Abstract
Promoter hypermethylation is a prevalent phenomenon, found in virtually all cancer types studied thus far, and accounts for tumor suppressor gene silencing in the absence of genetic mutations. The mechanism behind the establishment and maintenance of such aberrant hypermethylation has been under intense study. Here, we have uncovered a link between aberrant gene silencing associated with promoter CpG island DNA methylation and the siRNA/miRNA processing enzyme, DICER, in human cancer cells. By comparing demethylated HCT116 colon cancer cells with HCT116 cells genetically rendered hypomorphic for DICER, we identified a group of epigenetically silenced genes that became reactivated in the absence of functional DICER. This reactivation is associated with a dramatic loss of localized promoter DNA hypermethylation. Thus, intact DICER is required to maintain full promoter DNA hypermethylation of select epigenetically silenced loci in human cancer cells.
Introduction
Epigenetic gene silencing, which includes CpG island DNA cytosine methylation and histone modifications, has become widely accepted as a prominent alternative mechanism to genetic mutations by which cancer cells inactivate genes to facilitate the initiation, growth, and metastasis of tumors (1). DNA methylation refers to the covalent addition of a methyl group to the five position of cytosine residues, and this predominantly occurs in a CpG dinucleotide context in mammalian cells. Clusters of these CpG dinucleotides are present in the promoter regions of a large number of genes, and dense methylation, or hypermethylation, of these CpG islands are invariably associated with transcriptional silencing of the downstream gene (2). Although this phenomenon is widespread in human carcinogenesis, the specific mechanism(s) by which such epigenetic silencing is initiated in mammalian systems is largely unknown.
Recently, several groups have shown that synthetic small double-stranded RNAs homologous to gene promoter regions are able to induce transcriptional gene silencing at the targeted loci in human cells (3–6). This dsRNA-dependent Transcriptional Silencing (RdTS) was reproducibly found to associate with an enrichment of the silencing histone modification, H3 lysine 9 dimethylation (H3K9me2) but not DNA methylation in acute silencing events. However, H3K9me2 have been shown to precede DNA hypermethylation in the resilencing and remethylation of the tumor suppressor gene, p16ink4a, in HCT116 colon cancer cells (7), and therefore, it remains possible that RdTS may be a mechanism to initiate abnormal epigenetic silencing. Because other known RNA-mediated epigenetic silencing mechanisms in yeast (8, 9) and plants (10–12) all require the processing of RNA precursors by DICER, we investigated the possibility that DICER, an essential component of the RNAi pathway, is also required for the proper maintenance of epigenetic silencing in human cancer cells.
Materials and Methods
Cell culture and treatment
HCT116 parental cells and the isogenic DICERex5 cells were generous gifts from B. Vogelstein, Johns Hopkins University, School of Medicine, MD, and cultured in McCoy’s 5A medium supplemented with 10% fetal bovine serum. All other HCT116 derivative cells were also cultured in the same medium. HCT116 parental and DICERex5 cells were treated with DAC (Sigma) at 5 µmol/L in normal medium for 72 h to induce demethylation.
Microarray analysis
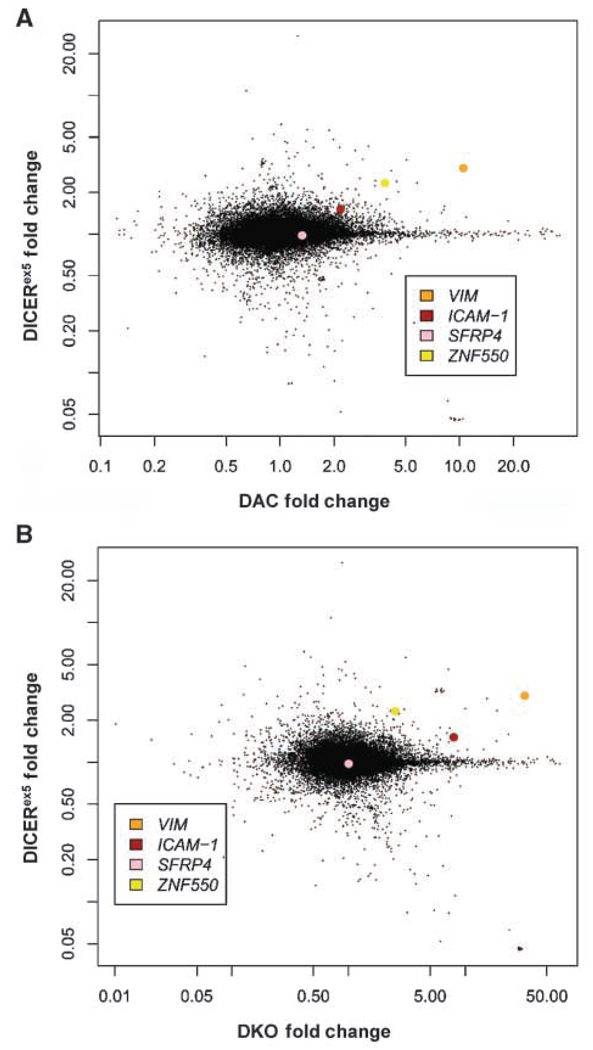
