Abstract
Although induced mutations in traditional laboratory animals have been valuable as models for human diseases, they have some important limitations. Here we propose a complementary approach to discover genes and mechanisms that might contribute to human disorders: the analysis of evolutionary mutant models whose adaptive phenotypes mimic maladaptive human diseases. If the type and mode of action of mutations favored by natural selection in wild populations are similar to those that contribute to human diseases, then studies in evolutionary mutant models have the potential to identify novel genetic factors and gene-by-environment interactions that affect human health and underlie human disease.
Mutant models for human disease: utility and limitations
Among the major biological insights of the late twentieth century was the discovery that a large portion of the genes and mechanisms that direct embryonic development are broadly conserved among metazoans [1]. This discovery galvanized the use of traditional animal models (from flies to mice) for the study of traits and phenomena relevant to human health. Although most organisms are not recognized as ‘model species’, the same conserved genetic features are likely to operate in an array of animal species that exhibit extensive phenotypic diversity, and therefore non-model species can provide important additional insights into human health and disease. Here we propose that the investigation of evolutionary mutant models (see Glossary) can enhance our basic understanding of the genetic and developmental basis for human diseases.
Mutations induced in forward genetic screens in laboratory animals provide highly useful models of human phenotypic variation and disease that have led to previously unsuspected insights into human pathology [2–4]. This approach, however, is not without limitations. Mutagenesis screens often identify phenotypes that are more severe and/or have earlier onset than the human diseases they model. Because researchers often identify the most visible mutations in laboratory screens, induced mutations are predominantly in the coding regions of genes and lead to the severe attenuation or complete abrogation of gene function. Furthermore, because forward genetic screens typically recover mutant animals with defects at the earliest developmental stage in which the gene provides an essential function, they often mask developmental pleiotropy. As a result, phenotype-driven mutation screens using chemical mutagens in zebrafish, Drosophila, and Caenorhabditis elegans generally yield early defects that preclude the examination of phenotypes at the later life stages that are relevant to many human diseases.
In contrast to mutants recovered in laboratory screens, many naturally occurring mutations in humans that either cause a disease or increase disease susceptibility under specific environmental conditions are caused by alterations in the regulatory regions of genes [5–10]. Because multiple promoters, enhancers, and silencers regulate the timing, abundance, and cell-specific localization of gene expression, alterations in cis-regulatory elements can result in normal gene expression early in development, but failed regulation of a gene’s activity in older individuals. For example, lactase persistence is a condition that results in an individual’s sustained ability to digest lactose through adulthood [11]. Most human babies produce intestinal lactase, an enzyme that digests the milk sugar lactose. In many human populations, however, lactase production decreases during teenage years, blocking the digestion of lactose and allowing it to be fermented by bacteria in the colon, which causes abdominal pain. The ability to maintain lactase production is inherited in an autosomal dominant fashion and is associated with non-coding variation upstream of the Lactase (LCT) gene that increases its expression, resulting in prolonged ability to digest lactose. Individuals lacking these variants gradually lose LCT activity and lack the ability to digest lactose as adults. Such regulatory mutations, when they occur in nature, are exposed to selection and can therefore be studied in evolutionary mutant models.
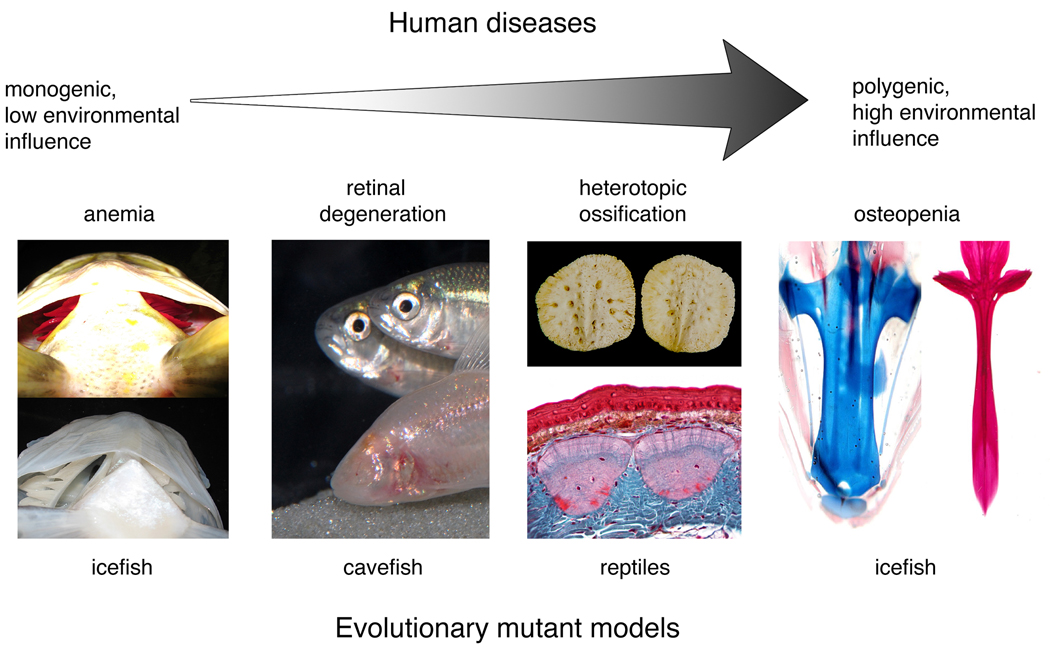
Human diseases lie along a continuum from ‘simple’ to ‘complex’. On one end of the spectrum are monogenic disease conditions that exhibit simple Mendelian inheritance and express little phenotypic variation in individuals with the same allele, such as albinism or cystic fibrosis. On the opposite end of the spectrum are polygenetic diseases, such as cancers or heart disease, whose expression and severity are highly variable and depend on an individual’s genotype at multiple loci as well as the environment. In the middle are disease traits that are affected by alleles of major effect, but whose expression is variable and affected by both genetic background and the environment, for example, cleft palate. While induced mutant models have been extremely useful in studying simple diseases, they have been less successful in deducing the etiology and pathophysiology of complex diseases. We propose that evolutionary mutant models can compliment traditional induced mutant models to understand the genetic basis of both simple and complex human diseases (Fig. 1).
