Abstract
The intracellular distribution of organelles is a crucial aspect of effective cell function. Chloroplasts change their intracellular positions to optimize photosynthetic activity in response to ambient light conditions. Through screening of mutants of Arabidopsis defective in chloroplast photorelocation movement, we isolated six mutant clones in which chloroplasts gathered at the bottom of the cells and did not distribute throughout cells. These mutants, termed chloroplast unusual positioning (chup), were shown to belong to a single genetic locus by complementation tests. Observation of the positioning of other organelles, such as mitochondria, peroxisomes, and nuclei, revealed that chloroplast positioning and movement are impaired specifically in this mutant, although peroxisomes are distributed along with chloroplasts. The CHUP1 gene encodes a novel protein containing multiple domains, including a coiled-coil domain, an actin binding domain, a Pro-rich region, and two Leu zipper domains. The N-terminal hydrophobic segment of CHUP1 was expressed transiently in leaf cells of Arabidopsis as a fusion protein with the green fluorescent protein. The fusion protein was targeted to envelope membranes of chloroplasts in mesophyll cells, suggesting that CHUP1 may localize in chloroplasts. A glutathione S-transferase fusion protein containing the actin binding domain of CHUP1 was found to bind F-actin in vitro. CHUP1 is a unique gene identified that encodes a protein required for organellar positioning and movement in plant cells.
INTRODUCTION
Organelles are likely to occupy spaces within the cell that facilitate and coordinate their functions. Although plants are static organisms, their organelles move actively within cells and take up positions that maximize metabolic activities. Therefore, the intracellular positioning and movement of organelles likely play a role in facilitating cell functions. Organelle movement in plant cells often is influenced by environmental stimuli such as light, gravity, and mechanical stress (Nagai, 1993; Williamson, 1993; Kagawa and Wada, 1995; Sack, 1997; Sato et al., 1999; Kagawa and Wada, 2002). For example, chloroplast photorelocation movement is a typical organellar response that is regulated by light. During photorelocation, the chloroplasts situate along the periclinal cell walls, optimizing their potential to harvest sufficient sunlight for optimal photosynthesis under low-light conditions. Under high light, the chloroplasts move away from the periclinal walls and toward the anticlinal walls, minimizing potential photodamage. We showed recently that in Arabidopsis mutants in which the chloroplasts do not redistribute normally in response to light intensity, the plastids suffer severe photodamage and become necrotic under continuous high light (Kasahara et al., 2002). From these observations, it is evident that the correct positioning of chloroplasts is very important for the survival of plants challenged with extreme light conditions.
Two cytoskeletal systems, actin filaments and microtubules, are critical for organelle movement and positioning. In plant cells, organelle movement appears to depend more on actin filaments than on microtubules, and actin filaments have been shown to be involved in the movement of chloroplasts (Kadota and Wada, 1992; Kandasamy and Meagher, 1999), mitochondria (Van Gestel et al., 2002), nuclei (Chytilova et al., 2000), peroxisomes (Collings et al., 2002; Jedd and Chua, 2002; Mano et al., 2002; Mathur et al., 2002), the endoplasmic reticulum (Williamson, 1993; Boevink et al., 1998), and the Golgi body (Boevink et al., 1998). A number of studies suggest that myosin plays a role in actin filament–based organelle movements as a motor protein (Williamson, 1993), although the components have yet to be identified.
Recently, we initiated a screen to identify the components involved in chloroplast movement in Arabidopsis and identified mutations in a gene encoding the photoreceptor that mediates the high-light–induced chloroplast avoidance movement (Kagawa et al., 2001). Here, we report the isolation of additional Arabidopsis mutants showing aberrant chloroplast positioning (termed chloroplast unusual positioning [chup]) using the same method and determined that in this case the defective gene (CHUP1) encodes a novel 112-kD protein. The domain structure predicted from the deduced amino acid sequence of CHUP1 indicates that this protein might be a factor involved in the actin-based cytoskeleton required for chloroplast positioning and movement. We discuss the possible functions of the CHUP1 protein.
RESULTS
Isolation of Chloroplast Movement Mutants
We screened 15,000 T-DNA lines and ∼100,000 ethyl methanesulfonate–mutagenized F2 Arabidopsis seeds for plants defective in chloroplast movement (see Methods) (Kagawa et al., 2001). Detached leaves were covered with a black plate with an open slit through which high-intensity white light was supplied for 1 h. The irradiated area of the wild-type leaf changed from green to pale green and was clearly distinguishable from the nonirradiated area (Figure 1). The irradiated area of chup1 leaves looked almost the same as the nonirradiated area. One mutant from T-DNA insertional mutant pools and five mutants from ethyl methanesulfonate–mutagenized mutant pools were allelic. They were all determined to contain single, nuclear, recessive mutations by complementation tests (data not shown).
Figure 1.
Leaves of Wild-Type and chup1-2 Plants before and after Partial Irradiation with High-Intensity White Light.
Leaves covered with a non-light-transmitting black plate with open slits were irradiated through the slits with high-intensity cool-white light for 1 h. The irradiated area of the wild-type (WT) leaf changed from dark green to pale green and was clearly distinguishable from the nonirradiated area. In the chup1-2 leaf, the irradiated area looked almost the same as the nonirradiated area.