Total RNA was harvested from log phase cells using Trizol (Invitrogen) and the RNeasy kit (Qiagen) according to manufacturer’s instructions, including a DNase digestion step. RNA was quantified using NanoDrop ND-100 followed by quality assessment with 2100 Bioanalyzer (Agilent Technologies). RNA concentrations for individual samples were >200 ng/µL, with 28 s/18 s ratios >2.2 and RNA integrity of 10 (10 scored as the highest). Sample amplification and labeling were performed using the Low RNA Input Fluorescent Linear Amplification kit (Agilent Technologies) according to manufacturer’s instructions. The labeled cRNA was purified using the RNeasy mini kit (Qiagen). RNA spike-in controls were added to RNA samples before amplification. Samples (0.75 µg) labeled with Cy3 or Cy5 were mixed with control targets, assembled on Oligo Microarray, hybridized, and processed according to the Agilent microarray protocol. Scanning was performed with the Agilent G2565BA microarray scanner using settings recommended by Agilent Technologies. Microarray data Gene Expression Omnibus accession numbers are GSM147895 and GSM147932 (dye swap). An identical experiment was performed using the Affymetrix platform according to the Affymetrix microarray protocol. The results for the Agilent microarray experiments are shown in Fig. 1.
Figure 1.
Microarray analysis of DICERex5, DKO, and DAC-treated HCT116 cells. A, comparison between gene expression level changes (in fold difference) in DICERex5 cells relative to parental HCT116 cells (Y-axis) and DAC-treated cells relative to parental HCT116 cells (X-axis). Each spot on the graph represents one probe on the microarray. SFRP4, ICAM-1, VIM, and ZNF550 are highlighted in different colors. B, comparison between gene expression level changes (in fold difference) in DICERex5 cells relative to parental HCT116 cells (Y-axis) and DKO HCT116 cells relative to untreated HCT116 cells (X-axis). Each spot on the graph represents one probe on the microarray.
Microarray data analysis
All arrays were subject to quality checks recommended by the manufacturer. Images were visually inspected for artifacts and distributions of signal. Background intensity of both red and green channels was examined to identify anomalous arrays. No irregularities were observed, and all arrays were retained. All calculations were performed using the R statistical computing platform and packages from Bioconductor bioinformatics software project. The log ratio of red signal to green signal was calculated after background-subtraction and LoEss normalization as implemented in the limma package from Bioconductor. Individual arrays were scaled to have the same interquartile range (75th–25th percentile). Log-fold changes were averaged over dye-swap replicate microarrays to produce a single set of expression values for each condition.
Reverse transcription-PCR
Total RNA were extracted from cell pellets with RNeasy Mini kit (Qiagen) and treated with DNase (Qiagen). Two micrograms of total RNA per sample were used in first-strand cDNA synthesis using Superscript 1st Strand Synthesis System (Invitrogen) with random hexamer primers. One microliter of cDNA were used for subsequent PCR reactions. PCR products were resolved on 2% agarose gels containing GelStar nucleic acid stain (Cambex) and visualized with Kodak Gel Logic 200 Imaging System. Real-time reverse transcription-PCR (RT-PCR) was carried out using Taq Man Gene Expression Assays (Applied Biosystems) and a 7900HT Fast Real-Time PCR System (Applied Biosystems) according to manufacturer’s instructions. SDS2.2.2 software (Applied Biosystems) was used to perform comparative δ cycle threshold analysis. GAPDH served as endogenous control. PCR conditions and primers used are available upon request.
DNA methylation analysis
Genomic DNA were extracted and treated with sodium bisulfite as previously described (13). Bisulite converted DNA was used in PCR reactions with nondiscriminatory bisulfite sequencing PCR primers. Bisulfite sequencing PCR primers and conditions are available upon request. The PCR products were subsequently cloned into the TOPO TA vector (Invitrogen) and sequenced with the M13R primer.