Figure 1. Examples of evolutionary mutant models for human disease.
Evolutionary mutant models have the potential to reveal novel insight into the genetic basis of an array of different types of human diseases, from simple to complex. An example of a natural system that models a simple human disease is anemia in icefish. Note that the icefish (bottom panel) lacks red blood cells (as seen in the gills) compared to the closely related rockcod (top panel). Evolutionary mutants can also model complex human diseases including osteopenia in icefish. Note that the base of the icefish neurocranium is cartilaginous (blue, left panel) compared to the closely related rockcod, which has a mineralized neurocranium (red, right panel). Evolutionary mutants will also be useful for diseases in the middle of the continuum that are affected by alleles of major effect, but whose expression is complex and variable. Diseases of this type include retinal degeneration and heterotopic ossification, which are modeled by blind cavefish and reptile osteoderms, respectively. Cavefish image reproduced with permission from R. Borowsky. Osteoderm image courtesy of M. Vickaryous.
Evolutionary mutants and the diseases they model
Occasionally, evolution by natural selection or genetic drift has resulted in populations with evolved phenotypes that mimic human disease, but are nevertheless adapted to their environment. Here we discuss selected examples of evolutionary mutant models that can inform our understanding of human disease. Although our focus will largely be on fish models, other metazoan systems will undoubtedly provide additional evolutionary mutant models for human disease phenotypes, and the principles discussed will apply across many species.
Antarctic icefish as a model of anemia
Anemias are diseases with diminished numbers of red blood cells, resulting in decreased oxygen delivery to tissues. The 16 “white-blooded” species of the Antarctic icefish family (Notothenioidei: Channichthyidae) are unique among vertebrates because they do not express the developmental program for erythrocyte formation. Icefish neither make hemoglobin nor do they produce erythrocytes, and therefore they model harmful human blood diseases, including anemias, hemoglobinopathies, and thalassemias. In a genome-wide scan of hematopoietic tissues comparing white-blooded icefish with red-blooded relatives, transcriptome-based representational difference analysis [cDNA RDA; 12] revealed that the novel gene bloodthirsty (bty) is expressed in the red-blooded relative but not in the white-blooded icefish [13]. The antisense knockdown of bty in zebrafish and the rescue of its expression by mRNA injection confirmed that bty plays a critical role in erythrocyte development [14]. The apparent human ortholog of bty belongs to the TRIM (tripartite motif) gene family, many of whose members encode E3 ubiquitin ligases (H. W. Detrich, unpublished). Thus, Bty/TRIM is likely to target a repressor of proerythroblast differentiation for degradation by the proteosome, thereby inducing terminal erythroid differentiation. These studies demonstrate that evolved genetic changes in icefish can provide a model for human anemias.
Antarctic icefish as a model of osteopenia
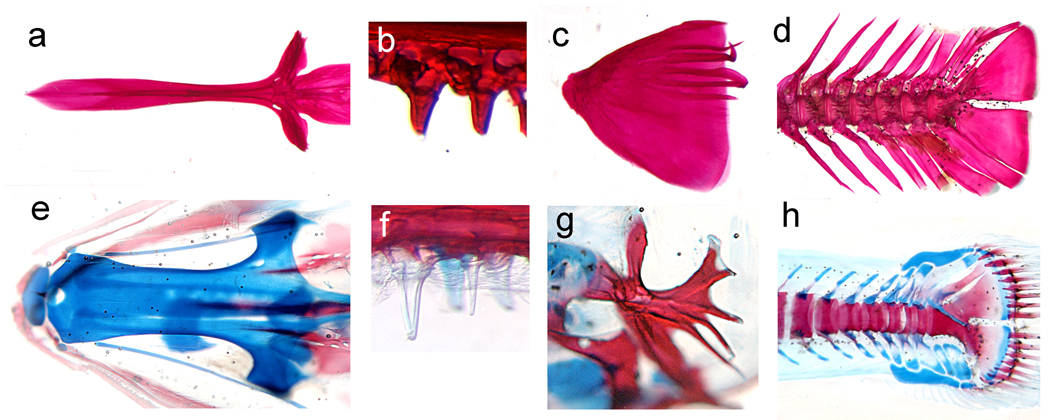
Osteopenia is a reduction in bone mineral density (BMD) that affects approximately 34 million American women and 12 million American men. It can lead to osteoporosis, a disease characterized by low bone mass, bone deterioration, bone fragility, increased susceptibility to fracture, and slow healing of bone fracture [15]. More than half of Americans over 50 years old have osteoporosis [16]. Because of their unique evolutionary history, certain lineages of Antarctic fish provide striking evolutionary mutant models of reduced bone density diseases. Specifically, in several notothenioid species, natural selection has favored structural changes in the musculoskeletal system to increase buoyancy, including the replacement of densely mineralized bone by connective tissue and the decreased ossification of the skeleton [17]. Figure 2 shows the dramatic difference in mineralization of the skeletons of an osteopenic juvenile icefish and a closely related robustly ossified species of comparable developmental stage. The identification of genetic factors that underlie these evolved differences has the potential to lead to a better understanding of the mechanisms that regulate bone density.
Figure 2. Osteopenia in Antarctic notothenioids.