Characterization of the Chloroplast Position in Leaf Cells of chup Plants
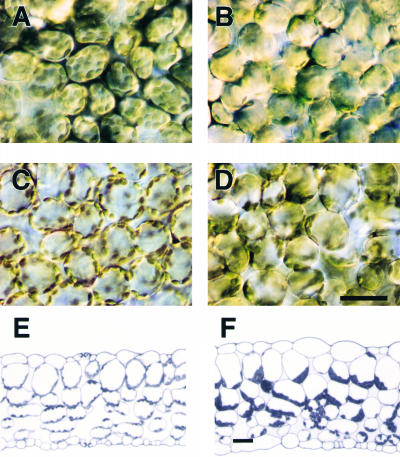
The effect of low and high light intensity on chloroplast positioning was observed in palisade cells. In wild-type cells, the chloroplasts accumulated along the periclinal cell walls under low light (Figure 2A) and along the anticlinal walls under high light (Figure 2C). In the mutant cells, chloroplast positioning was quite different from that in wild-type plants under both low-light and high-light conditions (Figures 2B and 2D), but although many chloroplasts gathered in parts of the mutant cells, it was difficult to determine the precise locations. To clarify the distribution patterns, leaves of mutant and wild-type plants were fixed and embedded, and cross-sections were observed in more detail. From these sections, it was clear that the chloroplasts in mutant plants accumulated at the bottom of palisade and spongy cells (i.e., the side farther from the adaxial surface of the leaf) (Figure 2F). Even when the mutant leaves were detached at their petioles and placed on the surface of an agar plate abaxial side up for 2 days, the positioning of the chloroplasts remained unchanged, suggesting that chloroplast accumulation at the bottom of the cells was not simply the result of gravity-dependent sedimentation. Although chloroplasts in wild-type cells changed their position depending on light intensity, chloroplast movement in the mutant cells was slight, if any movement occurred at all (data not shown).
Figure 2.
Distribution of Chloroplasts in the Mesophyll Cells of Wild-Type and chup1-2 Leaves.
(A) Low light–adapted wild-type cells.
(B) Low light–adapted chup1-2 cells.
(C) High light–treated wild-type cells.
(D) High light–treated chup1-2 cells.
(E) Cross-section of a wild-type leaf under low-light conditions.
(F) Cross-section of a chup1-2 leaf under low-light conditions.
Bars = 30 μm.
Observations of Other Organelles
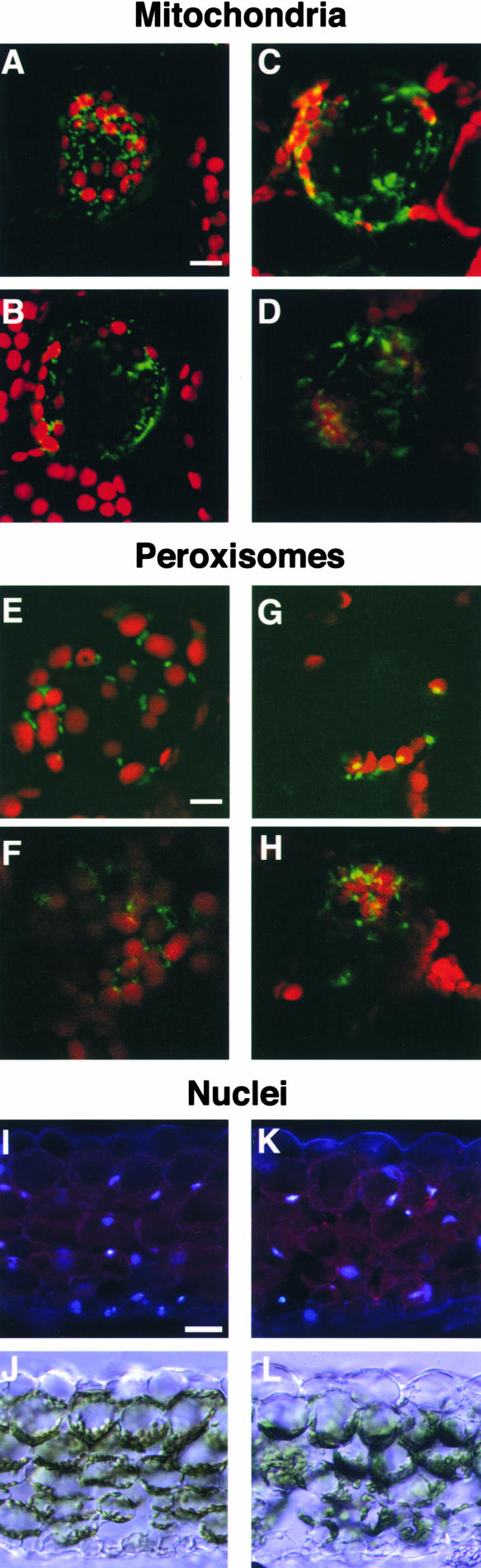
Next, the distribution of mitochondria and peroxisomes in wild-type and chup1 cells was observed using transient transformation with green fluorescent protein (GFP) fusions to mitochondrial or peroxisomal targeting sequences. Mitochondria were distributed throughout the cytoplasm, regardless of whether movement occurred. The types of mitochondrial activity observed can be divided into three classes: no movement, slow movement in random directions, and rapid linear, directional movement. The latter type of movement may reflect cytoplasmic streaming. Overall, there were no differences in mitochondrial distribution, number, or movement patterns between wild-type and chup1 cells (Figures 3A to 3D).
Figure 3.
Distribution of Mitochondria, Peroxisomes, and Nuclei in Wild-Type and chup1 Cells.
(A) to (D) Arabidopsis palisade mesophyll cells transiently expressing mitochondria-targeted GFP. Confocal microscopic images of the sections near the top ([A] and [C]) and the bottom ([B] and [D]) of wild-type and chup1-2 cells, respectively. The green and red signals show fluorescence from GFP and autofluorescence from chloroplasts, respectively. Bar = 10 μm.
(E) to (H) Arabidopsis palisade mesophyll cells transiently expressing peroxisome-targeted GFP. Confocal microscopic images of the sections near the top ([E] and [G]) and the bottom ([F] and [H]) of wild-type and chup1-2 cells, respectively. Fluorescent colors are the same as in (A) to (D). Bar = 10 μm.
(A) to (H) represent projections of seven optical 1.0-μm sections.
(I) to (L) Positions of nuclei in Arabidopsis leaves. Cross-sections of wild-type and chup1-2 leaves were stained with 4′,6-diamidino-2-phenylindole to visualize nuclei ([I] and [K], respectively), and the same cross-sections are shown for the transmission images ([J] and [L], respectively). Bar = 30 μm.
Most of the peroxisomes in palisade mesophyll cells were located adjacent to chloroplasts in both the wild type and the mutants and did not appear to move independently of chloroplast movement, in contrast to those in the mitochondria (Figures 3E to 3H). However, the distribution of peroxisomes in palisade mesophyll cells of chup1 differed from that of the wild type, because they remained at the bottom of the cells, along with the chloroplasts. Because peroxisomes usually localize close to chloroplasts, their abnormal distribution in chup1 leaves likely is a secondary effect of chloroplast mislocalization. In epidermal cells lacking chloroplasts, the peroxisomes moved actively around cells in both wild-type and chup1 leaves.
