Extracellular stimuli can induce localized actin rearrangements at the site of stimulation. To understand how this occurs, we have been studying enteropathogenic Escherichia coli (EPEC), a bacterial pathogen that induces formation of an actin-rich membrane pseudopod or pedestal beneath itself upon adherence to host intestinal epithelia1. Infection ultimately results in diarrhoea, which can cause death, especially among infants in developing countries1. Here we show that pedestal formation depends on localized recruitment and activation of two host-cell factors involved in actin polymerization: the heptameric Arp2/3 complex (Arp2/3c), which nucleates polymerization2, and members of the Wiskott–Aldrich syndrome (WAS) family of proteins (WASP and N-WASP)3, which bind to and activate Arp2/3c (ref. 2). Arp2/3c recruitment depends on WASP, and WASP recruitment depends on its GTPase-binding domain (GBD), suggesting involvement proximally of a Rho family GTPase. This is, to our knowledge, the first demonstration of cellular mediators of EPEC pedestal formation and of localized recruitment of WASP and Arp2/3c as part of a signalling cascade initiated at the cell surface.
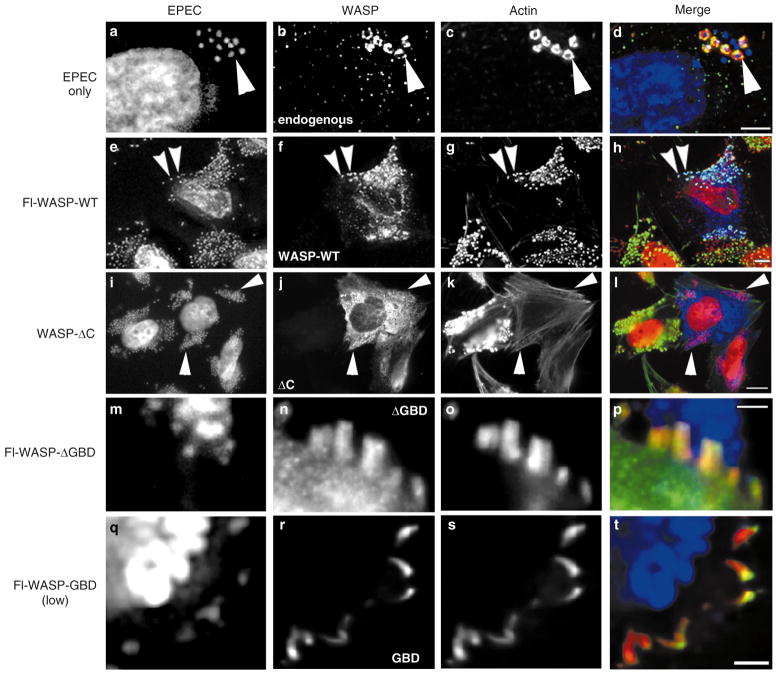
Exposure of HeLa cells to wild-type EPEC resulted in the formation of an actin-rich pedestal underneath the bacterium (see Supplementary Information). To determine whether endogenous WAS family proteins localized to the pedestals, HeLa cells were exposed to EPEC and then stained with a polyclonal antiserum that recognized WASP and N-WASP. At low magnification, pedestals are seen as punctate actin staining (Fig. 1c), directly apposed to the bacterium (Fig. 1a). The endogenous WASP-like protein (Fig. 1b) was enriched in the pedestal relative to the cell body.
Figure 1.
WASP is recruited through its GTPase-binding domain (GBD) to EPEC pedestals. a–d, Images of HeLa cells exposed to EPEC. Cells were stained with DAPI, to identify EPEC (a), anti-WASP polyclonal Ab (b) or rhodamine–phalloidin, to visualize actin (c). Arrowheads denote endogenous WASP-like protein localized in pedestals. In the merged images d, p and t, EPEC are pseudocoloured blue, WASP green, and actin red. In h and l, EPEC are pseudocoloured red, WASP blue, and actin green. e–h, Images of cells expressing Flag (Fl)-WASP-WT and exposed to EPEC. In this panel and in i–t, cells were stained with DAPI (e, i, m, q), anti-Flag mAb (f, j, n, r), and rhodamine–phalloidin (g, k, o, s). Arrowheads denote Fl-WASP-WT protein localized in pedestals. i–l, Images of cells expressing high levels of WASP-ΔC. Note that EPEC on expressing cells have no actin pedestals associated with them (arrowheads). m–p, Images of cell expressing Fl-WASP-ΔGBD. Note that WASP-ΔGBD was in pedestals but was not enriched relative to the cytoplasm. q–t, Images of cell expressing low levels of Fl-WASP-GBD. Note that the GBD protein localizes to pedestals. Scale bar represents 4 μm in d, 5 μm in h, 10 μm in l, and 2 μm in p, t.
