude that the expression of TPD1 is associated with the development of tapetal cells and microsporocytes.
TPD1 also was expressed in inflorescence meristems, floral meristems, carpel primordia (Figure 6B), and ovule primordia (Figure 6C), in which cells divide and differentiate actively. However, because no visible phenotype other than in the anthers was observed in the null tpd1 plants, its function remains unknown in tissues other than anthers.
TPD1 and EMS1/EXS Exhibit Partially Complementary Expression Patterns during Anther Development
From the same collection of Ds insertion lines, we identified a new ems1/exs allele, named ems1/exs1-2, which contained a Ds insertion into the kinase domain of EMS1/EXS (data not shown). Phenotypic comparisons revealed that tpd1 and ems1/exs exhibited similar phenotypes (Figures 7A and 7B). In addition, the tpd1 ems1/exs1-2 double mutant also exhibited the phenotype identical to tpd1 and ems1/exs1-2 single mutants (Figure 7C). These results suggested that TPD1 and EMS1/EXS might act in the same developmental process. To gain insight into the possible interaction mechanism between TPD1 and EMS1/EXS, we compared their expression patterns in developing anthers by RNA in situ hybridization. The expression patterns of TPD1 and EMS1/EXS in anthers overlapped with each other at stages 1 to 3. At late stage 4, TPD1 was expressed mostly in sporogenous cells, the microsporocyte precursors (Figure 7D), whereas EMS1/EXS RNA was detected mostly in primary tapetum (Figure 7E). At early stage 5, TPD1 RNA was found mostly in microsporocytes (Figure 7F), whereas EMS1/EXS RNA was found mostly in tapetal cells (Figure 7G). During stage 5, TPD1 RNA signal gradually became stronger in tapetal cells, and eventually it was predominant in both tapetal cells and microsporocytes at late stage 5 (Figure 7H), whereas the signal of EMS1/EXS RNA remained predominant in the tapetum (Figure 7I). These results indicate that TPD1 and EMS1/EXS exhibit complementary expression patterns during late stage 4 and early stage 5, the developmental stages at which the mutant phenotypes were first observed.
Figure 7.
Comparison of tpd1-1 and ems1/exs1-2 and Their Expression Patterns in Anthers by in Situ Hybridization.
(A) to (C) One of the four lobes from anther sections of tpd1-1 (A), ems1/exs1-2 (B), and tpd1-1 ems1/exs1-2 (C) at stage 5, showing that they exhibited similar phenotypes, as demonstrated by the formation of extra microsporocytes.
(D) and (E) Wild-type anthers at late stage 4.
(D) Predominant TPD1 expression in sporogenous cells.
(E) Predominant EMS1/EXS expression in the primary tapetum.
(F) and (G) Wild-type anthers at early stage 5.
(F) TPD1 RNA was detected predominantly in microsporocytes.
(G) EMS1 RNA was found mostly in tapetal cells.
(H) and (I) Wild-type anthers at late stage 5.
(H) TPD1 was expressed mostly in the tapetum and microsporocytes.
(I) EMS1 was expressed predominantly in the tapetum.
E, epidermis; En, endothecium; ML, middle layer; Msc, microsporocyte; PT, primary tapetum; SC, sporogenous cell; T, tapetum. Bars = 20 μm.
DISCUSSION
TPD1 Is Required for Tapetal Cell Fate
We report a novel gene, TPD1, that is involved in the regulation of cell fates in the Arabidopsis anther. Morphological analysis of tpd1 mutants showed that TPD1 is required for the formation of tapetal cells, as demonstrated by the absence of tapetum in the tpd1 anther. The fact that the precursors of tapetal cells developed into microsporocytes instead of tapetum in the tpd1 anther suggests that TPD1 is required for the developmental fates of tapetal precursors instead of their initiation.
In the wild type, primary tapetal cells originate from inner secondary parietal cells that are generated from primary parietal cells derived from archesporial cells (Yang et al., 1999). Although TPD1 is expressed in all cell types mentioned above, no morphological abnormality of the archesporial cells and parietal cells was observed in the tpd1 anther, indicating that the TPD1 product is not required for very early development. TPD1 shared identity in different regions with a group of unknown small proteins (Table 1). However, no evidence was obtained to indicate that TPD1 acts redundantly with other genes in those cell types. It is possible that TPD1 may have no function in those cell types other than in the tapetum and developing microsporocytes.