To observe nuclei, transverse sections of the leaves were made and stained with 4′,6-diamidino-2-phenylindole. The localization of nuclei in chup1 cells was similar to that in wild-type cells (Figures 3I to 3L). Overall, we conclude that the effects of chup1 are specific to chloroplast distribution and movement.
Cloning of CHUP1
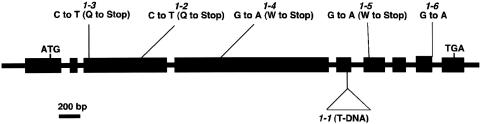
A segment of the DNA flanking the T-DNA insertion into chup1-1 was isolated by plasmid rescue and used to screen an Arabidopsis cDNA library. Because the cDNA clone isolated in this screen, which was a part of At3g25690, did not contain a full-length insert, 5′ rapid amplification of cDNA ends was used to obtain the full-length cDNA. The cDNA contained a single open reading frame of 3015 bp, encoding a deduced protein of 1004 amino acid residues with a predicted molecular mass of 112 kD. The corresponding CHUP1 gene contains nine exons and eight introns (Figure 4). The T-DNA insertion in the chup 1-1 allele was located in exon 5 (Figure 4). The molecular defects in the other alleles were characterized by sequencing genomic DNA fragments. All of the alleles tested carried mutations in the same gene. The chup1-2, -3, -4, and -5 mutants all have single base substitutions resulting in stop codons (Figure 4). The chup1-6 mutant has a single base substitution at the junction of exon 8 and intron 8. The presence of a lesion in each of the six independent mutants tested is consistent with the role of this gene in chloroplast positioning. Furthermore, it was confirmed that expression of a CHUP1-GFP gene (CHUP1 fused to GFP at the C terminus) driven by the 35S promoter of Cauliflower mosaic virus in chup1-2 complemented the mutant phenotype (data not shown). However, GFP fluorescence was not detected in the resulting transformant (data not shown).
Figure 4.
Structure of the CHUP1 Gene and Positions of chup1 Mutations.
The locations of start (ATG) and stop (TGA) codons are indicated. Nine exons (boxes) and eight introns (lines) were identified by comparing the genomic sequence with the cDNA sequence. Mutations in chup1-1, chup1-2, chup1-3, chup1-4, chup1-5, and chup1-6 are as follows: insertion of T-DNA into exon 5 in chup1-1; the codon for Gln-234 was changed to a stop codon in chup1-2; the codon for Gln-75 was changed to a stop codon in chup1-3; the codon for Trp-451 was changed to a stop codon in chup1-4; the codon for Trp-828 was changed to a stop codon in chup1-5; and the first base (G) of intron 8 was changed to A in chup1-6.
CHUP1 Encodes a Putative Actin Binding Protein with a Coiled-Coil Region, a Pro-Rich Region, and Two Leu Zipper Domains
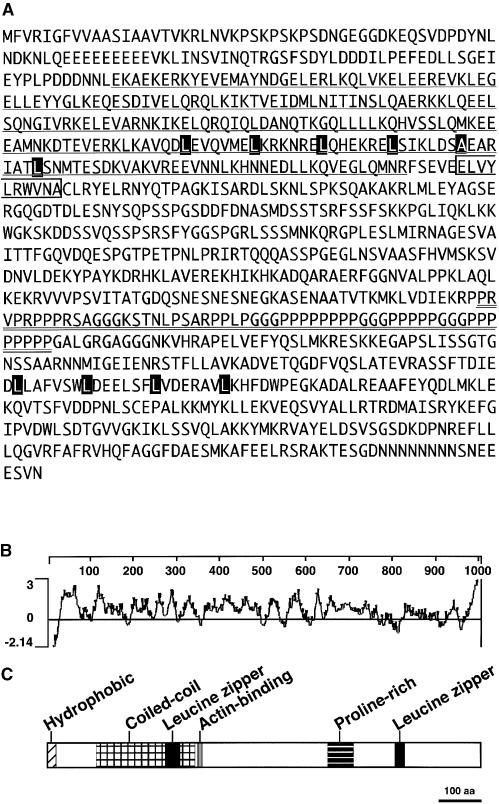
Comparison of the predicted amino acid sequence against sequences available in public databases indicated that CHUP1 contains some previously described domains, including an actin binding domain, a coiled-coil region, Pro-rich motifs, and Leu zipper motifs (Figure 5C).
Figure 5.
Structure of CHUP1.
(A) Amino acid sequence of CHUP1. The coiled-coil domain is underlined. The Pro-rich domain is double underlined. The actinin-like actin binding motif is indicated by an open box. Two Leu zipper motifs are indicated by series of black boxes.
(B) Hydrophilicity plot of the amino acid sequence of CHUP1. Hydrophilic indices were calculated using the Kyte-Doolittle scale (Kyte and Doolittle, 1982) and averaging over a window of 17 amino acid residues.
(C) Domain organization of CHUP1. The hydrophobic region, coiled-coil domain, actin binding motif, Pro-rich region, and two Leu zipper domains are indicated. aa, amino acids.
Coiled-Coil Region
The Paircoil program (Berger et al., 1995) predicted the presence of a coiled-coil region (Asp-112 to Arg-341) (Figure 5A, underline).
Actin Binding Domain
Comparison with PROSITE, a database of biologically significant sites (http://kr.expasy.org/prosite), patterns, and profiles, revealed the presence of an actinin-type actin binding domain signature in CHUP1 (Asp-346 to Ala-356) (Figure 5A, box).
Leu Zipper Motifs
Comparison with the PROSITE database sequences revealed the presence of two Leu zipper motifs (Leu-269 to Leu-304 and Leu-802 to Leu-823) (Figure 5A, black boxes). These regions may mediate protein–protein interactions with other proteins or with itself.