Chromatin immunoprecipitation assay
Cells were crosslinked and processed after the UpState Chromatin Immunoprecipitation (ChIP) Assay kit protocol (UpState). One hundred fifty micrograms of sonicated DNA were used for each immunoprecipiration (IP) reaction. Rabbit anti-H3 dimethyl-K4 (5 µg/IP; Upstate), rabbit anti-H3 dimethyl-K9, rabbit anti-H3 trimethyl-K9, rabbit anti-H3 dimethyl-K27, and rabbit anti-H3 trimethyl-K27 (10 µg/IP; gifts from T. Jenuwein of Research Institute of Molecular Pathology, Vienna, Austria) antibodies were used for the specific IP of the respective histone residues. No antibody and rabbit anti-HA antibody (10 µg/IP; Santa Cruz; SC805) were performed as controls. Fifty microliters of sonicated, pre-IP DNA from each sample were used as input controls. The final results for each sample were normalized to respective inputs. PCR conditions and primers used are available upon request.
McrBC digestion
Five micrograms of genomic DNA and 50 units of McrBC (New England Biolabs) enzyme were incubated with 1× NEBuffer, 100 µg/mL bovine serum albumin, and 1 mmol/L GTP for 12 h at 37°C. One third of each reaction was electrophoresed on 0.8% agarose gels containing GelStar nucleic acid stain (Cambex) and visualized with Kodak Gel Logic 200 Imaging System.
Western blotting
Twenty micrograms of whole cell lysates from each sample were resolved on 4% to 2% Bis-Tris gel and transferred onto a nitrocellulose membrane. The membrane was blocked in 10% milk in TTBS overnight at 4°C and incubated with 1:1,000 mouse anti-DICER1 (Imgenx), 1:500 rabbit anti-DNMT1 (Santa Cruz), and 1:10,000 mouse anti-βactin (Sigma) for 2 h at room temperature. The secondary antibodies are used at 1:1,000 for DICER1, 1:500 for DNMT1, and 1:10,000 for βactin.
Methyltransferase activity assay
DNA methyltransferase activity assays were performed as described previously (14). Briefly, 15 µg whole cell lysates were incubated with hemimethylated DNA oligos, S-adenosyl-l-[methyl-3H]methionine (3H-AdoMet; Amersham Biosciences), in reaction buffer [20 mmol/L Tris-HCl (pH 7.4), 1 mmol/L EDTA, 5% glycerol, and 1 mmol/L DTT]. After incubation at 37°C, reactions were stopped at various time points by adding one volume of 10 mmol/L nonradioactive S-adenosyl-l-methionine (Sigma). The reactions were bound to a SAM2 96 Biotin Capture plate (Promega). The plate was washed five times with PBS containing 2 mol/L NaCl and twice with dH2O to remove His6-DNMT and unreacted 3H-AdoMet. After drying the plate, 80 µL of Microscint-PS scintillation fluid (PerkinElmer) was added to each well, and tritium incorporation was quantified using the TopCount NXT liquid scintillation counter (PerkinElmer). All reactions were performed in sextuplicate. Data obtained were analyzed using the Enzyme Kinetics module of SigmaPlot (Systat Software).
Results and Discussion
First, to study the effects of loss of DICER on epigenetic silencing, we performed an expression microarray analysis comparing previously generated DICER helicase domain knockout (DICERex5) HCT116 human colon cancer cells (15) with wild-type parental HCT116 cells. Because DICER null cells are not viable (16), DICER gene exon 5, which encodes for the helicase domain, was deleted via homologous recombination to create the DICERex5 HCT116 cells (15). These DICERex5 cells were characterized to harbor extensive defects in microRNA processing but were never studied for changes in epigenetic silencing (15). The expression profile of the DICER-deficient cells was compared with that of parental HCT116 cells treated with the DNA demethylating agent 5-aza-2′-deoxycytidine (DAC; Fig. 1A) and HCT116 cells genetically deleted for the DNA methyltransferases, DNMT1 and 3b (DKO cells; Fig. 1B). Both the DAC-treated cells and the DKO cells exhibit dramatic loss of overall genomic cytosine methylation, loss of promoter CpG island hypermethylation, and re-expression of the demethylated genes (17). The striking change in gene expression profile in these two types of cells can be readily observed in our microarray analyses, displaying a large number of genes being reactivated in either setting (Fig. 1). Compared with the large number of genes reactivated in either DAC-treated or the DKO cells, only 31 genes were identified by the microarray study as substantially up-regulated in both the DICERex5 cells and the demethylated cells (Table 1).
