Abstract
Microorganisms can switch from a planktonic, free-swimming life style to a sessile, colonial state, called a biofilm, conferring resistance to environmental stress. Conversion between the motile and biofilm life style has been attributed to increased levels of the prokaryotic second messenger cyclic di-guanosine monophosphate (c-di-GMP), yet the signaling mechanisms mediating such a global switch are poorly understood. Here we show that the transcriptional regulator VpsT from Vibrio cholerae directly senses c-di-GMP to inversely control extracellular matrix production and motility, identifying VpsT as a master regulator for biofilm formation. Rather than being regulated by phosphorylation, VpsT undergoes a change in oligomerization upon c-di-GMP binding.
In V. cholerae, biofilm formation is facilitated by colonial morphotype variation (1–4). Rugose variants produce increased levels of extracellular matrix via the expression of Vibrio polysaccharide (vps) genes and genes encoding matrix proteins. vps expression is under the control of two positive transcriptional regulators, VpsT and VpsR (5, 6). VpsT is a member of the FixJ/LuxR/CsgD family of prokaryotic response regulators, typically effectors in two-component signal transduction systems that use phosphoryl transfer from upstream kinases to modulate response regulator protein activity (7–9). Although the putative phosphorylation site is conserved in VpsT’s receiver domain, other residues crucial for phosphotransfer-dependent signaling are not and no cognate kinase has been identified to date (Fig. S1). Regulation by VpsT and VpsR has been linked to signal transduction utilizing the bacterial second messenger c-di-GMP (10, 11) (Fig. S2), yet little is known about the direct targets of the nucleotide. A riboswitch has been identified as a c-di-GMP-target regulating gene expression of a small number of genes, but is unlikely to account for the global change in transcriptional profile required for biofilm formation (12). Neither do PilZ domain-containing proteins, potential c-di-GMP effectors, affect rugosity since a V. cholerae strain lacking all five PilZ domain-containing proteins retains its colony morphology and ability to overproduce vps gene products (13).
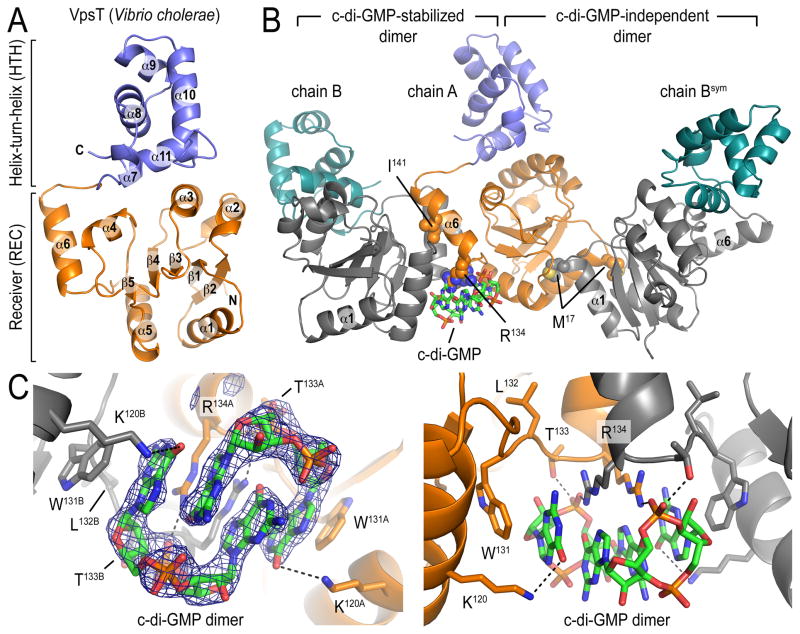
VpsT consists of an N-terminal receiver (REC) and a C-terminal helix-turn-helix (HTH) domain, with the latter mediating DNA binding (Fig. 1A; see Supplemental Information for details). Unlike other REC domains, the canonical (α/β)5-fold in VpsT is extended by an additional helix at its C-terminus (Fig. 1A; helix α6). The HTH domain buttresses against an interface formed by helices 3 and 4 of the N-terminal regulatory domain. There are two non-overlapping dimerization interfaces between non-crystallographic VpsT protomers (chain A-chain B and chain A-chain Bsym; Fig. 1B). The c-di-GMP-independent interface involves interactions mediated by a methionine residue (M17) located at the beginning of α1 and a binding pocket that extends into the putative phosphorylation site of the REC domain (Fig. S3A). The second interface involves α6 of the REC domain, in contrast to canonical response regulators such as CheY and PhoB that utilize a surface formed by α4-β5-α5 for dimerization (9). The binding of two intercalated c-di-GMP molecules to the base of α6 stabilizes VpsT dimers utilizing this interface (Fig. 1 and S3B).