Pro-Rich Motifs
The region from Pro-648 to Pro-705 is abundant in Pro residues (Figure 5A, double underline). A Pro-rich motif (PRM) can be classified by comparison with consensus sequences derived from several different PRM binding domains (Holt and Koffer, 2001). The PRM of CHUP1 is most similar to PRM1, which has been identified as a profilin binding motif.
Hydrophobic Region
A hydrophilicity plot revealed the presence of a short hydrophobic region at the N-terminal end of the protein (Figure 5B). Comparison of the CHUP1 sequence with sequences in the SignalP database, which enables prediction of the presence and location of signal peptides (Nielsen et al., 1997), led to the prediction that the hydrophobic region may function as a signal peptide. The hydrophobic region could play a role in membrane anchoring. We further investigated the possible roles of this region, and these efforts are described in more detail below.
Proteins That Are Similar to the C-Terminal Region of CHUP1 in Arabidopsis
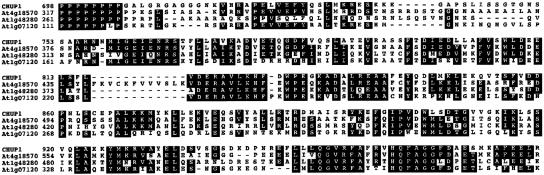
No sequences similar to the full-length CHUP1 sequence were found in the Arabidopsis database (TAIR; http://www.arabidopsis.org). However, in Arabidopsis, there are three genes that encode proteins strikingly similar to the C-terminal region (from the Pro-rich region to the end) of CHUP1 (Figure 6). The functions of these genes are not known. We were unable to find any additional sequence homology with other known proteins or functional domains. Although the function of this C-terminal region is not known, it is likely to play a key role in the molecular function of CHUP1.
Figure 6.
Alignment of the C-Terminal Region of CHUP1 with Those of Arabidopsis At4g18570, At1g48280, and At1g07120 Gene Products.
Amino acid residues identical in at least three sequences are indicated by black boxes. Numbers given are amino acid positions for each protein sequence.
Localization of the CHUP1–N-Terminal Hydrophobic Region–GFP Fusion Protein
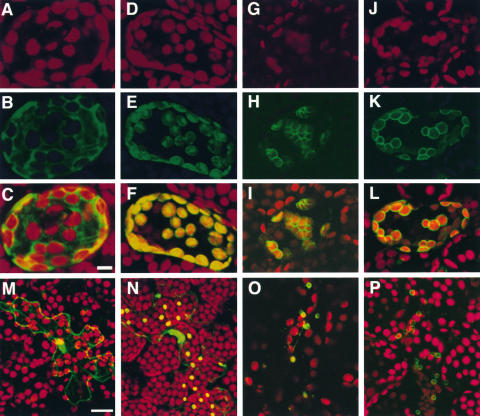
GFP was fused to the C-terminal end of the N-terminal hydrophobic region of CHUP1 (Met-1 to Pro-25) to allow the study of the possible roles of this N-terminal region in intact cells. The construct was introduced into leaf cells by particle bombardment to allow the transient expression of the fusion protein (N-CHUP-GFP). In the transformants, the outlines of chloroplasts were stained strongly with green fluorescence in the cells expressing N-CHUP-GFP, whereas GFP was distributed uniformly in the cytosol (Figure 7). The localization of N-CHUP-GFP was significantly similar to that of AtOEP7-GFP, which is targeted to chloroplast envelope membrane (Lee et al., 2001), rather than to that of pt-sGFP, a fusion of the transit peptide of the ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit and GFP that is targeted to chloroplast stroma (Niwa et al., 1999). Thus, N-CHUP-GFP probably was targeted to chloroplast and plastid envelopes in mesophyll and epidermal cells, respectively (Figure 7). These results suggest that the N-terminal region may target and anchor CHUP1 in the chloroplast envelope membrane.
Figure 7.
Localization of N-CHUP-GFP.
(A) to (L) Confocal microscopic images of optical sections of Arabidopsis palisade mesophyll cells transiently expressing GFP ([A] to [C]), pt-sGFP ([D] to [F]), AtOEP7-GFP ([G] to [I]), and N-CHUP-GFP ([J] to [L]). The green and red signals show fluorescence from GFP and autofluorescence from chloroplasts, respectively. Merged images of the green and red channels are shown ([C], [F], [I], and [L]). These images represent projections of 17 optical 1.25-μm sections. Bar = 10 μm.
(M) to (P) Confocal microscopic images of optical sections of Arabidopsis epidermal cells transiently expressing GFP (M), pt-sGFP (N), AtOEP7-GFP (O), and N-CHUP-GFP (P). Merged images of the green and red channels are shown. These images represent projections of eight optical 1.5-μm sections. Bar = 20 μm.
Expression of CHUP1
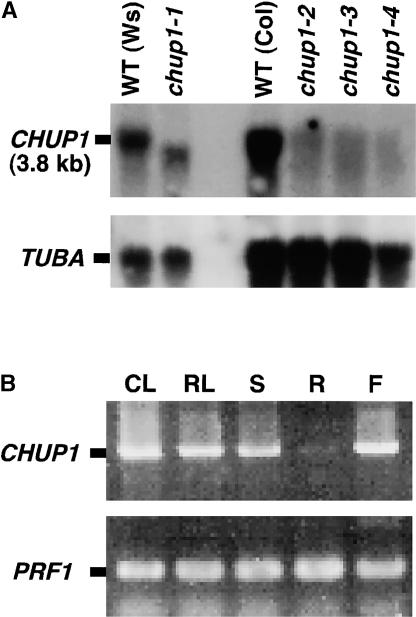
A 3.8-kb transcript was detected in wild-type plants using the full-length CHUP1 cDNA sequence, and the expression levels were reduced greatly in all of the mutants tested (Figure 8A). The gene was expressed in cauline leaves, rosette leaves, stems, and inflorescences (flowers), but not in roots. If the CHUP1 protein plays specific roles in determining the intracellular distribution of chloroplasts, it is reasonable to expect that the gene would be expressed in organs that contain chloroplasts.
Figure 8.
CHUP1 Gene Expression.