Table 1.
Candidate genes up-regulated in both DICERex5 and either DKO- or DAC-treated HCT-116 cells
| Symbol | Location | CpG island | Expression change | Methylation in HCT116 | Demethylation in DICERex5 |
|---|---|---|---|---|---|
| SFRP4 | 7p14.1 | Yes | Yes | Yes | Yes |
| ICAM-1 | 19p13.3-p13.2 | Yes | Yes | Yes | Yes |
| RASGRP1 | 15q15 | Yes | Yes | Yes | Yes |
| RTN1 | 14q23.1 | Yes | Yes | Yes | Yes |
| SLC4A4 | 4q21 | Yes | Yes | Yes | Yes |
| SLC40A1 | 2q32 | Yes | Yes | Yes | Yes |
| ZNF550 | 19q13.43 | Yes | Yes | Yes | Yes |
| VIM | 10p13 | Yes | Yes | Yes | Yes |
| LHFP | 13q12 | Yes | Yes | Yes | No |
| LIPG | 18q21.1 | Yes | Yes | Yes | No |
| RGC32 | 13q14.11 | Yes | Yes | Yes | No |
| CXCR4 | 2q21 | Yes | Yes | No | |
| HOXB2 | 17q21-q22 | Yes | Yes | No | |
| CPEB2 | 4p15.33 | Yes | Yes | No | |
| PELI2 | 14q21 | Yes | Yes | No | |
| SERTAD4 | 1q32.1-q41 | Yes | Yes | No | |
| PANK3 | 5q34 | Yes | No | ||
| NANOS1 | 10q26.11 | Yes | No | ||
| CYR61 | 1p31-p22 | Yes | No | ||
| KRTAP2-4 | 17q12-q21 | No | |||
| TRIM6 | 11p15.4 | No | |||
| SPARC | 5q31.3-q32 | No | |||
| DHRS2 | 14q11.2 | No | |||
| FLJ36492 | 17p11.2 | No | |||
| RAFTLIN | 3p24.3 | No | |||
| APOBEC3G | 22q13.1-q13.2 | No | |||
| C10orf10 | 10q11.21 | No | |||
| DKK4 | 8p11.2-p11.1 | No | |||
| GBP3 | 1p22.2 | No | |||
| GNG2 | 14q21 | No | |||
| SERPINA1 | 14q32.1 | No | |||
| TM4SF1 | 3q21-q25 | No |
NOTE: These genes were first sorted by whether or not there is a CpG island in their promoters. Next, based on either existing literature or empirical DNA methylation analyses, the genes containing promoter CpG islands were categorized based on their promoter hypermethylation status in the parental HCT116 cells. SFRP4, ICAM-1, VIM, and ZNF550 were all identified as genes hypermethylated in the parental HCT116 cells.
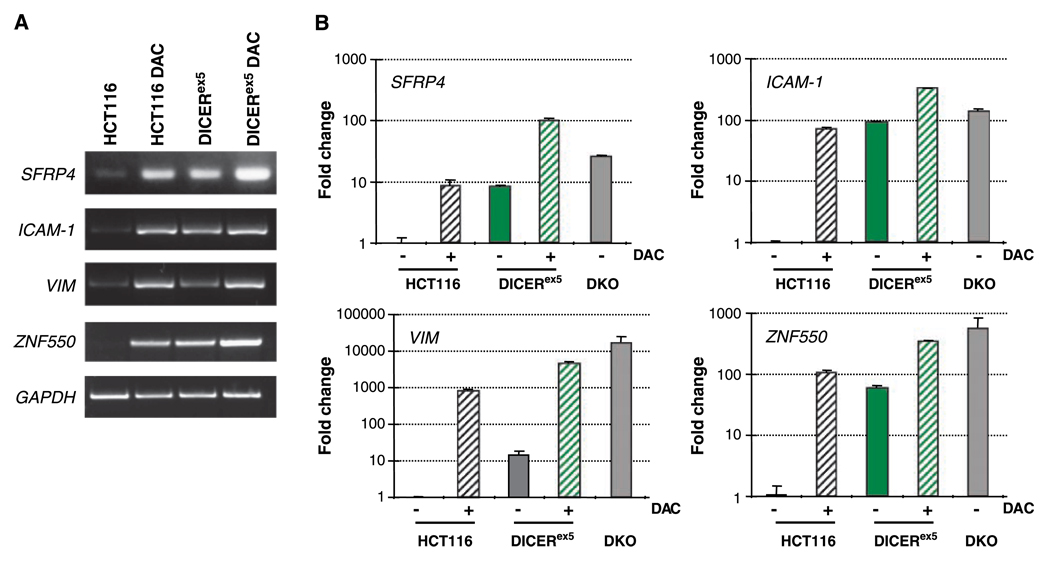
We next determined, as defined by a high stringency criteria (18), that 18 of the 31 candidate genes for regulation by DICER contained promoter CpG islands (Table 1). We then verified by qualitative gel-based RT-PCR that 15 of these 18 candidates were indeed re-expressed in the DICERex5 cells (Supplementary Fig. S1). We also checked expression levels of genes known to be aberrantly silenced and promoter DNA hypermethylated in the parental HCT116 cells but which did not seem to be re-expressed in the DICERex5 cells as negative controls (Supplementary Fig. S1). To our surprise, SFRP4, one of the genes analyzed as a negative control, was actually up-regulated in DICERex5 cells and was