Figure 1. Crystal structure of VpsT.
(A) Structure of a VpsT protomer. (B) Structure of a crystallographic trimer representing two potentially relevant, non-overlapping dimerization interfaces. Cyclic di-GMP molecules are shown as sticks, key residues mediating ligand binding and interprotomer interactions are shown as spheres. (C) Closeup view of the nucleotide binding pocket with residues involved in coordinating the ligand shown as sticks. A (|Fo|-|Fc|) electron density map contoured at 3.6σ is shown as calculated from a model prior to inclusion of c-di-GMP.
The binding motif for c-di-GMP in VpsT consists of a 4-residue-long, conserved W[F/L/M][T/S]R sequence (Fig. S1). The side chains of the tryptophan and arginine form π-stacking interactions with the purine rings of the nucleotide (Fig. 1C). While the hydrophobic residue in the second position plays a structural role being buried in the REC domain, the threonine residue at position 3 forms a hydrogen bond with the phosphate moiety of c-di-GMP. A subclass of VpsT/CsgD homologs exists with a proline substitution in position 3 (W[F/L/M]PR). Although CsgD is also functionally linked to c-di-GMP signaling in E. coli and Salmonella (14, 15), its binding pocket appears to be distinct from that of VpsT since it displays a highly conserved YF[T/S]Q motif that is unlikely to accommodate c-di-GMP (Fig. S3B).
The apparent affinity of VpsT for c-di-GMP, determined by isothermal titration calorimetry, is 3.2 μM with 1:1 stoichiometry, consistent with a dimer of c-di-GMP binding to a dimer of VpsT (Fig. S4A). Single-point mutations in the conserved c-di-GMP binding motif (VpsTR134A, VpsTW131F or VpsTT133V) or in the isoleucine in α6 of the c-di-GMP-stabilized REC dimerization interface (VpsTI141E) abolished c-di-GMP binding, indicating that dimeric REC domains are required for binding (Fig. S4B). Conversely, mutation of a key residue in the nucleotide-independent interface (VpsTM17D) had no effect on c-di-GMP binding. Based on static multi-angle light scattering, VpsTM17D exists as a monomeric species in the absence of c-di-GMP, whereas intermediate molecular weights for the wild-type VpsT and the mutants VpsTR134A and VpsTI141E indicated fast exchange between monomers and dimers, presumably through the c-di-GMP-independent interface (Fig. S5 and Table S2). Addition of c-di-GMP increases the molecular weight of VpsTM17D and wild-type VpsT (Fig. S5 and S6) while the oligomeric state of VpsTR134A and VpsTI141E is insensitive to the nucleotide.
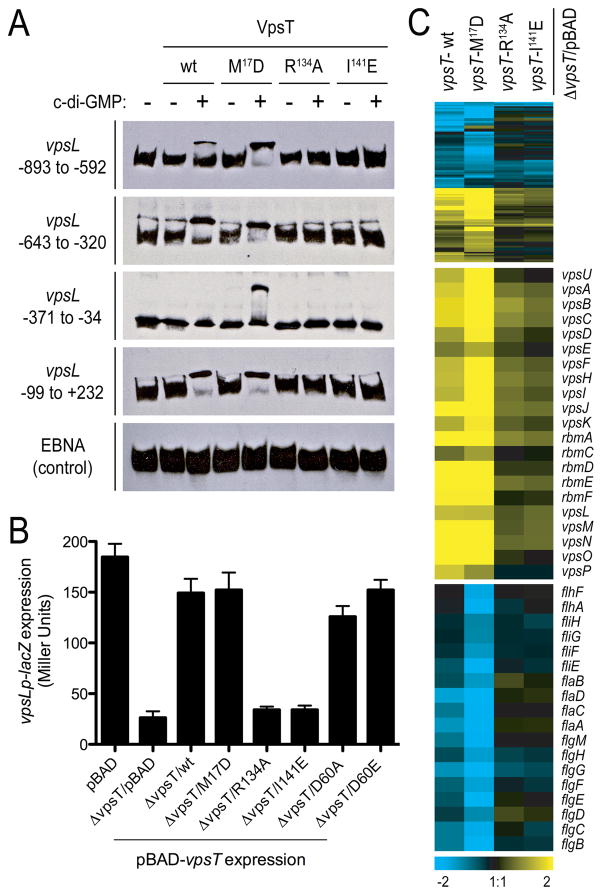
The role of c-di-GMP recognition and the relevance of the two dimer interfaces in DNA-binding and VpsT-regulated gene expression was assessed by using c-di-GMP binding (R134) and dimerization (I141 or M17) mutants (Fig. 2). In electromobility shift assays we used regulatory sequences upstream of vpsL, a gene under positive control of VpsT (Fig. 2A) (6). DNA mobility shifts were observed only for the wild-type (wt) and VpsTM17D forms, where the effect was protein-specific and c-di-GMP-dependent. In addition, nucleotide-dependent DNA binding of VpsT was observed to multiple and relatively remote sites in the regulatory region of vpsL.