(A) RNA gel blot hybridization was performed with the 32P-radiolabeled CHUP1 or TUBA cDNA as a probe against total RNA. Total RNA samples were prepared from wild-type (WT) ecotype Wassilewskija (Ws), wild-type ecotype Colombia (Col), and four mutant alleles of chup1. The respective wild-type parental ecotype of each mutant was included for comparison (Ws for chup1-1 and Col for chup1-2, chup1-3, and chup1-4). TUBA encoding tubulin was used as a control.
(B) CHUP1 mRNA accumulation was analyzed by reverse transcriptase–mediated PCR. PRF1 encoding profilin was amplified as a control. CL, cauline leaves; RL, rosette leaves; S, stems; R, roots; F, flowers.
Actin Binding Activity
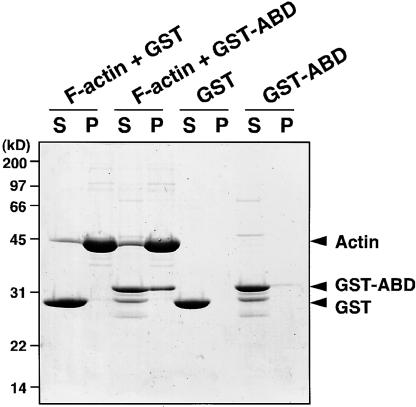
Because the domain organization of CHUP1 included an actin binding site, we studied the binding activity of this gene product to actin. Attempts to express full-length CHUP1 as a glutathione S-transferase (GST)–tagged protein in Escherichia coli were unsuccessful, because the level of expression was too low to be useful and all of the expressed protein became insoluble (data not shown). Hence, we instead expressed a truncated form of CHUP1 consisting of the putative actin binding domain as a GST fusion protein, GST-CHUP1-ABD. The fusion protein coprecipitated with F-actin (Figure 9, lane P of F-actin + GST-ABD), whereas the control GST lacking the CHUP1 domain did not (Figure 9, lane P of F-actin + GST). These results demonstrate that the putative actin binding motif in CHUP1 can function as an actin binding site.
Figure 9.
Actin Binding Activity of CHUP1.
GST or GST-ABD was incubated with or without F-actin at 25°C for 1 h and then centrifuged at 100,000g for 1 h. Supernatant fraction (S) and precipitant fraction (P) were loaded on an SDS-PAGE gel. The gel was stained with Coomassie Brilliant Blue R 250. The arrowheads indicate the positions of actin, GST-ABD, and GST. Molecular mass standards are indicated at left.
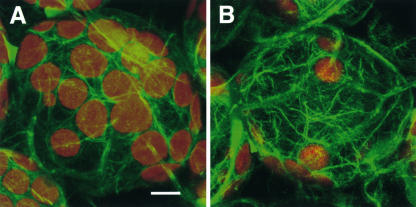
Actin Cytoskeleton in chup1 Cells
To visualize actin filaments, wild-type and chup1-2 plants were transformed with a construct to express GFP–mouse talin (GFP-mTalin), a marker for the actin cytoskeleton in live plant cells (Kost et al., 1998). The actin filaments were distributed over the cell cortex even around the chloroplasts in chup1 cells and wild-type cells (Figure 10). There was no apparent difference between chup1 and wild-type cells in terms of the pattern of actin filaments.
Figure 10.
Actin Cytoskeleton in Palisade Mesophyll Cells of the Wild Type and chup1-2.
The actin filaments were visualized in transgenic Arabidopsis plants by expressing GFP-mTalin with a confocal microscope in wild-type (A) and chup1-2 (B) backgrounds. The green and red signals show fluorescence from GFP and autofluorescence from chloroplasts, respectively. The images represent projections of 20 optical 0.95-μm sections. Bar = 10 μm.
DISCUSSION
In this study, we describe the isolation of a novel gene, CHUP1, required for the positioning and movement of chloroplasts. The predicted CHUP1 protein sequence contains multiple functional domains (Figure 5C), and of these, we demonstrate that the actin binding motif can bind F-actin (Figure 9). Because CHUP1 contains an actin binding domain, it is logical to assume that the function of CHUP1 will include interaction with the actin-based cytoskeleton.
There are multiple systems for targeting proteins to the chloroplast outer envelope membrane (Keegstra and Cline, 1999). The protein encoded by pea OEP14, the ortholog of Arabidopsis OEP7 (AtOEP7), is targeted to the chloroplast outer envelope membrane using one of the targeting systems (Li and Chen, 1996). The N-terminal hydrophobic segment of OEP14 has been shown to play roles in localization to the chloroplast outer envelope membrane and anchoring to the membrane (Li and Chen, 1996). Although the transit peptide required to transport protein into chloroplasts is cleaved after targeting, the hydrophobic segment of OEP14, which likely works as a targeting signal in this system, is not cleaved. In Figure 7, we showed that the N-terminal hydrophobic segment of CHUP1 has the ability to target GFP into the chloroplast envelope. It is difficult to determine what components of the chloroplast envelope (e.g., the outer envelope membrane, the inner envelope membrane, or the space between these membranes) N-CHUP-GFP localizes at in this experiment. However, by analogy with OEP14, N-CHUP-GFP is likely to localize at the chloroplast outer membrane using the same targeting system as OEP14. Thus, CHUP1 could function at the periphery of the chloroplast outer membrane.
To date, the majority of studies attempting to elucidate the mechanisms of organelle movement have involved physiological and histochemical approaches using cytoskeleton-disrupting drugs and fluorescent probes to stain cytoskeleton components (Kandasamy and Meagher, 1999; Sato et al., 2001; Collings et al., 2002; Jedd and Chua, 2002; Mano et al., 2002; Mathur et al., 2002; Van Gestel et al., 2002). The outcomes of these experiments suggest that the actin- and/or tubulin-based cytoskeleton and their motor proteins play crucial roles in the positioning and movement of organelles. It also is likely that chloroplast positioning and movement involve the actin-based cytoskeleton but not the tubulin-based cytoskeleton (Kadota and Wada, 1992; Tlalka and Gabrys, 1993; Kandasamy and Meagher, 1999). Myosin has been proposed to be a motor protein for chloroplast movement based on inhibitor experiments using the fern Adiantum (Kadota and Wada, 1992; Sato et al., 1999) and is the best candidate for the motor protein driving chloroplast movement in Arabidopsis cells. However, CHUP1 does not show any similarity to the consensus sequence for myosin motor domains.