subsequently included in our analyses of candidate genes (Table 1; Fig. 2A and B). Representative RT-PCR analyses for SFRP4, ICAM-1, VIM, and ZNF550 are shown (Fig. 2A), and the positions of the genes on the microarray comparisons are indicated in Fig. 1. Similar data were obtained for all other candidate genes (Supplementary Fig. S1). SFRP4, or Secreted Frizzle-related Protein 4, belongs to a family of soluble WNT signaling pathway antagonists. Several members of this family, including SFRP4, have been shown to be silenced by promoter CpG island hypermethylation in various tumors and cancer cell lines (19). ICAM-1 encodes for a cell adhesion molecule whose expression is important for the tumor metastasis process and has been shown to be silenced by promoter hypermethylation as well (20). VIM and ZNF550 are newly identified in this study to be potentially regulated by promoter DNA methylation. The expression changes in these genes were also independently verified by quantitative Taq man RT-PCR (Fig. 2B). Basal expressions are either silent or very low in the parental HCT116 cells but significantly up-regulated in the DICERex5 cells.
Figure 2.
Validation of microarray expression data by RT-PCR. A, SFRP4, ICAM-1, VIM, and ZNF550 mRNA levels in parental HCT116 and DICERex5 cells were assessed by RT-PCR. RT-PCR for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was performed as a control to show that equal amounts of cDNA were used for each sample. DAC-treated parental and DICERex5 cells were used as positive controls for gene expression for all four genes. B, quantitative Taq Man RT-PCR for SFRP4, ICAM-1, VIM, and ZNF550. Expression levels of each gene in each sample were normalized to the expression level detected in the untreated parental HCT116 cells and expressed as fold change. DAC-treated parental and DICERex5 cells and DKO cells were included as positive controls for gene expression.
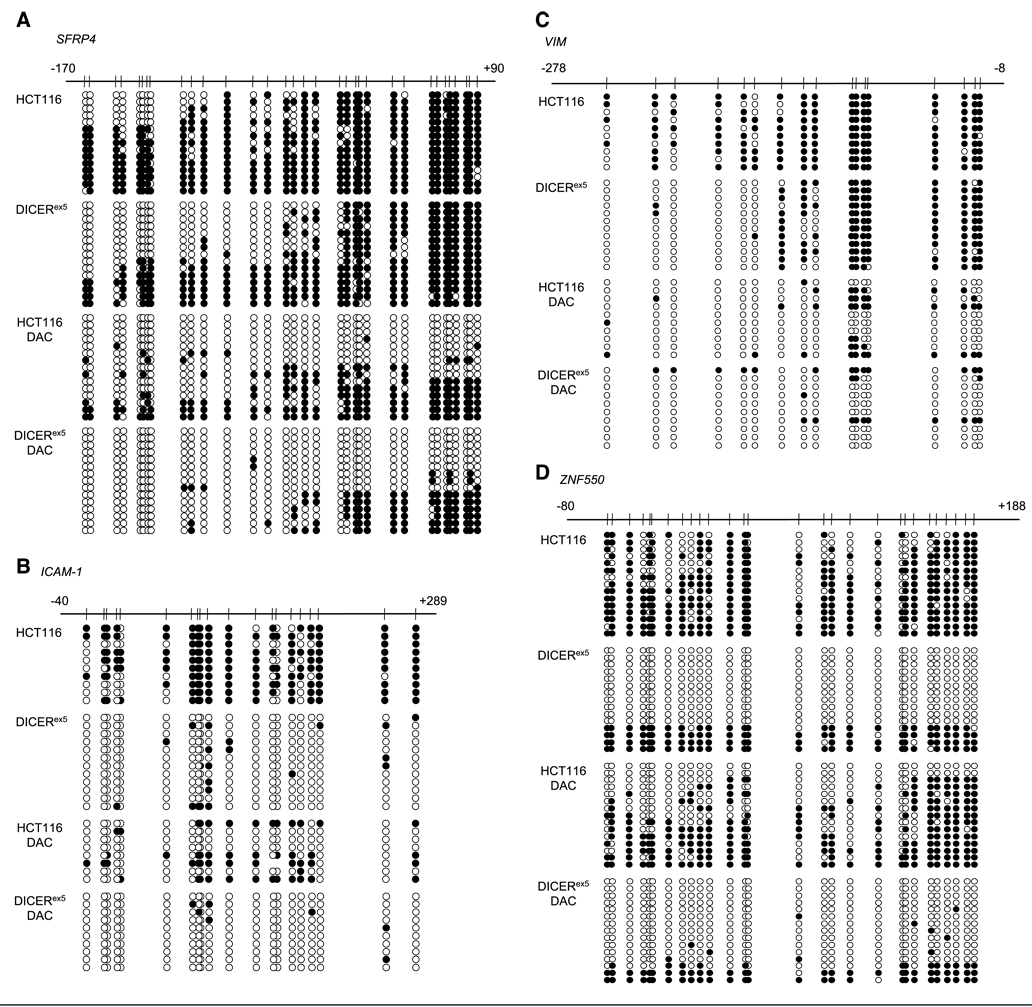
Next, we investigated the possibility that the observed derepression of genes in the DICERex5 cells was associated with changes in their promoter DNA hypermethylation. We identified that 11 promoters of the 16 re-expressed genes had CpG island hypermethylation in the parental HCT116 cells, and 8 of the 11 became demethylated in the DICERex5 cells (Fig. 3; Supplementary Fig. S2). We comprehensively evaluated changes in promoter hypermethylation by bisulfite sequencing (13). As clearly illustrated for SFRP4 (Fig. 3A), ICAM-1 (Fig. 3B), VIM (Fig. 3C), and ZNF550 (Fig. 3D), these gene promoters were densely hypermethylated in the parental HCT116 cells but became significantly demethylated in the DICERex5 cells.