We next asked whether exogenous WAS family proteins affected pedestal formation or localized to pedestals. Flag-tagged wild-type WASP (WASP-WT) was expressed in HeLa cells, and the cells were exposed to EPEC. The EPEC are shown in Fig. 1e and the WASP-WT, detected by staining with the Flag antibody, in Fig. 1f. WASP-WT had no effect on pedestal formation, as measured by actin staining (Fig. 1g). Like the endogenous WAS-like protein, WASP-WT concentrated in pedestals (Fig. 1e–h, arrowheads). Haemagglutinin (HA)-tagged N-WASP behaved identically to WASP-WT (data not shown). Localization of transfected WASP-WT to pedestals was specific: green fluorescent protein (GFP) fluorescence was not enriched in pedestals relative to the cell body (data not shown).
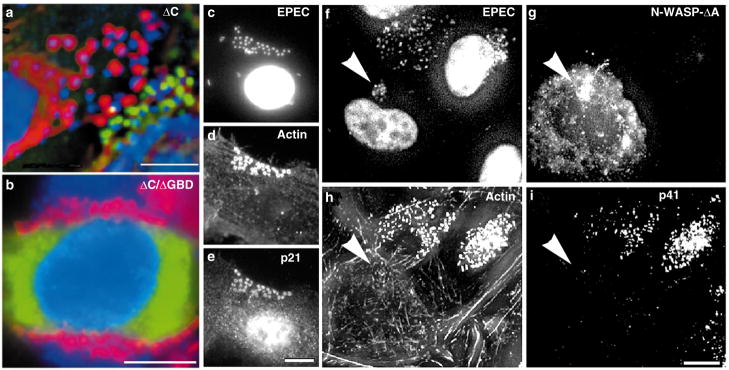
We determined whether WASP was required for pedestal formation by expressing various domains of WASP or N-WASP. Deletion of the WASP carboxy terminus (ΔC) results in pronounced defects in the capacity of the protein to polymerize actin4. Expression of WASP-ΔC blocked pedestal formation in a dominant-negative fashion (Fig. 1i–l). Pedestals, as measured by actin staining (Fig. 1k), were not evident beneath attached bacteria (Fig. 1i arrowheads) in cells expressing WASP-ΔC (Fig. 1j). Blockade occurred even in cells where the construct was expressed at low levels (Fig. 2a cell on right). In such cells WASP-ΔC protein (green, Fig. 2a) localized directly adjacent to the bacteria (blue, Fig. 2a), but no pedestals (red) were evident (compare the cell on the right with the non-expressing cell on the left in Fig. 2a). The C terminus by itself neither localized nor blocked pedestal formation (data not shown).
Figure 2.
Pedestal formation and localization of Arp2/3c to pedestals require the WASP acidic domain. a, Image of HeLa cell expressing low levels of WASP-ΔC and exposed to EPEC. Cells were stained with DAPI, anti-Flag, and rhodamine–phalloidin. In this merged image and in b, EPEC are pseudocoloured blue, actin red and WASP green. The cell on the right expresses low levels of WASP-ΔC protein which localizes next to the bacteria (blue) and blocks pedestal formation. The cell on the left did not express the protein and develops pedestals (red). b, Image of cell expressing Fl-WASP-ΔC/ΔGBD and exposed to EPEC. Cells were stained and the image pseudocoloured as in a. In the absence of C and GBD, pedestals formed and the protein was not localized exclusively within them. Thus, the GBD is required for WASP-ΔC to act as a dominant negative. Unmerged images are provided in Supplementary Information. c–e, Images of HeLa cell stained for Arp2/3c subunit p21 and exposed to EPEC. The cell was stained with DAPI (c), anti-actin mAb and Cy-5-conjugated secondary antibody (d), and anti-p21 antiserum and FITC secondary antibody (e). Note the recruitment of p21 to pedestals. f–i, Images of cell expressing N-WASP-ΔA and exposed to EPEC. The cell was stained with DAPI (f), anti-HA mAb and FITC secondary antibody, to recognize the N-WASP-ΔA (g), rhodamine–phalloidin (h), and anti-p41 antibody and Cy-5 secondary antibody (i). Note that p41 fails to accumulate beneath EPEC in the cell expressing N-WASP-ΔA, and that the cell fails to make pedestals. Arrowheads denote the lack of p41 accumulation in the cell expressing N-WASP-ΔA. All scale bars represent 10 μm.
The WASP C terminus contains a WH2 domain, a cofilin domain and an acidic (A) domain (see Supplementary Information). N-WASP alleles with mutations in the cofilin domain can act in a dominant-negative fashion5. However, we could detect no effect of N-WASP-Δcofilin on pedestal formation or localization, even when the protein was expressed at high levels. In contrast, WASP protein lacking the WH2 domain continued to localize beneath the bacteria but blocked pedestal formation only when expressed at high levels (data not shown). The WASP WH2 domain expressed independently neither blocked nor localized. N-WASP-ΔA and N-WASP-ΔA/Δcofilin both blocked when expressed at low levels and were indistinguishable from WASP-ΔC (Fig. 2f–h).