TPD1 is expressed mostly in developing microsporocytes during late stage 4 and early stage 5, the developmental stages at which the mutant phenotypes were first observed. However, the tpd1 mutation affected tapetum formation instead of microsporocyte formation. One possible reason for this finding is that the expression of TPD1 in the developing tapetal cells may be too weak to ensure their autonomous development, and the development of tapetal cells may depend mainly on the expression of TPD1 in the developing microsporocytes. If so, TPD1 may be involved in the interaction between developing tapetal cells and microsporocytes. In addition, the development of the tapetal precursors into microsporocytes in tpd1 indicated that microsporocyte fate might be a default pathway (Zhao et al., 2002) and that the specialization of tapetal cell fate requires a pathway that is dependent on TPD1.
The tpd1 Mutation Blocks Microspore Formation
Somatic tapetal cells are very important to pollen formation because they nourish the developing pollen (Pacini et al., 1985; Scott et al., 1991; Goldberg et al., 1993; McCormick, 1993). Tapetal cells produce and provide callase and other proteins important for the release of microspores from tetrads and chemicals important for the formation of pollen outer walls (Mepham, 1970; Izhar and Frankel, 1971; Stieglitz, 1977; Gori, 1982). The essential role of tapetal cells in pollen formation was demonstrated primarily by the selective destruction of tapetal cells, which resulted in the failure of pollen development (Mariani et al., 1990, 1992; Denis et al., 1993). Recently, more information was obtained from analyses of several mutations that affected microspore formation as a result of the defects of tapetal cells in Arabidopsis. For example, the MALE STERILILTY1 (MS1) gene was expressed specifically in tapetal cells; however, the ms1 mutation caused both abnormal vacuolation of tapetal cells and defects in microspores (Dawson et al., 1993; Wilson et al., 2001; Ito and Shinozaki, 2002). TPD1 acts early relative to MS1. No tapetum was formed in tpd1, but its anther still was able to produce morphologically normal microsporocytes.
This finding indicates that the formation of microsporocytes is independent of the developing tapetum. In the tpd1 anther at stage 7, no tetrad was formed, although the meiotic nuclear division was normal (Figure 3H). Obviously, microspore formation was disrupted before the meiotic cytokinesis was completed. One possible explanation for this finding is that the TPD1 product may be required directly for meiotic cytokinesis, because it also was expressed in the meiocytes at stage 6 (Figure 6I). Alternatively, meiosis may be disrupted indirectly by the absence of the tapetum (i.e., meiotic cytokinesis may require the support of tapetal cells).
TPD1 May Function in Tapetum Formation in Coordination with EMS1/EXS
The tpd1 and ems1/exs mutants exhibited highly similar phenotypes. The tpd1 ems1/exs1-2 double mutant appeared phenotypically nearly identical to tpd1 and ems1/exs1-2. These data suggest that TPD1 and EMS1/EXS may be involved in the same developmental pathway. During anther development, EMS1/EXS was expressed mostly in developing tapetal cells and their precursors. This finding suggests that EMS1/EXS receptor protein kinase likely perceives a signal received by immature tapetal cells. Our previous studies with the spl mutant led us to suggest that signal from developing microsporocytes is required for the proper differentiation of somatic cell types, including the tapetum in anther walls (Yang et al., 1999). Therefore, it is possible that the signal perceived by EMS1/EXS is from developing microsporocytes. TPD1 was expressed more strongly in developing microsporocytes than in developing tapetal cells, but the tpd1 mutations affected tapetum formation rather than microsporocyte formation. Although the tpd1 microsporocytes appeared morphologically normal, we cannot exclude the possibility that they had specific physiological defects caused by the tpd1 mutation, which disrupted the interaction between the developing microsporocytes and tapetal cells. Nevertheless, our results suggest that the TPD1 gene product may be involved in the transduction of a signal from developing microsporocytes to developing tapetal cells, which then is perceived by EMS1/EXS.
In conclusion, we have shown that the novel TPD1 gene in Arabidopsis is required for the specialization of tapetal cells and consequently for microspore formation. In the absence of TPD1, tapetal cell precursors adopt a microsporocyte fate. Double mutant analysis and comparison of the TPD1 and EMS1/EXS expression patterns suggest that the TPD1 product plays an important role in signaling tapetal cell specialization in coordination with EMS1/EXS1 receptor protein kinase during anther development.