The evolutionary history of certain Antarctic notothenioid fish species has led to changes in the musculoskeletal system to increase buoyancy. Among other adaptations, several species have evolved bone loss, and now possess greatly reduced bony skeletons. These evolutionary adaptations model human diseases of decreased bone mineralization including osteopenia. An example of an osteopenia notothenioid species is Pseudochaenichthys georgianus (e–h), which, compared to the closely related but robustly mineralized species, Notothenia rossii (a–d), shows reduced levels of bone mineralization in the base of the skull (a,e), oral teeth (b,f), opercular bone (c,g), and caudal skeleton. Images show the staining of cartilage with alcian blue and bone with alizarin red in juvenile fish.
Blind cavefish as a model of retinal degeneration
Many cave-dwelling species evolve degenerated lenses and retinas, similar to humans with retinal degeneration diseases [18, 19]. The Mexican tetra (Astyanax mexicanus) has both surface populations with eyes and cave populations without eyes, providing an ideal opportunity to study the genetic basis of evolved eye loss. Relative to surface forms, expression of pax6, which when mutated in human and mouse causes malformation of iris and lens [20, 21], was shown to be down-regulated in the developing optic field of blind cavefish [22]. Furthermore, the experimental over-expression of two upstream regulators of eye development, shha (sonic hedgehog a) and shhb, in surface fish resulted in the down-regulation of pax6 and reduction of the optic cup, which suggests that the hedgehog signaling pathway is important in the evolution of cavefish eye degeneration [23]. It was therefore surprising that a recent mapping study showed that the causative genetic variants for eye loss in blind cavefish are not linked to shha, shhb, or pax6 [19], implicating other factors in the evolution of this phenotype. At least four to six unlinked loci are involved in cavefish eye loss [24], and different cavefish populations show varying degrees of eye degeneration, underscoring the complexity of this trait [25]. The molecular genetic factors that actually cause the evolution of eye loss in blind cavefish have yet to be identified, but their subsequent localization may provide novel candidate genes and pathways for human retinal degeneration. The regressive evolution of eye loss was likely a gradual process during the transition from a surface to cave environment, and the genetic changes that occurred over evolutionary time might mimic changes in gene regulation that occur over developmental time in genetically complex degenerative eye diseases of old age.
East African cichlid fish as a model of craniofacial disease
Craniofacial diseases involve distortions in the construction of the face and jaw. An emerging challenge for developmental biologists is to learn the molecular determinants of craniofacial shape, and the challenge for physicians is to convert that knowledge into therapies and preventative practices. Closely related cichlid fishes from lakes in the East African Rift Valley have undergone extensive evolutionary modifications of their oral jaws and faces, providing an array of evolutionary mutant models for medically important human craniofacial variation.
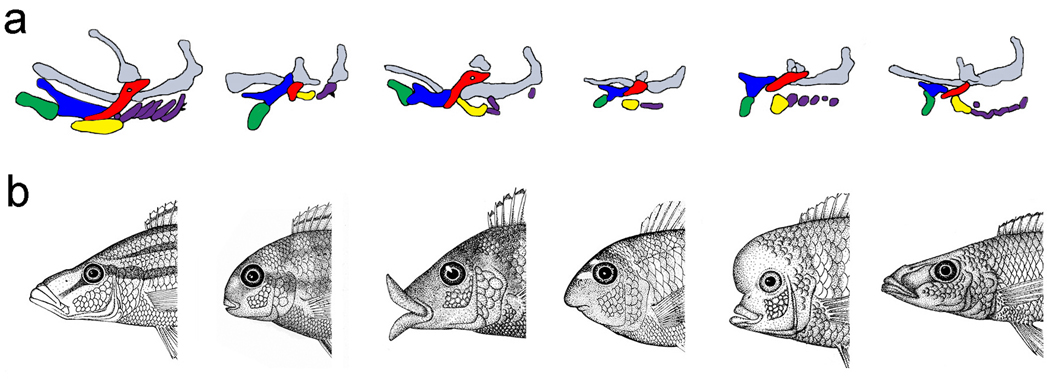
Just as chemically induced zebrafish mutants have been used to study craniofacial patterning, natural populations of cichlids can be used to study the development of craniofacial shape (Fig. 3). Diversity in cichlid tooth size, shape, and number, for example, provides a useful model for human hyperdontia (i.e., Apert syndrome), hypodontia (i.e., Crouzon syndrome), and macrodontia (i.e., hemifacial hyperplasia) [27]. Some cichlids eat the scales of other fish and are genetically specialized to feed from the prey’s left or right side [28]. Many of these scale-eating species have evolved skeletal and dental asymmetries similar in presentation to individuals with hemifacial microsomia and hemifacial hyperplasia [29]. Both in the adaptive situation in the fish and the disease state in humans, the ventral derivatives of the first and second pharyngeal arches are affected, and the asymmetries have a directional bias. Because few laboratory mutants are characterized by craniodental asymmetries [but see 30], analyses of laterality in scale-eating cichlids have the potential to reveal novel factors involved in regulating craniofacial symmetry. Compared to laboratory-based mutagenesis in a few distantly related model organisms (e.g., zebrafish and mouse), a comparative approach to jaw development in a group of closely related, but phenotypically divergent, cichlid species provides a robust opportunity to estimate the number, type, and mode of action of genes involved in jaw morphogenesis. Continued work on cichlids will further our understanding of the genetic changes and the gene-by-environment interactions that affect evolutionary adaptation, as well as provide new targets for the prediction and treatment of complex human craniofacial disorders.
Figure 3. Induced versus evolutionary mutant models.