The observation that N-WASP-ΔA blocked pedestal formation led us to consider whether Arp2/3c, which binds to the acidic domain of WASP2, localizes to pedestals. Subunits p21 (Fig. 2c–e) and p40 (data not shown) of the endogenous Arp2/3c, as well as transfected Arp3-GFP (data not shown), were recruited to the pedestals. As noted above, WASP-ΔC and N-WASP-ΔA blocked pedestal formation but continued to localize beneath EPEC (Fig. 2a for WASP-ΔC; Fig. 2g (arrowheads) for N-WASP-ΔA). WASP-ΔC or N-WASP-ΔA also prevented localization of p41 (Fig. 2f–i, arrowheads), and of p40 and GFP-Arp3 (data not shown), beneath EPEC. We conclude that recruitment of Arp2/3c depends on the WASP acidic domain and that both a WASP-like protein and Arp2/3c are required for pedestal formation. Our in vivo results complement in vitro studies which show that the C terminus of WASP associates with and potentiates the nucleating activity of Arp2/3c (ref. 2). Whereas the acidic domain directly binds the Arp2/3c subunit p21, both the acidic and WH2 domains are required for activation.
To determine which domains of WASP were required for localization in the pedestal, we expressed WASP proteins containing deletions that disrupt domains outside the C terminus. Deletion of the GBD prevented WASP from localizing specifically in the pedestals (Fig. 1m–p). Quantitation of fluorescence intensity in the pedestal and cell body showed that although WASP-ΔGBD was present in the pedestals, it was not enriched relative to the cell body. The effect was specific: WASP proteins with deletions in the amino terminus encompassing the pleckstrin-homology (PH) and WASP-homology 1 (WH1) domains, or in the polyproline (PP) or N-WASP-Δcofilin domains all localized in a manner comparable to WASP-WT. Thus, the GBD is necessary for recruitment of WASP to pedestals.
When expressed alone at low levels, the WASP GBD domain localized to the pedestals (Fig. 1q–t). When expressed at high levels, WASP GBD blocked pedestal formation but continued to localize beneath the bacterium (data not shown). We surmise that when highly expressed, the GBD can competitively inhibit binding of an endogenous WASP-like protein, but that this domain alone is not as effective a competitor as WASP-ΔC, possibly because of the effects of additional domains in WASP. The effect of expressing this domain alone was specific. Expression of PH/WH1 domain or PP domain was without effect on pedestal formation and neither protein localized. Moreover, a GBD from the kinase PAK3, which shares 70% amino-acid homology with the WASP GBD, neither blocked pedestal formation nor localized to pedestals. Thus WASP GBD was necessary and sufficient for localization to pedestals. However, we cannot rule out the possibility that additional domains contribute to localization.
We next determined whether WASP activity was dependent on localization of WASP to pedestals. To do this we assessed whether the dominant-negative effects of the WASP-ΔC required an intact GBD. Expression of mutant WASP with deletions of both the C terminus and the GBD (ΔC/ΔGBD) neither blocked pedestal formation nor localized beneath the bacterium (Fig. 2b). Thus the GBD is required for dominant-negative effects, and localization of WASP through the GBD is required to recruit Arp2/3c and for pedestal formation.
The requirement for WASP GBD suggested that, in pedestal formation, a GTPase both recruits and activates WASP. The Cdc42 GTPase interacts with the WASP GBD5, but pedestal formation is insensitive to the effects of Clostridium dificile toxin B (ToxB)6, which inactivates Rho family members, including Cdc42. We provide evidence elsewhere that a novel Cdc42 homologue, called Chp7, is ToxB-insensitive and may mediate EPEC signalling to WASP (D.K. et al., unpublished observations).
Recent evidence has also implicated Arp2/3c, WASP and Rho family GTPases in spatial control of actin polymerization. First, Arp2/3c redistributes to the up-gradient surface of the plasma membrane in neutrophils responding to chemoattractant8. Second, artificial targeting of WASP or Cdc42 to a localized site on the plasma membrane is sufficient to generate actin-rich filopodia-like structures at that site9. Accordingly, our data suggest that EPEC triggers localized recruitment and activation of WASP and Arp2/3c in vivo, leading to localized actin rearrangements.