Figure 3.
Methylation analysis of SFRP4, ICAM-1, VIM, and ZNF550 promoters in parental HCT116 and DICERex5 cells. A, bisulfite sequencing results for SFRP4. Thirty-three CpG sites within the SFRP4 CpG island were examined. The position of each CpG site is marked relative to transcription start site and is represented by a circle. ○, unmethylated CpG sites; ●, methylated CpG sites. B, bisulfite sequencing results for ICAM-1. Twenty CpG sites within the ICAM-1 CpG island were examined. Representation of the data are same as in A. C, bisulfite sequencing results for VIM. Seventeen CpG sites within the VIM CpG island were examined. Representation of the data are same as in A. D, bisulfite sequencing results for ZNF550. Twenty-eight CpG sites within the ZNF550 CpG island were examined. Representation of the data are same as in A.
It is formally possible that the observed demethylation at the candidate promoters is due to overall genomic demethylation in the DICERex5 cells rather than a locus-specific demethylation and reactivation. However, for genes such as SFRP1 and SFRP2, which did not show re-expression in the DICERex5 cells, their promoters remained hypermethylated (Supplementary Fig. S3). Moreover, overall genomic DNA methylation, as analyzed by restriction enzyme digests of total DNA with the methylation sensitive McrBC, was unaltered in the DICERex5 cells (Supplementary Fig. S4A). These results suggest that the demethylation of candidate genes in the DICERex5 cells is specific to these promoters. We also tested whether the demethylation was due to global down-regulation of the major enzyme contributing to overall DNA methyltransferase activity in mammalian cells, DNMT1, possibly as a consequence of disrupted micorRNA regulation in the DICERex5 cells. We found that this enzyme was expressed at comparable levels in the parental and the DICERex5 cells (Supplementary Fig. S4B). We further verified this by DNA methyltransferase activity assays to assess the total enzyme activity and found this to be comparable in the parental and DICERex5 cells (Supplementary Fig. S4C). Finally, according to our microarray data, the mRNA levels for all the DNMTs remained the same in DICERex5 cells when compared with the parental cells (data not shown). Based on these results, we concluded that intact DICER function is essential for the maintenance of full promoter hypermethylation and transcriptional repression at the identified loci.