(a) Chemically induced zebrafish craniofacial mutants (wild-type configuration is to the left). Shaded shapes represent different pharyngeal cartilage elements depicted in the lateral view with the rostral-caudal axis running from left to right. Induced laboratory mutants have been useful for deducing the factors that pattern the embryonic craniofacial skeleton, including (left to right following the wild-type configuration) low/tfap2a, fla/pold1, bab/rerea, and her [26]. Note differences in the number, size, and orientation of elements. Gray elements depict the skull cartilages, green is the lower jaw, blue in the upper jaw, red in the hyosympletic cartilage, yellow is the ceratohyal cartilage, and purple elements are the posterior gill cartilages. Reproduced with permission of the Company of Biologists. (b) A series of evolutionary mutants illustrates the striking difference in craniofacial shape among closely related cichlid species. Here, natural selection has screened random mutations for differences in adult feeding morphology. These evolutionary mutants model both normal and clinical variation in human craniofacial architecture. Cichlid illustrations were drawn by R.C.A.
Reptiles as models of heterotopic ossification
Heterotopic ossification (HO) refers to the formation of bone in soft tissues. In humans this condition is a frequent complication of injuries to the central nervous system, hip surgeries and burns [31], and some people may be genetically predisposed to HO after injury [32]. There are also several rare inherited forms of HO, including fibrodysplasia ossificans progressiva (FOP), progressive osseous heteroplasia (POH), and Albright’s hereditary osteodystrophy [33]. Several recent studies implicate altered BMP (bone morphogenetic protein) signaling as a primary cause of FOP [34, 35], and decreased expression of the alpha subunit of the stimulatory G protein (GNAS) of adenylyl cyclase has been linked to POH [36]. In spite of this progress, the pathophysiology of acquired and inherited forms of HO remains unclear [31]. In HO, the process of bone formation itself appears to be normal, but the temporal and spatial patterns of skeletogenesis are misregulated. Osteoderm development in squamate (scaled) reptiles could provide a model to better understand the onset and progression of these diseases. Osteoderms are secondary dermal ossifications that fortify the integument and serve to protect, camouflage and ornament organisms that possess them. Among tetrapods, osteoderms have evolved independently in reptiles (i.e., gecko lizards), amphibians (i.e., horned frogs) and mammals (i.e., armadillos) [37]. Although the specific mode of osteoderm ossification varies, all osteoderms develop from within connective tissues that possess latent skeletogenic potential [38]. In reptiles, osteoderms form when connective tissues transform into bone without cellular dedifferentiation, osteoblasts or a periosteum, similar to FOP [39]. Whether the extensive variation in osteoderm size, shape and number found among closely related lizard species involves the same genes implicated in human HO pathologies, or as yet undiscovered genes, remains to be seen (Box 1). What is clear is that these evolutionary novelties mimic HO in both anatomy and developmental sequence. In humans, HO is progressive and degenerative and is subject to environmental and genetic factors, whereas in reptiles it is restricted and adaptive.
Box 1. Does natural selection target ‘disease genes’?
The argument that analyses in evolutionary mutant models may lead to a better understanding of the genetic nature of human disease traits relies on the assumption that the same genes are implicated in both human diseases and adaptive phenotypes. Work in several evolutionary mutant models suggests that evolutionary divergence sometimes involve genes previously implicated in human diseases. For example, various populations of blind cavefish (A. mexicanus) have evolved pigment loss by the recurrent fixation of mutations in the coding region of Ocular and Cutaneous Albinism-2 (OCA2), the most commonly mutated gene in cases of human albinism [40]. The evolution of reduced pigmentation has also occurred in different populations of threespine stickleback, and a recent study has shown that cis-regulatory changes in the KIT ligand gene (kitl/KITLG) are responsible for differences in pigmentation in both stickleback and human populations [41]. In domestic dogs, a candidate gene approach was used to search for genes that may be involved in the diversification of skull shapes [42, 43]. One such study focused on the gene that causes Treacher Collins Syndrome (TCS: OMIM 154500), TCOF1 (Treacher Collins-Franceschetti syndrome 1), an autosomal dominant disease that affects craniofacial development and leads to variable facial anomalies, including malformations of the outer ears, cleft palate, and mandibular hypoplasia [44]. In a survey of dog breeds, distinct haplotypes of the canine homolog of TCOF1 were shown to be associated with differences in head shape, particularly in the length and width of the skull [42]. In a similar study, allelic variation in IGF1 (Insulin-like Growth Factor-1), a gene linked to congenital growth defects in humans [45], was shown to be responsible for differences in the size of dogs [46]. Finally, turtle shells form through intramembranous ossification of the dermis [47], and whereas the turtle carapace is distinct from osteoderms, it represents another elaboration of the dermal skeleton through heterotopic bone formation. Similar to patients with FOP, BMP signaling in the intercostal dermis has been implicated in this type of heterotopic ossification [47]. These studies show that genes known to play a role in human diseases are also involved in evolutionary change, which supports the hypothesis that evolutionary mutants are appropriate models for the study of human disease.
The nature of genetic variation in the wild and the tools to study it
The molecular genetic nature of phenotypic variation in evolutionary mutant models differs considerably from that of mutants induced in the laboratory. Laboratory mutagenesis screens use a mutagen to ‘break’ genes, which perturbs normal development and leads to large phenotypic effects that investigators easily identify and sort. In contrast, natural selection and genetic drift seal the fate of novel mutations in the wild, often leading to the accumulation of many alleles with small to moderate effect on phenotypes. To be maintained over evolutionary time, new natural genetic variation must also properly integrate into established developmental and physiological systems. Evolutionary mutant models are therefore more likely than induced mutations to harbor genetic variation that affects the regulation of tissue-specific, post-embryonic, and/or adult phenotypes. Thus, differences between evolutionary mutant models and mutants induced in the lab include (1) the degree of exhibited phenotypic variation, (2) the numbers of genes involved in producing this variation, (3) the magnitude of mutational effects, (4) the parts of the gene affected by mutations, and (5) the age of onset of allelic effects. Tools for the genetic analysis of evolutionary mutant models must be able to accommodate these differences (Box 2).