In summary, we present a model for EPEC pedestal formation (see Supplementary Information). EPEC adheres to the outside of the host cell and inserts the virulence factor Tir into the host plasma membrane using a type III secretion system1. Tir then binds the EPEC surface protein intimin, and directly or indirectly recruits and activates a host-cell GTPase of the Chp subfamily. The Chp-like GTPase recruits WASP by binding the GBD, thereby exposing the WH2 and acidic domains in the WASP C terminus. The WASP C terminus in turn recruits and activates Arp2/3c, thereby stimulating actin nucleation and polymerization. Actin polymerization drives membrane protrusion and pedestal formation.
According to this model, WASP-ΔC or N-WASP-ΔA act in a dominant-negative fashion because they compete efficiently with an endogenous WASP-like protein for recruitment, but neither mutant activates Arp2/3c. Notably, the C terminus of WASP blocks activation of Arp2/3c in other systems10 but does not block EPEC pedestal formation when expressed at similar levels to WASP-ΔC. We suggest that this difference in potency results because WASP-ΔC competes for a limited number of EPEC-generated recruitment sites, whereas the C terminus must titrate the relatively abundant Arp2/3c. Indeed, this capacity makes WASP-ΔC a generally useful probe of WASP function.
In conclusion, WASP and Arp2/3c are the first identified and among the most distal mediators of a signalling cascade initiated at the cell surface by EPEC and culminating in actin polymerization and pedestal formation. An eventual understanding of how EPEC interfaces with the mammalian cellular signalling machinery will provide insight into the molecules that link physiological receptors at the cell surface with reorganization of the actin cytoskeleton.
Methods
HeLa cells were grown on glass coverslips in Dulbecco’s Modified Eagles medium (DMEM) supplemented with 10% fetal calf serum and incubated for 6–8 h at 37 °C with WT EPEC at a multiplicity of infection of 10. For some experiments cells were transfected with plasmid vectors using calcium phosphate precipitation or Fugene-6 (Boehringer) 3 days before infection. Constructs are described in Supplementary Information. Cells were processed for immunocytochemistry as described previously8. EPEC was recognized by staining with 4′6-diamidino-2-phenylindole (DAPI; 1 μg ml−1; Sigma). Before staining, some polyclonal rabbit antisera were incubated for 20 min with EPEC previously fixed in formaldehyde, and then centrifuged. This procedure removed serum contaminants that nonspecifically bound EPEC. The primary antibodies and concentrations used in this study were as follows: 9E10 monoclonal antibody (mAb) (ascites, 1:200 dilution), anti-Flag mAb (0.1 μg ml−1, Sigma), anti-WASP polyclonal antibody (affinity purified, 1:200 dilution), and anti-HA mAb (3F10; 0.1μg ml−1, Boehringer; 1:500 dilution). Secondary antibodies were obtained from Jackson Immunochemicals. Images were acquired with a scientific-grade cooled charge-coupled device on a multi-wavelength wide-field three-dimensional microscopy system8. Samples were imaged in successive 0.25-μm focal planes, and out-of-focus light was removed with a constrained iterative deconvolution algorithm8.
Supplementary Material
Acknowledgments
The authors thank A. Mallavarapu and J. Taunton for helpful discussions, S. Keshavarzi for assistance, M. Welch for Arp2/3c antibodies, H. Miki for N-WASP cDNA, and H. Bourne, A. Johnson and J. Engel for commenting on the manuscript. The work was supported by the H.H.M.I. (O.D.W and B.B.F.), the N.S.E.R.C., Canada (D.L.G), and grants from the M.R.C., Canada (B.B.F), from the N.I.H. (J.W.S. and A.A.), and from the N.C.I. (J.M.B.).
Footnotes
Supplementary information is available on Nature Cell Biology’s World-Wide Web site (http://cellbio.nature.com) or as paper copy from the London editorial office of Nature Cell Biology.
References
- 1.Goosney DL, de Grado M, Finlay B. Trends Cell Biol. 1999;9:1–4. doi: 10.1016/s0962-8924(98)01418-4. [DOI] [PubMed] [Google Scholar]
- 2.Machesky LM, Insall RH. J Cell Biol. 1999;146:267–292. doi: 10.1083/jcb.146.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abo A. Cell Mol Life Sci. 1998;54:1145–1153. doi: 10.1007/s000180050242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Symons M, et al. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- 5.Miki H, Sasaki T, Takai Y, Takenawa T. Nature. 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Ami G, et al. Infect Immun. 1998;66:1755–1758. doi: 10.1128/iai.66.4.1755-1758.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aronheim A, et al. Curr Biol. 1998;8:1125–1128. doi: 10.1016/s0960-9822(98)70468-3. [DOI] [PubMed] [Google Scholar]
- 8.Weiner OD, et al. Nature Cell Biol. 1999;1:75–81. doi: 10.1038/10042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castellano F, et al. Curr Biol. 1999;9:351–360. doi: 10.1016/s0960-9822(99)80161-4. [DOI] [PubMed] [Google Scholar]
- 10.Machesky L, Insall RH. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.