DICER-mediated epigenetic gene silencing has been described in both plants and yeast. In plants, a RNA-directed DNA Methylation pathway uses RNA species to direct DNA cytosine methylation and repressive histone modifications to homologous DNA sequences to initiate transcriptional silencing (11, 12). Furthermore, RNA-dependent heterochromatin assembly has also been described in yeast for normal silencing of centromeric repeats (8, 9). Although DNA methylation is absent from the yeast system, histone tail modifications usually associated with DNA methylation, such as histone H3 lysine 9 and lysine 27 methylation, are recruited to the silenced yeast loci by double-stranded RNA molecules with sequence homology (21). Both of the above processes lead to the epigenetically silenced state, which resembles that observed for epigenetically silenced genes in cancer.
Given the above, we thus examined SFRP4 and ICAM-1 promoters by ChIP analyses for chromatin modifications that might provide insight into mechanisms that define the DICER-targeted genes. We observed some changes in chromatin modifications at DICER regulated loci that seem to be distinctive from other hypermethylated genes studied thus far (Supplementary Fig. S5; ref. 22). The demethylated and transcriptionally active SFRP4 and ICAM-1 promoters in the DICERex5 cells exhibited an increase in the enrichment of histone H3 lysine 4 dimethylation (H3K4me2) and general decreases in the enrichment of histone H3K9me2 (Supplementary Fig. S5A). These changes are identical to those seen in multiple other DNA hypermethylated genes when induced to demethylate by DAC, or in DKO cells (summary for these other genes in Supplementary Fig. S5B; ref. 22). However, unlike these previously characterized DNA hypermethylated promoters, histone H3 lysine 27 tri-methylation (H3K27me3) showed a small but consistent decrease across the entire promoter regions at the DICER-regulated SFRP4 and ICAM-1 promoters. All other DNA demethylated promoters evaluated to date seemed to retain or augment H3K27me3 (Supplementary Fig. S5B; ref. 22). This pattern of increase in H3K4me2 and decrease in H3K27me3 was not observed for the unresponsive loci, SFRP1, SFRP2, GATA4, and GATA5, in the DICERex5 cells. This suggested that the observed changes in chromatin modifications could be characteristic of the DICER-target genes.
The high degree of similarity between the RNA-mediated epigenetic gene silencing mechanisms in diverse systems, including what has been observed in human cancer with synthetic RNA oligonucleotides, suggest an evolutionarily conserved mechanism. In the present study, we established that DICER, an essential protein of the RNAi pathway in all organisms studied thus far, was indispensable for the full maintenance of promoter DNA hypermenthylation and transcriptional repression of select endogenous loci in cancer cells. We attempted to verify whether DICER is also responsible for the initiation of epigenetic silencing at these target loci by ectopically expressing functional DICER in the DICERex5 cells. However, the re-expression of DICER seemed to be lethal, and therefore, this point requires further investigation (data not shown). To our knowledge, this is the first observation directly linking DICER and mammalian epigenetic silencing of endogenous protein coding loci. It is suggestive of RdTS being a mechanism involved in aberrant hypermethylation in human cancer cells. Alternatively, the derepression of these genes could be a secondary effect due to the loss of miRNA repression of upstream common transcription factors/repressors as a result of dysregulation of the miRNA pathway.
We only observed such loss of epigenetic silencing at a subset of the many genes known to be hypermethylated and silenced in colon cancer cells. We may have missed other target genes in our initial microarray screening because SFRP4 is clearly a target locus that lacked change of expression detectable by the microarray. Also, because the DICERex5 allele is only hypomorphic, we might not have observed the fullest effects of loss of DICER function relating to epigenetic silencing. Alternatively, our more favored hypothesis is that this pathway is only responsible for select epigenetically controlled genes. DICER processes only a subset of noncoding RNA, such as miRNA, but not all noncoding RNA that could influence epigenetic silencing. For instance, the antisense RNA involved in the epigenetic silencing of the p15 locus does not require DICER (23). What differentiates DICER-regulated genes from the rest of the epigenetically controlled loci remains to be determined. The histone modification changes we observed for these loci may be the first step toward characterizing these genes. Because H3K27me3 is thought to be specifically mediated by Polycomb group (PcG) proteins, the observed changes in H3K27me3 modification unique to SFRP4 and ICAM-1 could indicate that DICER-mediated epigenetic silencing requires PcG proteins to maintain DNA methylation. This hypothesis is in line with observations in Drosophila, where full PcG-mediated long-range gene silencing required RITS components, including dcr-2 (24). Exactly which and how PcG proteins may participate in this DICER-mediated epigenetic silencing in human cancer cells should be further examined.