Box 2. Tools for the analysis of evolutionary mutant models
The genetic analysis of traits in evolutionary mutant models is similar to that used for induced mutations in that both require the recovery of offspring from defined matings, which can best occur in controlled crosses in the laboratory. However, in contrast to genetic mapping of a single induced mutation with high penetrance, complex traits in natural populations are mapped using Quantitative Trait Locus (QTL) analysis. In the former, the correlation between phenotype (mutant vs. wild type) and genetic markers is generally perfect, and the precision of genetic mapping is limited only by the number of available recombination events. In QTL mapping, the association becomes explicitly statistical because many different combinations of factors (genetic and environmental) can lead to similar phenotypes. Moreover, for quantitative traits the expectation of uniform expressivity of mutations across genetic and environmental variation does not hold, which is similar to many human diseases. For example, the rate of diabetes in Pima Indians living in Mexico (7%) is less than one-fifth that found in genotypically similar Pima Indians living in the United States (38%) [48], a dramatic effect of environment on phenotypic expression of a single genotype. Thus, QTL mapping is not simply a way of mapping traits in the absence of a mutagen, rather it allows for the characterization of gene function within a variable genetic and environmental background.
Despite the strengths of pedigree mapping in evolutionary mutant models, some organisms cannot be readily crossed in the laboratory (e.g., birds, reptiles, and large mammals). Association mapping in natural populations provides an additional way to correlate phenotypic variation with specific genomic regions. This technique relies on the extent of linkage disequilibrium, which should be much smaller in natural populations, with their numerous generations of recombination, than in laboratory crosses [49]. The decreasing costs of massively parallel sequencing, as well as a plethora of new genotyping technologies, is making association and population genomic mapping quite feasible even for non-model systems [50].
Evolutionary mutant models can often involve differences in gene expression (as opposed to complete loss-of-function mutations identified in the lab), especially those mutations that affect regulatory regions of specific genes. Therefore, in addition to the genetic mapping of causal alleles, identification of variation in levels of transcription or translation through mRNA or protein abundance, respectively, can provide hints pointing to evolving pathways or be used as confirmatory evidence for one of a set of candidates in a QTL region. In systems where QTL mapping is not possible, expression analysis may also be used to identify candidate genes and pathways for evolved phenotypic differences. Methods to detect variation in transcription can be performed at the level of individual genes (e.g., qPCR), or on a genome-wide scale [51]. A particularly useful synthesis of technologies combines whole genome transcriptional profiling with genetic mapping to identify expression QTL (eQTL) [52].
Once candidate genes for the phenotypic trait in the evolutionary mutant model have been mapped using laboratory crosses or association analysis and verified by differences in expression, one ultimately wants to identify the particular nucleotide changes responsible for observed evolved phenotypic change. If sequencing rules out causative genetic change in the coding regions of evolutionary mutant models, thereby implicating regulatory elements, then a potentially fruitful approach is to search for Conserved NonCoding (CNC) sequences near the identified gene of interest through multiple alignment analysis [53]. Definitive proof of genotype-phenotype relationships, however, must come from manipulative studies that show that specific nucleotide substitutions cause observed phenotypic change. This last “proof of principle” step will not be possible in many evolutionary mutant models, but in some cases the same approaches used in laboratory models will be useful for confirming the genetic basis for evolutionary mutant phenotypes, including allelic introgression, gene knockdown, phenocopies, and phenotype rescue experiments.
Choosing evolutionary models: Practical considerations
Although the main consideration for choosing an evolutionary mutant model will be its phenotypic similarity to a given human disease, several practical considerations come into play. First, the most useful evolutionary models will likely be those that have evolved rapidly. Rapid evolution provides the opportunity to make genetic crosses among variant populations to dissect the identity, number, and mode of action of loci affecting complex traits. Reproductive barriers in rapidly evolving systems are often pre-zygotic, which makes it possible to obtain fertile, interspecific hybrids by in vitro fertilization.
Second, the magnitude of phenotypic divergence among closely related species is important because it affects the statistical power for the detection of QTL in line-cross experiments [54]. Among vertebrates, bony fishes are exemplary evolutionary models [55]. No other vertebrate class exhibits comparable levels of phenotypic diversity or rates of speciation, and as described above, investigators can use this diversity to screen a wide spectrum of traits.
A third important criterion is an understanding of the biogeography and phylogeny of an evolutionary model system. Studying organismal design in the context of a well-resolved phylogeny facilitates insights into the patterns and tempo of evolutionary change, which in turn informs an understanding of how phenotypes evolve in response to environmental change. Are evolutionary shifts incremental or has adaptation involved saltatory events? Do distinct traits evolve independently or as units? The answer to these and related questions can help us deduce the underlying genetic complexity of, and linkage between, complex phenotypes. The turtle shell and the bat wing, for example, evolved without obvious evolutionary intermediates, suggesting that the appearance of these morphological novelties was sudden and may have involved relatively simple molecular genetic changes and heterotopic shifts of pre-existing regulatory networks [47,56,57]. In cichlids, analyses of the evolutionary diversification of dentition have revealed a link between tooth size, shape and number such that selection on one results in coordinated changes in the others [26]. Interestingly, genetic changes that control the periodic patterning of the odontogenic program appear to be relatively simple [58,59]. These findings are novel and relevant to craniodental development in general. Phylogenetic reconstructions can also reveal the direction of evolutionary change, and whether different taxa have similar traits due to independent evolutionary events or shared ancestry.