METHODS
Plant Material and Mutant Isolation
All Arabidopsis thaliana plants used in this study were in the Landsberg erecta background. The seeds were pregerminated on MS agar plates (Murashige and Skoog, 1962) with or without 50 μg/mL kanamycin at 22°C under a 16-h-light/8-h-dark cycle. The plants were grown in soil at 22°C under the same light cycle for germination. The generation of Ds insertion lines and screening of mutants were performed as described by Sundaresan et al. (1995). The selected mutant plants were crossed with wild-type plants to purify the tpd1 mutations. The F3 plants with a single Ds insertion linked to the tpd1 phenotype then were selected for further phenotypic characterization.
Characterization of the tpd1 Mutant Phenotype
For sectioning, wild-type and mutant flowers at different stages were fixed overnight in 50% ethanol, 5% acetic acid, and 3.7% formaldehyde in water, dehydrated through an ethanol series, embedded into Historesin (Leica, Wetzlar, Germany), and sectioned with a Leica microtome. Serial sections (2 to 3 μm thick) were mounded on slides and stained with 0.25% toluidine blue O (British Drug House). The slides were observed with a Leica DMRB microscope and photographed with a Nikon digital camera (COOLPIX995; Tokyo, Japan). The selected images were compiled using the Adobe Photoshop (Mountain View, CA) and Microsoft Powerpoint (Redmond, WA) programs. Meiotic observation was performed as described by Ross et al. (1996).
Generation of the tpd1-1 ems1/exs1-2 Double Mutant
A tpd1-1 homozygous plant was used as the female parent in a cross with an ems1/exs1-2 heterozygous plant. To generate the tpd1-1 ems1/exs1-2 double mutant, tpd1-1 ems1/exs1-2 double heterozygous F1 plants were selected by the presence of PCR products of the Ds-gene junction sequences corresponding to the Ds insertion sites in both mutants. The tpd1 ems1/exs1-2 plants then were screened from a population of 79 F2 male-sterile plants by the absence of the PCR products of the wild-type EMS1/EXS and TPD1 sequences corresponding to the Ds insertion sites. The selected double mutant plants were used for further characterization. In addition, the double mutants were confirmed genetically by crossing with tpd1-1 ems1/exs1-2 double heterozygous plants.
Molecular Cloning of TPD1 and Sequencing
Isolation of the flanking sequences adjacent to the Ds element by thermal asymmetric interlaced PCR (Liu et al., 1995; Grossniklaus et al., 1998) was performed as described previously (Yang et al., 1999) with the tpd1 genomic DNAs and Ds3/AD2 or Ds5/AD4 primers. The full-length TPD1 cDNA was amplified using the Access reverse transcriptase–mediated PCR system (Promega) with the gene-specific primers 5′-AGACGAGAGTCCTCAAATCCA-3′ and 5′-TCCGTATCTAGTAAGACCCTC-3′. The 3′ end sequence of TPD1 cDNA was confirmed by 3′ end rapid amplification of cDNA ends (RACE) performed with the 5′/3′ cDNA RACE kit (Roche, Mannheim, Germany). All DNA fragments obtained were cloned into pGEM-T vector or pGEM-T Easy vector (Promega). Sequencing was performed using the ABI PRISM Rhodamine Terminator Thermal Cycling Sequencing Ready Reaction kit (Perkin-Elmer Applied Biosystems) with gene-specific primers or vector-derived M13 and T7 primers.
TPD1 Complementation Experiment
A 5.281-kb TPD1 genomic fragment was amplified using the ACCuTaq LA DNA polymerase PCR kit (Sigma) with the gene-specific primers 5′-CGAACACGATCGATGATCTGT-3′ and 5′-ATCACGCGATAACACGAGAA-3′ and cloned into pGEM-T vector. After verification by sequencing, the fragment was subcloned into pCAMBIA1300 Ti-derived binary vector (CAMBIA; www.cambia.org.au) (Ying and Wang, 2000) and introduced into tpd1-1 heterozygous plants using the infiltration method. The transformants were selected using 20 mg/L hygromycin and 50 mg/L kanamycin.