CHUP1 is likely to be a factor required to spread chloroplasts around cells close to the plasma membrane, because chloroplasts gather at the bottom of chup1 cells. Interestingly, chloroplasts aggregate aberrantly when actin filaments are depolymerized using the actin-disrupting drug latrunculin B in Arabidopsis cells (Kandasamy and Meagher, 1999). The aggregation pattern induced by this drug is somewhat similar to the aberrant positioning of chloroplasts in chup1 cells. It is likely that the proper distribution of actin filaments and the presence of functional CHUP1 are required for normal chloroplast positioning. Although it is premature to attempt to describe precisely the function of CHUP1, it is possible to infer some general roles from our observations. CHUP1 might function as a chloroplast-anchoring factor, binding between chloroplasts and actin filaments. Alternatively, although the major actin filaments are likely to be normal in chup1 cells (Figure 10), it remains possible that CHUP1 may serve to build the kind of actin filaments required for chloroplast positioning and movement, which might be so unstable or very thin that they could not be visualized using transformants expressing GFP-mTalin. In this regard, the Pro-rich region of CHUP1 could recruit profilactin, a complex of G-actin and profilin, to provide the actin monomers required for F-actin polymerization, as many proteins with Pro-rich regions do (Holt and Koffer, 2001). Analysis of the various CHUP1 subdomains would be the next logical step in the further investigation of the molecular function of CHUP1.
In seed plants, it has been proposed that peroxisomes (Jedd and Chua, 2002) and mitochondria (Van Gestel et al., 2002) move within cells using an actomyosin system similar to that used by chloroplasts. To determine if this is the case, the distribution of mitochondria and peroxisomes was visualized using GFP fused to localization signals for these organelles. Our observations revealed that CHUP1 likely has a function specific for chloroplast movement. There are three genes with sequence similarity to the CHUP1 C-terminal region in Arabidopsis (Figure 6). Although we have already isolated six different mutant alleles of chup1 in our screen, mutants defective in these similar genes have yet to be identified. It is not known whether these gene products play roles in the movement of other organelles. If they do, mutants may not have been obtained yet because insufficient mutations have been screened or because the candidate genes have redundant functions.
Under low-light conditions, chloroplasts accumulate in areas irradiated with light, presumably to optimize photosynthesis (“accumulation movement”), although conversely, they avoid areas irradiated with high-intensity light, minimizing photodamage of the photosynthetic apparatus (“avoidance movement”). When chup1 plants were exposed to continuous high-intensity light, the leaves were severely bleached and became necrotic (Kasahara et al., 2002). Therefore, the correct relocation of chloroplasts in response to changing light conditions is critical for plant survival. This could be why the phenomenon of chloroplast photorelocation movement is found in all of the chloroplast-containing organisms examined to date, from algae to seed plants. We recently identified the photoreceptors responsible for the regulation of chloroplast photorelocation movement, phot1 and phot2 (Jarillo et al., 2001; Kagawa et al., 2001; Sakai et al., 2001). Arabidopsis phot mutants are defective in accumulation movement and/or avoidance movement because of disrupted regulation of the light signal transduction pathways. The CHUP1 protein described in this study probably functions as part of the machinery needed for chloroplast positioning and movement, rather than in the signal transduction pathway. Therefore, CHUP1 represents another category of gene involved in chloroplast movement. Further investigation of the functions of CHUP1 is likely to reveal the mechanism of chloroplast movement and confirm the importance of proper organelle distribution in plant cells.
METHODS
Plant Materials and Growth Conditions
Seeds of Arabidopsis thaliana (Columbia and Wassilewskija ecotypes) were sown on one-third-strength Murashige and Skoog (1962) medium containing 1% (w/v) sucrose with 0.8% (w/v) agar, incubated at 4°C for 2 days, and then grown under light conditions of 16 h of light (≈70 μmol·m−2·s−1) and 8 h of dark at 25°C.
Isolation of Mutants
Leaves of 2-week-old plants mutagenized by ethyl methanesulfonate (Lehle Seeds, Round Rock, TX) or T-DNA insertion (ABRC, Columbus, OH) were removed and placed adaxial side up on the surface of agar plates solidified with 1% agar. These leaves were covered by a black plate with 1-mm-wide slits and irradiated through these slits with strong white light (≈30 W/m2) for 1.5 h using a metal halide lamp. The color of leaves irradiated with strong light through these slits changed from green to pale green as a result of chloroplast avoidance movement. Mutants were identified as plants that did not change their leaf color in the slit region after irradiation with high-intensity light.
Observation of Organelle-Targeted GFP
Arabidopsis leaves were bombarded with gold particles coated with plasmids containing mitochondrion-targeted GFP gene fusions (Niwa et al., 1999) or peroxisome-targeted GFP gene fusions (Mathur et al., 2002) using a Bio-Rad PDS-1000/He particle-delivery system. Bombarded leaves were maintained at 25°C for at least 12 h until microscopic observation, and then GFP-expressing cells were imaged using a confocal laser scanning microscope (LSM model 410; Zeiss, Jena, Germany).
Cloning of CHUP1
For plasmid rescue, DNA was prepared from chup1-1 Arabidopsis plants and digested with SalI. After phenol/chloroform extraction, the digested DNA was self-ligated at 16°C overnight and introduced into Escherichia coli (DH5α). Rescued plasmid DNA was sequenced using a T-DNA left border primer (5′-TTTAAGACGCTATTCCTCAACTTCC-3′). A Basic Local Alignment Search Tool (BLAST) search confirmed that the flanking sequence of the T-DNA was identical to part of the genomic sequence in BAC T5M7.
An Arabidopsis cDNA library was screened with a probe of the DNA fragment described above, and a cDNA clone containing the open reading frame of CHUP1 was isolated. Because the clone did not contain the 5′ end of CHUP1, 5′ rapid amplification of cDNA ends was performed using a kit (Invitrogen, Carlsbad, CA) to obtain the full-length sequence of CHUP1.