Finally, systems that have repeated independent convergence on a phenotype are particularly useful since they offer the opportunity to study the genetic basis of complex phenotypes in replicate natural experiments, which models the repeated occurrence of complex human diseases in different populations [e.g., cleft palate; 60]. Furthermore, a trait’s genetic architecture and propensity for molecular convergence will provide insights into the complexity of the regulatory networks that underlie its development. For example, although oca2 is implicated in the convergent evolution of albinism in different cavefish populations (see Box 1) [40], multiple genetic factors are likely to be involved in convergent loss of cavefish eyes [61]. The oca2 gene appears to be a “large” target for evolutionary change in pigmentation, and one might predict that the evolution of albinism in other cave-dwelling species (e.g., amphibians, shrimp, spiders) might also be due to mutations in oca2. That a similarly “large” target has not been found for eye loss underscores the anatomical, developmental, and genetic complexity of this organ. In a similar way, the many factors implicated in human cleft palate highlight the complexity of palatal development [reviewed in 62]. Compared to albinism in blind cavefish, genetic analyses of eye loss in different cavefish populations have the potential to reveal a multitude of targets for human degenerative eye disease.
Conclusions
Human diseases lie along a continuum from ‘simple’ to ‘complex’, and the utility of evolutionary mutant models should extend to both ends of this spectrum. For example, whereas adaptive traits that phenocopy simple human disease states may be polygenic (e.g., eye/lens degeneration in humans and cavefish), knowledge of these additional loci can provide insight into the biology and expressivity of the human disease trait, pinpoint candidate genes and pathways that can be examined functionally in more traditional laboratory models, and provide potential targets for therapeutics. Studies of evolutionary mutant models also promise a much better understanding of the combined polygenic and environmental components of many complex disease traits including osteoporosis, heart disease or diabetes. While the majority of examples discussed in this article are of more simple disease models, examples of evolutionary mutant models for complex diseases also exist and include hypertension in giraffes [63], as well as osteopenia and anemia in Antarctic notothenioid fish (discussed above). Examples of evolutionary mutant models for human diseases are few because this line of inquiry is in its infancy and the tools and resources for developmental genetic analysis of organisms in natural populations have only recently become available. Nevertheless, the studies highlighted here attest to the utility of this approach, and with subsequent technological advances we expect that evolutionary mutant models will rapidly become a valuable tool for deciphering the interacting genetic and environmental risk factors of many human diseases.
Acknowledgements
Thanks to S. Bassham, W.J. Copper, B. Calvi and T. Starmer for commenting on early versions of this manuscript, and to the editor and reviewers for their insightful critiques of the ideas and hypotheses presented above. Work on red blood cell formation in Antarctic notothenioid fish is supported by grants OPP-0336932 and ANT-0635470 from the National Science Foundation. Work on osteopenia in Antarctic notothenioids is supported by Award Number R01AG031922 from the National Institute On Aging. W. Cresko's laboratory is supported by grants from NSF (IOS-0642264) and NIH (1R24GM079486). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Science Foundation, the National Institute On Aging, the National Institute of General Medical Sciences, or the National Institutes of Health.
Glossary
- Association mapping
A method for mapping traits without a pedigree by scoring variation segregating within natural populations. Association mapping is based on the concept of linkage, such that individuals that inherit a functional mutation should also inherit alleles at nearby loci. Because population-level genealogies are much deeper than pedigrees and hence have experienced meioses over many generations, association mapping can usually map traits to a finer interval than pedigree mapping.
- Conserved NonCoding (CNC) sequence
A DNA sequence that does not contribute to the mature transcript of a gene but is highly conserved across distantly related taxa. CNCs can contain tens to hundreds of base pairs and many are considered to be putative regulatory regions of gene activity.
- Evolutionary mutant models
An extant assemblage of related organisms in which certain variants (populations/species) express a phenotype that mimics the clinical features of a human disease. The gene pool and genomic organization of evolutionary mutant models result from many generations of natural selection and genetic drift.
- eQTL
A quantitative trait locus that affects gene expression. The search for eQTLs uses transcriptional profiling as a phenotype that can be mapped to assess how segregating allelic variation effects transcriptional regulation.
- Genetic architecture
A set of genetic loci and their interactions that account for an expressed phenotype.
- Linkage disequilibrium
The non-random association of alleles at two or more loci. The rate of recombination, rate of mutation, genetic drift, non-random mating, and population structure can all affect the degree of linkage disequilibrium.
- Quantitative trait loci (QTL) analysis
A search for loci that affect variation of a quantitative trait. It is a statistical analysis of genotype-by-phenotype correlations using pedigrees of known structure, where individuals are genotyped at marker loci spread across the genome, and then the inheritance of each marker is compared to that of a phenotype. A QTL is localized to a particular region of the genome when the segregation of marker loci within that region matches the segregation of phenotypic variation.