RNA Gel Blot and DNA Gel Blot Hybridizations
Total RNAs were extracted using the TRIzol reagent kit (Gibco BRL) as described by the supplier. Poly(A)+ RNAs were purified from total RNAs using the Oligotex mRNA Midi kit (Qiagen, Valencia, CA) as described by the supplier. The extraction of the plant genomic DNAs was performed as described by Yang et al. (1999). Poly(A)+ RNAs and a restricted DNA sample were fractionated on a 1% agarose gel. For the TPD1 probe, a genomic fragment was amplified by PCR using the primers 5′-TTCCGGTGTTAGTCACATCGA-3′ and 5′-ACGAAAATGGAGGAACCAAGA-3′. For the Ds probe, a 755-bp 5′ end fragment of the Ds element was amplified from plasmid pWS32 (Sundaresan et al., 1995) using the primers 5′-CTCACAGCACTTAGCAGTACA-3′ and 5′-CATACATCCGATGTGCACTTC-3′. The TPD1 and Ds fragments obtained were cloned into pGEM-T vector and verified via sequencing. All RNA and DNA probes were labeled using the digoxigenin (DIG) RNA labeling kit (Roche) or the PCR DIG Probe Synthesis Kit (Roche) as described by the supplier. RNA and DNA gel blotting and hybridization were performed according to the instructions in the “DIG System and DIG Application Manual” provided by the supplier.
In Situ Hybridization
RNA in situ hybridization on flower buds was performed according to the protocol provided by the Cold Spring Harbor Laboratory Arabidopsis Molecular Genetics Course organizers (http://www.arabidopsis.org/cshl-course/5-in-situ.html). For the TPD1 probe, a 672-bp TPD1 cDNA fragment was amplified using the primers 5′-GAAGACGAGAGTCAGCTTCAT-3′ and 5′-TCCGTATCTAGTAAGACCCTC-3′. For the SDS probe, a 998-bp cDNA fragment was amplified from SDS cDNA using the primers 5′-TTGAGATAGTCGGATGCGTCT-3′ and 5′-ACGAGTGCAGCTGCTACAGGT-3′ (Azumi et al., 2002). For the ATA7 probe, a 500-bp cDNA fragment was amplified from ATA7 using the primers 5′-TCACCTTCGCTAGCTAGTGTA-3′ and 5′-GTTACAAGGCTTCCCTTC-3′ (Rubinelli et al., 1998). The EMS1 probe was prepared as described by Zhao et al. (2002). The DNA fragments obtained were cloned into pGEM-T vectors and verified by sequencing. The antisense and sense RNA probes for in situ hybridization were generated and labeled using the DIG RNA labeling kit (Roche) as described by the supplier.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact De Ye, yede@imcb.a-star.edu.sg.
Accession Numbers
The GenBank accession numbers for the sequences mentioned in this article are as follows: AL049657 (BAC F6I7), AF412108 (a 580-bp EST sequence), AY394846 (a 935-nucleotide cloned cDNA), AC092387 (a putative rice protein), AAN74746 (M2D3.4 from Marchantia polymorpha), and AAD19962 (LGC1 from Lilium longiflorum).
Acknowledgments
We thank Srinivasan Ramachandran and Qi Xie for their critical comments on the manuscript. This work was supported by research grants from the National Science and Technology Board of Singapore and the Agency for Science, Technology, and Research of Singapore.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.016618.
References
- Azumi, Y., Liu, D., Zhao, D.Z., Li, W., Wang, G.F., Hu, Y., and Ma, H. (2002). Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLA DANCERS gene encoding a novel cyclin-like protein. EMBO J. 21, 3081–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales, C., Bhatt, A.M., Scott, R., and Dickinson, H. (2002). EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 12, 1718–1727. [DOI] [PubMed] [Google Scholar]
- Dawson, J., Wilson, Z.A., Aarts, M.G.M., Braithwaite, A.F., Briarty, L.G., and Mulligan, N.J. (1993). Microspore and pollen development in six male-sterile mutants of Arabidopsis thaliana. Can. J. Bot. 71, 629–638. [Google Scholar]
- Denis, M., Delourme, R., Gourret, J.P., Mariani, C., and Renard, M. (1993). Expression of engineered nuclear male sterility in Brassica napus. Plant Physiol. 101, 1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, R.B., Beals, T.P., and Sanders, P.M. (1993). Anther development: Basic principles and practical application. Plant Cell 5, 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori, P. (1982). Accumulation of polysaccharide in the anther cavity of Allium sativum, clone Piemonte. J. Ultrastruct. Res. 81, 158–162. [DOI] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science 280, 446–450. [DOI] [PubMed] [Google Scholar]