RNA Gel Blot and Reverse Transcriptase–Mediated PCR Analyses
Total RNA was isolated from 4-week-old plants. Ten micrograms of total RNA was electrophoresed on an agarose gel and blotted onto a nylon membrane (Hybond-N+; Amersham). After hybridization with a probe (a PCR product of the full-length CHUP1), the blot was exposed to x-ray film (X-Omat; Kodak).
Localization of N-CHUP1-GFP
To make a construct to express a fusion protein of the N-terminal hydrophobic region of CHUP1 with GFP, a SalI site and an NcoI site were introduced immediately upstream and downstream of the codons for Met-1 and Pro-25 of CHUP1, respectively, using PCR. The two primers used were 5′-AAGTCGACATGGGAAAAACTTCGGGA-3′ and 5′-AACCATGGGTTTAACGTTGAGCCGCTTAA-3′. PCR was performed with the primers using CHUP1 cDNA as a template. The PCR product was digested with SalI and NcoI and cloned into the SalI-NcoI site of the CaMV35SΩ-sGFP(S65T) plasmid (Chiu et al., 1996; Niwa et al., 1999). The resulting plasmid was named pN-CHUP-GFP.
To make a construct to express the AtOEP7-GFP fusion protein, we followed the method described by Lee et al. (2001). GFP was fused to the C terminus of the full-length AtOEP7. A SalI site and an NcoI site were introduced immediately upstream and downstream of the codons for Met-1 and Leu-64 of AtOEP7, respectively, using PCR. The PCR product was digested with SalI and NcoI and cloned into the SalI-NcoI site of the CaMV35SΩ-sGFP(S65T) plasmid (Chiu et al., 1996; Niwa et al., 1999). The resulting plasmid was named pAtOEP7-GFP.
Arabidopsis leaves were bombarded with gold particles coated with plasmids, the CaMV35SΩ-sGFP(S65T) plasmid, the plastid-targeted pt-sGFP(S65T) plasmid (Chiu et al., 1996; Niwa et al., 1999), pN-CHUP-GFP, or pAtOEP7-GFP using a Bio-Rad PDS-1000/He particle-delivery system. Bombarded leaves were maintained at 25°C for at least 12 h until microscopic observation, and then GFP-expressing cells were imaged using a confocal laser scanning microscope (LSM model 410; Zeiss).
Construction, Expression, and Purification of GST-CHUP1-ABD
A BamHI site and an EcoRI site were introduced immediately upstream and downstream of the codons for Lys-332 and Asn-364, respectively, using PCR. The two primers used were S1-F (5′-TTGGATCCAAGCAAGTGGAAGGGCTT-3′) and S1-R (5′-TTGAATTCGTTTCTTAACTCATACCG-3′). PCR was performed with the primers using CHUP1 cDNA as a template. The PCR product was digested with BamHI and EcoRI and cloned into the BamHI-EcoRI site of pGEX-2T (Amersham). E. coli BL21(DE3) pLysS was transformed with the resulting plasmid, pGEX-CHUP1-ABD, and was grown in Luria-Bertani medium (400 mL) at 25°C. The recombinant protein, GST-CHUP1-ABD, was produced by inducing exponentially growing cells with 1 mM isopropylthio-β-galactoside. Cells were harvested by centrifugation after 5 h of cultivation, frozen at −20°C, thawed at room temperature, resuspended in 25 mL of 50 mM Tris-HCl buffer, pH 8.0, containing 10% (w/v) glycerol, 150 mM NaCl, 2 mM EDTA, 0.1% (w/v) Triton X-100, and 10 mM 2-mercaptoethanol (buffer A), and then disrupted by sonication. The cell extract was centrifuged at 16,000g for 30 min and applied to a glutathione-Sepharose column (0.5-mL bed volume). The column was washed with 20 mL of buffer A and 10 mL of buffer B (50 mM Tris-HCl, pH 8.0, and 10% [w/v] glycerol). The protein was eluted with buffer B supplemented with 10 mM glutathione.
Actin Binding Assay
One milligram of actin (A-2522; Sigma, St. Louis, MO) was dissolved in 0.5 mL of 2 mM Tris-HCl buffer, pH 8.0, containing 0.1 mM CaCl2, 0.2 mM ATP, and 1 mM 2-mercaptoethanol (G buffer). The actin solution was centrifuged at 100,000g for 1 h at 4°C. A supernatant fraction (0.45 mL) was recovered and added to 50 μL of 10 × F buffer (0.5 M Tris-HCl, pH 8.0, 1 M KCl, 20 mM MgCl2, 10 mM EGTA, 2 mM ATP, and 10 mM 2-mercaptoethanol) to polymerize actin protein and centrifuged at 100,000g for 1 h at 4°C. The supernatant fraction was discarded to remove unpolymerized actin fraction. The precipitated fraction was dissolved in 100 μL of G buffer and used for the actin binding assay. GST or GST-CHUP1-ABD was mixed with or without the actin solution. The mixture was added to one-tenth final volume of 10 × F buffer, incubated at 25°C for 1 h, and centrifuged at 100,000g for 1 h at 4°C. Both the supernatant and the precipitated fractions were analyzed using SDS-PAGE.
Transformation of Arabidopsis with a GFP-mTalin–Expressing Construct
We constructed a transformation plasmid containing the gene that encodes GFP-mTalin (Kost et al., 1998) under the control of the 35S promoter of Cauliflower mosaic virus. Arabidopsis ecotype Columbia (gl1) and the chup1-2 mutant were transformed with the GFP-mTalin construct using the in planta transformation method (Bechtold et al., 1993).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Masamitsu Wada, wada-masamitsu@c.metro-u.ac.jp.
Accession Numbers
The sequence data for CHUP1 has been deposited in the DDBJ/GenBank/EBI Data Bank with accession number AB087408. The sequence information for At4g18570, At1g48280, and At1g07120 shown in Figure 6 was obtained from the TAIR World Wide Web site (http://www.arabidopsis.org).