References
- 1.Carroll SB, et al. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design, 2nd edition. Malden, MA, USA: Blackwell; 2005. [Google Scholar]
- 2.Donovan A, et al. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature. 2000;403:776–781. doi: 10.1038/35001596. [DOI] [PubMed] [Google Scholar]
- 3.Montosi G, et al. Autosomal-dominant hemochromatosis is associated with a mutation in the ferroportin (SLC11A3) gene. J. Clin. Invest. 2001;108:619–623. doi: 10.1172/JCI13468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamason RL, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 5.Sabherwal N, et al. Long-range conserved non-coding SHOX sequences regulate expression in developing chicken limb and are associated with short stature phenotypes in human patients. Hum. Mol. Genet. 2007;16:210–222. doi: 10.1093/hmg/ddl470. [DOI] [PubMed] [Google Scholar]
- 6.Lauderdale JD, et al. 3' deletions cause aniridia by preventing PAX6 gene expression. Proc. Natl. Acad. Sci. USA. 2000;97:13755–13759. doi: 10.1073/pnas.240398797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 8.Loots GG, et al. Genomic deletion of a long-range bone enhancer misregulates sclerostin in Van Buchem disease. Genome Res. 2005;15:928–935. doi: 10.1101/gr.3437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Velagaleti GV, et al. Position effects due to chromosome breakpoints that map approximately 900 Kb upstream and approximately 1.3 Mb downstream of SOX9 in two patients with campomelic dysplasia. Am. J. Hum. Genet. 2005;76:652–662. doi: 10.1086/429252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kikuta H, et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 2007;17:545–555. doi: 10.1101/gr.6086307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olds LC, Sibley E. Lactase persistence DNA variant enhances lactase promoter activity in vitro: functional role as a cis regulatory element. Hum. Mol. Genet. 2003;12:2333–2340. doi: 10.1093/hmg/ddg244. [DOI] [PubMed] [Google Scholar]
- 12.Hubank M, Schatz DG. cDNA representational difference analysis: a sensitive and flexible method for identification of differentially expressed genes. Methods Enzymol. 1999;303:325–349. doi: 10.1016/s0076-6879(99)03021-9. [DOI] [PubMed] [Google Scholar]
- 13.Detrich HW, Yergeau DA. Comparative genomics in erythropoietic gene discovery: synergisms between the Antarctic icefishes and the zebrafish. In: Detrich HW, Westerfield M, Zon LI, editors. Methods in Cell Biology, The Zebrafish, 2nd edition: Genetics, Genomics, and Informatics. Vol. 77. Academic Press; 2004. pp. 475–503. [DOI] [PubMed] [Google Scholar]
- 14.Yergeau DA, et al. bloodthirsty, an RBCC/TRIM gene required for erythropoiesis in zebrafish. Dev. Biol. 2005;283:97–112. doi: 10.1016/j.ydbio.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 15.NIH Consensus Development Panel on Osteoporosis Prevention. Osteoporosis prevention, diagnosis, and therapy. Jama. 2001;285:785–795. doi: 10.1001/jama.285.6.785. [DOI] [PubMed] [Google Scholar]
- 16.National Osteoporosis Foundation. About Osteoporosis. 2008 www.nof.org.
- 17.Eastman JT. Antarctic Fish Biology: Evolution in a Unique Environment. Academic Press; 1993. [Google Scholar]
- 18.Jeffery WR. Adaptive evolution of eye degeneration in the Mexican blind cavefish. J. Hered. 2005;96:185–196. doi: 10.1093/jhered/esi028. [DOI] [PubMed] [Google Scholar]
- 19.Protas ME, et al. Regressive evolution in the Mexican cave tetra, Astyanax mexicanus. Curr. Biol. 2007;17:452–454. doi: 10.1016/j.cub.2007.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitzpatrick DR, van Heyningen V. Developmental eye disorders. Curr. Opin. Genet. Dev. 2005;15:348–353. doi: 10.1016/j.gde.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 21.Schmahl W, et al. Defects of neuronal migration and the pathogenesis of cortical malformations are associated with Small eye (Sey) in the mouse, a point mutation at the Pax-6 locus. Acta. Neuropathol. 1993;86:126–135. doi: 10.1007/BF00334879. [DOI] [PubMed] [Google Scholar]
- 22.Strickler AG, et al. Early and late changes in Pax6 expression accompany eye degeneration during cavefish development. Dev. Genes Evol. 2001;211:138–144. doi: 10.1007/s004270000123. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto Y, et al. Hedgehog signaling controls eye degeneration in blind cavefish. Nature. 2004;431:844–847. doi: 10.1038/nature02864. [DOI] [PubMed] [Google Scholar]
- 24.Wilkens H. Evolution and genetics of epigean and cave Astyanax fasciatus (Characidae, Pisces) Evol. Biol. 1988;23:271–367. [Google Scholar]
- 25.Jeffery WR. Cavefish as a model system in evolutionary developmental biology. Dev. Biol. 2001;231:1–12. doi: 10.1006/dbio.2000.0121. [DOI] [PubMed] [Google Scholar]
- 26.Schilling TF, et al. Jaw and branchial arch mutants in zebrafish I: branchial arches. Development. 1996;123:329–344. doi: 10.1242/dev.123.1.329. [DOI] [PubMed] [Google Scholar]
- 27.Streelman JT, Albertson RC. Evolution of Novelty in the Cichlid Dentition. J. Exp. Zool. B: Mol. Dev. Evol. 2006;306B:216–226. doi: 10.1002/jez.b.21101. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi S, Hori M. Coexistence of competing species by the oscillation of polymorphisms. J. Theor. Biol. 2005;235:591–596. doi: 10.1016/j.jtbi.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Gougoutas AJ. Hemifacial microsomia: clinical features and pictographic representations of the OMENS classification system. Plast. Reconstr. Surg. 2007;120:112e–120e. doi: 10.1097/01.prs.0000287383.35963.5e. [DOI] [PubMed] [Google Scholar]
- 30.Albertson RC, Yelick PC. Roles for fgf8 signaling in left-right patterning of the visceral organs and craniofacial skeleton. Dev. Biol. 2005;283:310–321. doi: 10.1016/j.ydbio.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 31.Vanden Bossche L, Vanderstraeten G. Heterotopic ossification: a review. J. Rehabil. Med. 2005;37:129–136. doi: 10.1080/16501970510027628. [DOI] [PubMed] [Google Scholar]
- 32.Kluger G, et al. Heterotopic ossification in childhood and adolescence. J. Child. Neurol. 2001;15:406–413. doi: 10.1177/088307380001500610. [DOI] [PubMed] [Google Scholar]