In summary, we have shown that at least some genes that become DNA hypermethylated and silenced in cancer cells require DICER to maintain this epigenetic status. The only well-defined role for DICER is to process dsRNAs, and therefore, RNA molecules may be involved in the initiation and/or maintenance in this novel silencing pathway. Therefore, it is imperative that we continue working on identifying the specific mechanism of this DICER-dependent epigenetic silencing, its relationship to the RdTS phenomenon, and ultimately, its role in cancer progression and normal development. Current therapies aimed at reversing epigenetic defects focus heavily on DNMT and histone-modifying enzyme inhibitors. The study of this DICER-mediated gene silencing will help enrich our understanding of the nature of aberrant epigenetic silencing in cancers and may prove useful in revealing new targets for cancer therapies aimed at circumventing epigenetic defects.
Acknowledgments
Grant support: This study was partially supported by NIEHS grant ES011858.
We thank Drs. Bert Vogelstein and Judith Bender for helpful discussions and critical reading of the manuscript, Dr. Wayne Yu for technical assistance with the microarray studies, and Dr. Thomas Jenuwein (Research Institute of Molecular Pathology, Vienna, Austria) for the generous gifts of antibodies against the various histone modifications.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome-components and functional correlates. Genes Dev. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 3.Ting AH, Schuebel KE, Herman JG, Baylin SB. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat Genet. 2005;37:906–910. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castanotto D, Tommasi S, Li M, et al. Short hairpin RNA-directed cytosine (CpG) methylation of the RASSF1A gene promoter in HeLa cells. Mol Ther. 2005;12:179–183. doi: 10.1016/j.ymthe.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Janowski BA, Corey DR. Inhibiting transcription of chromosomal DNA using antigene RNAs. Nucleic Acids Symp Ser (Oxf) 2005;49:367–368. doi: 10.1093/nass/49.1.367. [DOI] [PubMed] [Google Scholar]
- 6.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 7.Bachman KE, Park BH, Rhee I, et al. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell. 2003;3:89–95. doi: 10.1016/s1535-6108(02)00234-9. [DOI] [PubMed] [Google Scholar]
- 8.Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin-and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 9.Grewal SI, Rice JC. Regulation of heterochromatin by histone methylation and small RNAs. Curr Opin Cell Biol. 2004;16:230–238. doi: 10.1016/j.ceb.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Mette MF, van der Winden J, Matzke MA, Matzke AJ. Production of aberrant promoter transcripts contributes to methylation and silencing of unlinked homologous promoters in trans. EMBO J. 1999;18:241–248. doi: 10.1093/emboj/18.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelissier T, Thalmeir S, Kempe D, Sanger HL, Wassenegger M. Heavy de novo methylation at symmetrical and non-symmetrical sites is a hallmark of RNA-directed DNA methylation. Nucleic Acids Res. 1999;27:1625–1634. doi: 10.1093/nar/27.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wassenegger M, Heimes S, Riedel L, Sanger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 13.Frommer M, McDonald LE, Millar DS, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee BH, Yegnasubramanian S, Lin X, Nelson WG. Procainamide is a specific inhibitor of DNA methyltransferase 1. J Biol Chem. 2005;280:40749–40756. doi: 10.1074/jbc.M505593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cummins JM, He Y, Leary RJ, et al. The colorectal microRNAome***. Proc Natl Acad Sci U S A. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 17.Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki H, Watkins DN, Jair KW, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 20.Hellebrekers DM, Castermans K, Vire E, et al. Epigenetic regulation of tumor endothelial cell anergy: silencing of intercellular adhesion molecule-1 by histone modifications. Cancer Res. 2006;66:10770–10777. doi: 10.1158/0008-5472.CAN-06-1609. [DOI] [PubMed] [Google Scholar]
- 21.Shankaranarayana GD, Motamedi MR, Moazed D, Grewal SI. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol. 2003;13:1240–1246. doi: 10.1016/s0960-9822(03)00489-5. [DOI] [PubMed] [Google Scholar]
- 22.McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- 23.Yu W, Gius D, Onyango P, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]