Figure 2. Transcriptional regulation by VpsT.
(A) Electromobility shift assays with purified proteins and biotin labeled fragments tiling the vpsL promoter region. Numbers indicate position relative to the open reading frame start. (B) vpsL gene expression in different genetic backgrounds harboring a single-copy chromosomal vpsLp-lacZ fusion. Data are mean of 8 replicates −/+ SD. (C) Whole genome expression profiling. Top, compact heatmap in a log2-based pseudocolor scale (yellow, induced; blue, repressed) comparing a total of 108 differentially expressed genes in a ΔvpsT strain expressing wild-type (wt) or mutated VpsT versions compared to the vector control (midpanel, expression profiles of genes located in and between the vps-I and vps-II clusters; bottom, expression profiles of flagellar biosynthesis genes).
To evaluate the functional importance of VpsT oligomers and c-di-GMP binding in cells, we measured transcription of vps genes by using a chromosomal vpsLp-lacZ transcriptional fusion in the ΔvpsT strain harboring wild-type VpsT, VpsT point mutants (VpsTM17D, VpsTR134A or VpsTI141E) or the insert-less expression vector (pBAD) (Fig. 2B). The presence of wild-type VpsT and VpsTM17D resulted in increased vpsL expression, similar to the wild-type rugose strain carrying vector only, while ΔvpsT strains with VpsTR134A, VpsTI141E or the empty vector did not exhibit such an increase. These data confirm that c-di-GMP-mediated oligomerization is critical for VpsT function. Mutations in the putative phosphorylation site designed to produce a constitutively inactive or active state, VpsTD60A or VpsTD60E, respectively, did not alter the efficiency of VpsT significantly. Hence, regulation of gene expression is presumably independent of phosphorylation of VpsT (see also Fig. S7).
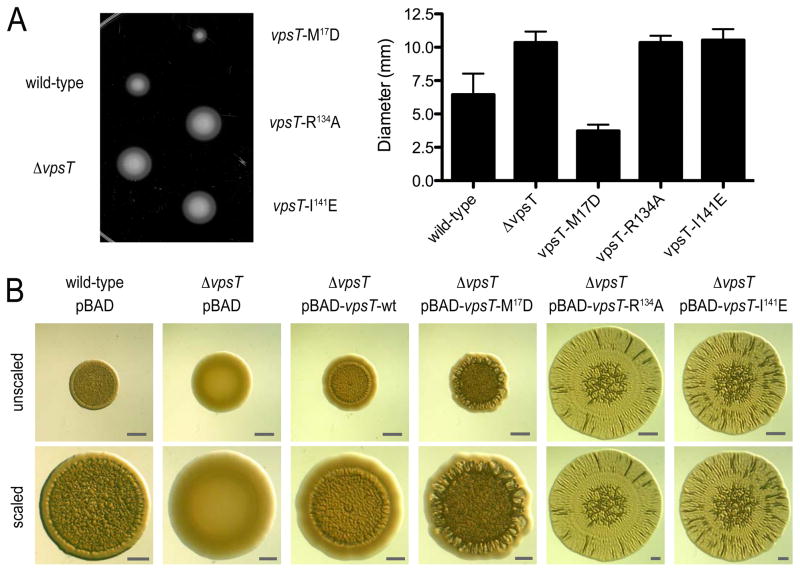
Next, we determined the gene regulatory potential as a function of c-di-GMP binding and oligomerization through whole genome expression profiling by comparing a ΔvpsT strain harboring either wild-type VpsT or VpsT point mutants (VpsTM17D, VpsTR134A or VpsTI141E) to that of cells harboring the pBAD vector alone (Fig. 2C; Table S3). Genes located in the vps-I and vps-II clusters, as well as the vps intergenic region were strongly induced upon expression of wild-type VpsT and VpsTM17D, and significantly less so in the strains expressing VpsTR134A or VpsTI141E. We also observed that the expression of several genes encoding flagellar proteins was decreased in cells expressing wild-type VpsT and VpsTM17D but not in cells expressing VpsTR134A or VpsTI141E, suggesting that VpsT inversely regulates motility and matrix production in a c-di-GMP-dependent manner (Fig. 2C and S7). These results were corroborated in motility assays, in which a ΔvpsT strain or strains expressing c-di-GMP-binding mutants showed increased migration on soft agar plates compared to rugose strains that express VpsT forms that are competent of c-di-GMP-dependent dimerization (Fig. 3A).