- 33.Job-Deslandre C. Inherited ossifying diseases. Joint Bone Spine. 2004;71:98–101. doi: 10.1016/s1297-319x(03)00130-1. [DOI] [PubMed] [Google Scholar]
- 34.Kan L, et al. Transgenic mice overexpressing BMP4 develop a fibrodysplasia ossificans progressiva (FOP)-like phenotype. Am. J. Pathol. 2004;165:1107–1115. doi: 10.1016/S0002-9440(10)63372-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serrano de la Peña L, et al. Fibrodysplasia ossificans progressiva (FOP), a disorder of ectopic osteogenesis, misregulates cell surface expression and trafficking of BMPRIA. J. Bone Miner. Res. 2005;20:1168–1176. doi: 10.1359/JBMR.050305. [DOI] [PubMed] [Google Scholar]
- 36.Shore EM, et al. Paternally inherited inactivating mutations of the GNAS1 gene in progressive osseous heteroplasia. N. Engl. J. Med. 2002;346:99–106. doi: 10.1056/NEJMoa011262. [DOI] [PubMed] [Google Scholar]
- 37.Hill RV. Integration of morphological data sets for phylogenetic analysis of Amniota: the importance of integumentary characters and increased taxonomic sampling. Syst. Biol. 2005;54:530–547. doi: 10.1080/10635150590950326. [DOI] [PubMed] [Google Scholar]
- 38.Vickaryous MK, Hall BK. Osteoderm morphology and development in the nine-banded armadillo, Dasypus novemcinctus (Mammalia, Xenarthra, Cingulata) J. Morphol. 2006;267:1273–1283. doi: 10.1002/jmor.10475. [DOI] [PubMed] [Google Scholar]
- 39.Vickaryous MK, Hall BK. Development of the dermal skeleton in Alligator mississippiensis (Archosauria, Crocodylia) with comments on the homology of osteoderms. J. Morphol. 2007;269:398–422. doi: 10.1002/jmor.10575. [DOI] [PubMed] [Google Scholar]
- 40.Protas ME, et al. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat. Genet. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- 41.Miller CT, et al. cis-Regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2008;131:1179–1189. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haworth KE, et al. Canine TCOF1; cloning, chromosome assignment and genetic analysis in dogs with different head types. Mamm. Genome. 2001;12:622–629. doi: 10.1007/s00335-001-3011-0. [DOI] [PubMed] [Google Scholar]
- 43.Fondon JW, 3rd, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc. Natl. Acad. Sci. USA. 2004;101:18058–18063. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dixon MJ. Treacher Collins syndrome. Hum Mol Genet. 1996;5:1391–1396. doi: 10.1093/hmg/5.supplement_1.1391. [DOI] [PubMed] [Google Scholar]
- 45.Woods KA, et al. Insulin-like growth factor I gene deletion causing intrauterine growth retardation and severe short stature. Acta. Paediatr. Suppl. 1997;423:39–45. doi: 10.1111/j.1651-2227.1997.tb18367.x. [DOI] [PubMed] [Google Scholar]
- 46.Sutter NB, et al. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cebra-Thomas J, et al. How the turtle forms its shell: a paracrine hypothesis of carapace formation. J. Exp. Zoolog. B Mol. Dev. Evol. 2005;304:558–569. doi: 10.1002/jez.b.21059. [DOI] [PubMed] [Google Scholar]
- 48.Schulz LO, et al. Effects of traditional and western environments on prevalence of type 2 diabetes in Pima Indians in Mexico and the U.S. Diabetes Care. 2006;29:1866–1871. doi: 10.2337/dc06-0138. [DOI] [PubMed] [Google Scholar]
- 49.Luikart G, et al. The power and promise of population genomics: from genotyping to genome typing. Nat. Rev. Genet. 2003;4:981–994. doi: 10.1038/nrg1226. [DOI] [PubMed] [Google Scholar]
- 50.Kruglyak L. The road to genome-wide association studies. Nat. Rev. Genet. 2008;9:314–318. doi: 10.1038/nrg2316. [DOI] [PubMed] [Google Scholar]
- 51.Torres TT, et al. Gene expression profiling by massively parallel sequencing. Genome Res. 2008;18:172–177. doi: 10.1101/gr.6984908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gibson G, Weir B. The quantitative genetics of transcription. Trends Genet. 2005;21:616–623. doi: 10.1016/j.tig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 53.Hardison RC. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 2000;16:369–372. doi: 10.1016/s0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- 54.Lynch M, Walsh B. Genetics and analysis of quantitative traits. Sinauer; 1998. [Google Scholar]
- 55.Streelman JT, et al. Developmental Genetics of Adaptation in Fishes: The Case for Novelty. Ann. Rev. Ecol. Evol. Syst. 2007;38:655–681. [Google Scholar]
- 56.Sears KE, et al. Development of bat flight: morphologic and molecular evolution of bat wing digits. Proc. Natl. Acad. Sci. USA. 2006;103:6581–6586. doi: 10.1073/pnas.0509716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weatherbee SD, et al. Interdigital webbing retention in bat wings illustrates genetic changes underlying amniote limb diversification. Proc. Natl. Acad. Sci. USA. 2006;103:15103–15107. doi: 10.1073/pnas.0604934103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Albertson RC, et al. Directional selection has shaped the oral jaws of Lake Malawi cichlid fishes. Proc. Natl. Acad. Sci. USA. 2003;100:5252–5257. doi: 10.1073/pnas.0930235100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fraser GJ, et al. A periodic pattern generator for dental diversity. BMC Biol. 2008;6:32. doi: 10.1186/1741-7007-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jugessur A, Murray JC. Orofacial clefting: recent insights into a complex trait. Curr. Opin. Genet. Dev. 2005;15:270–278. doi: 10.1016/j.gde.2005.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bororsky R. Restoring sight in blind cavefish. Curr Biol. 2008;18:R23–R24. doi: 10.1016/j.cub.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 62.Murray JC, Schutte BC. Cleft palate: players, pathways, and pursuits. J. Clin. Invest. 2004;113:1676–1678. doi: 10.1172/JCI22154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paton JF, et al. Harvey Cushing and the regulation of blood pressure in giraffe, rats and man: introducing Cushing's mechanism. Exp Physiol. 2008 Sep 26; doi: 10.1113/expphysiol.2008.043455. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]