- Ishizaki, K., Shimizu-Ueda, Y., Okada, S., Yamamoto, M., Fujisawa, M., Yamato, K.T., Fukuzawa, H., and Ohyama, K. (2002). Multicopy genes uniquely amplified in the Y chromosome-specific repeats of the liverwort Marchantia polymorpha. Nucleic Acids Res. 30, 4675–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., and Shinozaki, K. (2002). The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger motif, is expressed in the tapetal cells and is required for pollen maturation. Plant Cell Physiol. 43, 1285–1292. [DOI] [PubMed] [Google Scholar]
- Izhar, S., and Frankel, R. (1971). Mechanism of male sterility in Petunia: The relationship between pH, callase activity in the anthers, and the breakdown of microsporogenesis. Theor. Appl. Genet. 44, 104–108. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Mitsukawa, N., Oosumi, T., and Whitier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Mariani, C., Beuckeleer, M.D., Truettner, J., Leemans, J., and Goldberg, R.B. (1990). Induction of male sterility in plants by a chimaeic ribonuclease gene. Nature 347, 737–741. [Google Scholar]
- Mariani, C., Beuckeleer, M.D., Truettner, J., Leemans, J., and Goldberg, R.B. (1992). A chimaeric ribonuclease-inhibitor gene restores fertility to male sterile plants. Nature 357, 384–387. [Google Scholar]
- McCormick, S. (1993). Male gametophyte development. Plant Cell 5, 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mepham, R.M. (1970). Development of pollen grain wall: Further work with Tradescantia bracteata. Protoplasma 69, 39–54. [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Pacini, E., Franchi, G.C., and Hesse, M. (1985). The tapetum: Its form, function, and possible phylogeny in Embryophyta. Plant Syst. Evol. 149, 155–185. [Google Scholar]
- Ross, K.J., Fransz, P., and Jones, G.H. (1996). A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res. 4, 507–516. [DOI] [PubMed] [Google Scholar]
- Rubinelli, P., Hu, Y., and Ma, H. (1998). Identification, sequence analysis and expression studies of novel anther-specific genes of Arabidopsis thaliana. Plant Mol. Biol. 37, 607–619. [DOI] [PubMed] [Google Scholar]
- Sanders, P.M., Anhthu, Q.B., Weterings, K., McIntire, K.N., Hsu, Y., Lee, P.Y., Troung, M.T., Beals, T.P., and Goldberg, R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11, 297–322. [Google Scholar]
- Schiefthaler, U., Balasubramantan, S., Sieber, P., Chevalier, D., Wisman, E., and Schneitz, K. (1999). Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, R., Hodge, R., Paul, W., and Draper, J. (1991). The molecular biology of anther differentiation. Plant Sci. 80, 167–191. [Google Scholar]
- Stieglitz, H. (1977). Role of β-1,3-glucan postmeiotic microspore release. Dev. Biol. 57, 87–97. [DOI] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797–1810. [DOI] [PubMed] [Google Scholar]
- Taylor, P.E., Glover, J.A., Lavithis, M., Craig, S., Singh, M.B., Knox, R.B., Dennis, E.S., and Chaudhury, A.M. (1998). Genetic control of male fertility in Arabidopsis thaliana: Structural analyses of postmeiotic developmental mutants. Planta 205, 492–505. [DOI] [PubMed] [Google Scholar]
- Wilson, Z.A., Morroll, S.M., Dawson, J., Swarup, R., and Tighe, P.J. (2001). The Arabidopsis MALE STERILITY 1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 28, 27–39. [DOI] [PubMed] [Google Scholar]
- Yang, W.-C., and Sundaresan, V. (2000). Genetics of gametophyte biogenesis in Arabidopsis. Curr. Opin. Plant Biol. 3, 53–57. [DOI] [PubMed] [Google Scholar]
- Yang, W.-C., Ye, D., Xu, J., and Sundaresan, V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13, 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying, Z., and Wang, G.-L. (2000). Evidence of multiple complex patterns of T-DNA integration into the rice genome. Theor. Appl. Genet. 100, 461–470. [Google Scholar]
- Xu, H., Swoboda, I., Bhalla, P.L., and Singh, M.B. (1999). Male gametic cell-specific gene expression in flowering plants. Proc. Natl. Acad. Sci. USA 96, 2554–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, D.-Z., Wang, G.-F., Speal, B., and Ma, H. (2002). The EXCESS MICROSPOROCYTE1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16, 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]