Acknowledgments
We are grateful to Jane Silverthorne of the University of California, Santa Cruz, for critical reading of the manuscript. This work was partly supported by BRAIN (Program for the Promotion of Basic Research Activities for Innovative Biosciences), by a Grant-in-Aid for Scientific Research (on Priority Areas, No. 13139203, and A, No. 13304061) from the Ministry of Education, Sports, Science, and Technology of Japan (to M.W.) and by a grant from PRESTO, the Japan Science and Technology Corporation (to T. Kagawa). K.O. and N.S. were supported by fellowship from the Japan Society for the Promotion of Science for Young Scientists.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.016428.
References
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad Sci. Paris 316, 1194–1199. [Google Scholar]
- Berger, B., Wilson, D.B., Wolf, E., Tonchev, T., Milla, M., and Kim, P.S. (1995). Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. USA 92, 8259–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink, P., Oparka, K., Santa Cruz, S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 15, 441–447. [DOI] [PubMed] [Google Scholar]
- Chiu, W.-I., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. [DOI] [PubMed] [Google Scholar]
- Chytilova, E., Macas, J., Sliwinska, E., Rafelski, S.M., Lambert, G.M., and Galbraith, D.W. (2000). Nuclear dynamics in Arabidopsis thaliana. Mol. Biol. Cell 11, 2733–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collings, D.A., Harper, J.D.I., Marc, J., Overall, R.L., and Mullen, R.T. (2002). Life in the fast lane: Actin-based motility of plant peroxisomes. Can. J. Bot. 80, 430–441. [Google Scholar]
- Holt, M.R., and Koffer, A. (2001). Cell motility: Proline-rich proteins promote protrusions. Trends Cell Biol. 11, 38–46. [DOI] [PubMed] [Google Scholar]
- Jarillo, J.A., Gabrys, H., Capel, J., Alonso, J.M., Ecker, J.R., and Cashmore, A.R. (2001). Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature 410, 952–954. [DOI] [PubMed] [Google Scholar]
- Jedd, G., and Chua, N.-H. (2002). Visualization of peroxisomes in living plant cells reveals acto-myosin-dependent cytoplasmic streaming and peroxisome budding. Plant Cell Physiol. 43, 384–392. [DOI] [PubMed] [Google Scholar]
- Kadota, A., and Wada, M. (1992). Photoorientation of chloroplasts in protonemal cells of the fern Adiantum as analyzed by use of a video-tracking system. Bot. Mag. Tokyo 105, 265–279. [Google Scholar]
- Kagawa, T., Sakai, T., Suetsugu, N., Oikawa, K., Ishiguro, S., Kato, T., Tabata, S., Okada, K., and Wada, M. (2001). Arabidopsis NPL1: A phototropin homolog controlling the chloroplast high-light avoidance response. Science 291, 2138–2141. [DOI] [PubMed] [Google Scholar]
- Kagawa, T., and Wada, M. (1995). Polarized light induces nuclear migration in prothallial cells of Adiantum capillus-veneris L. Planta 196, 775–780. [Google Scholar]
- Kagawa, T., and Wada, M. (2002). Blue light-induced chloroplast relocation. Plant Cell Physiol. 43, 367–371. [DOI] [PubMed] [Google Scholar]
- Kandasamy, M.K., and Meagher, R.B. (1999). Actin-organelle interaction: Association with chloroplast in Arabidopsis leaf mesophyll cells. Cell Motil. Cytoskeleton 44, 110–118. [DOI] [PubMed] [Google Scholar]
- Kasahara, M., Kagawa, T., Oikawa, K., Suetsugu, N., Miyao, M., and Wada, M. (2002). Chloroplast avoidance movement reduces photodamage in plants. Nature 420, 829–832. [DOI] [PubMed] [Google Scholar]
- Keegstra, K., and Cline, K. (1999). Protein import and routing systems of chloroplasts. Plant Cell 11, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.-H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16, 393–401. [DOI] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle, R.F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Lee, Y.J., Kim, D.H., Kim, Y.-W., and Hwang, I. (2001). Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13, 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.-M., and Chen, L.-J. (1996). Protein targeting and integration signal for the chloroplastic outer envelope membrane. Plant Cell 8, 2117–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano, S., Nakamori, C., Hayashi, M., Kato, A., Kondo, M., and Nishimura, M. (2002). Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: Dynamic morphology and actin-dependent movement. Plant Cell Physiol. 43, 331–341. [DOI] [PubMed] [Google Scholar]
- Mathur, J., Mathur, N., and Hülskamp, M. (2002). Simultaneous visualization of peroxisomes and cytoskeletal elements reveals actin and not microtubule-based peroxisome motility in plants. Plant Physiol. 128, 1031–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nagai, R. (1993). Regulation of intracellular movements in plant cells by environmental stimuli. Int. Rev. Cytol. 145, 251–310. [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Niwa, Y., Hirano, T., Yoshimoto, K., Shimizu, M., and Kobayashi, H. (1999). Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J. 18, 455–463. [DOI] [PubMed] [Google Scholar]
- Sack, F.D. (1997). Plastids and gravitropic sensing. Planta 203, S63–S68. [DOI] [PubMed] [Google Scholar]
- Sakai, T., Kagawa, T., Kasahara, M., Swartz, T.E., Christie, J.M., Briggs, W.R., Wada, M., and Okada, K. (2001). Arabidopsis nph1 and npl1: Blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. USA 98, 6969–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, Y., Kadota, A., and Wada, M. (1999). Mechanically induced avoidance response of chloroplasts in fern protonemal cells. Plant Physiol. 121, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, Y., Wada, M., and Kadota, A. (2001). Choice of tracks, microtubules and/or actin filaments for chloroplast photo-movement is differentially controlled by phytochrome and a blue light receptor. J. Cell Sci. 114, 269–279. [DOI] [PubMed] [Google Scholar]
- Tlalka, M., and Gabrys, H. (1993). Influence of calcium on blue-light-induced chloroplast movement in Lemna trisulca L. Planta 189, 491–498. [Google Scholar]
- Van Gestel, K., Kohler, R.H., and Verbelen, J.P. (2002). Plant mitochondria move on F-actin, but their positioning in the cortical cytoplasm depends on both F-actin and microtubules. J. Exp. Bot. 53, 659–667. [DOI] [PubMed] [Google Scholar]
- Williamson, R.E. (1993). Organelle movements. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 181–202. [Google Scholar]