Figure 3. Functional characterization of wild-type rugose, ΔvpsT strains, and ΔvpsT strains expressing wild-type or mutant forms of VpsT.
(A) Motility phenotypes on semisolid LB agar plates. For strains expressing mutants of VpsT, single chromosomal insertion mutants are shown. The graph shows the mean migration zone diameter of each strain. Data are mean of 11 replicates −/+ SD. (B) Spot morphologies. A wild-type rugose strain carrying the vector (pBAD) and ΔvpsT strains carrying the vector or plasmids containing wild-type or mutant vpsT are shown (top, unscaled; bottom, scaled to similar diameter; bars=1 mm).
The strain harboring VpsTM17D had a similar expression profile to the strains harboring wild-type VpsT however with increased magnitude, indicating that c-di-GMP-independent dimerization could be inhibitory or regulatory (Fig. 2A and 2C). In contrast, the c-di-GMP-dependent interaction between two VpsT monomers is sufficient and necessary for DNA recognition and transcriptional regulation.
The corrugated appearance of rugose colonies can be attributed largely to increased levels of exopolysaccharides, which are induced by VpsT (6). As a consequence, V. cholerae mutants lacking vpsT produce smooth and flat colonies (Fig. 3B). To elucidate phenotypic consequences of mutations abolishing c-di-GMP binding and/or dimerization of VpsT, we compared the colony morphology of a ΔvpsT strain harboring wild-type VpsT or one of the point mutants described above. Expression of wild-type VpsT and VpsTM17D resulted in smooth-to-rugose conversion, where spot corrugation was greater in a ΔvpsT strain harboring VpsTM17D compared to a strain with wild-type VpsT. Introduction of VpsTR134A, VpsTI141E or a double-mutant VpsTM17D/R134A failed to promote the smooth-to-rugose switch, but led to a distinct phenotype, characterized by increased spot diameter and weak corrugation with a notable radial pattern (Fig. 3B and S8).
Cyclic di-GMP in the rugose variant is required for increased vps and VpsT gene expression (10, 11), suggesting that VpsT is involved in a positive feedback loop that integrates c-di-GMP to produce a robust transcriptional response. Robust matrix and biofilm formation relies on the mutual dependence of VpsT and VpsR, with VpsT introducing c-di-GMP-sensitivity to the regulatory network. In contrast, the transcriptional regulator FleQ from Pseudomonas aeruginosa, a distant VpsR-homolog, appears to directly sense c-di-GMP independently of a VpsT-homolog by using a distinct c-di-GMP binding motif (16).
Taken together, we establish VpsT as a transcriptional regulator that inversely regulates biofilm formation and motility by directly integrating c-di-GMP signaling. Cyclic di-GMP-driven dimerization is mediated by an extension of the canonical receiver domains, a structural motif that defines a wide-spread class of response regulators including CsgD and other LuxR family proteins. While some mechanisms may only pertain to close homologs of VpsT such as c-di-GMP-dependent dimerization, the general mode of action involving dimerization accompanied with changes in the relative orientation of the DNA binding domains is likely to be relevant for the large family of homologous transcription factors.
Supplementary Material
Footnotes
Author contributions: P.V.K., F.H.Y. and H.S. designed research; P.V.K., J.C.N.F., N.J.S. and S.B. performed research; P.V.K., J.C.N.F., N.J.S., M.V.A.S.N., F.H.Y., and H.S. analyzed data; and P.V.K., F.H.Y. and H.S. wrote the paper.
Publisher's Disclaimer: This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References
- 1.Mizunoe Y, Wai SN, Takade A, Yoshida SI. Infect Immun. 1999;67:958. doi: 10.1128/iai.67.2.958-963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yildiz FH, Schoolnik GK. Proc Natl Acad Sci USA. 1999;96:4028. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fong JC, Karplus K, Schoolnik GK, Yildiz FH. J Bacteriol. 2006;188:1049. doi: 10.1128/JB.188.3.1049-1059.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fong JC, Yildiz FH. J Bacteriol. 2007;189:2319. doi: 10.1128/JB.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beyhan S, Bilecen K, Salama SR, Casper-Lindley C, Yildiz FH. J Bacteriol. 2007;189:388. doi: 10.1128/JB.00981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casper-Lindley C, Yildiz FH. J Bacteriol. 2004;186:1574. doi: 10.1128/JB.186.5.1574-1578.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chirwa NT, Herrington MB. Microbiology. 2003;149:525. doi: 10.1099/mic.0.25841-0. [DOI] [PubMed] [Google Scholar]
- 8.Romling U, Bian Z, Hammar M, Sierralta WD, Normark S. J Bacteriol. 1998;180:722. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao R, Stock AM. Annu Rev Microbiol. 2009 doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim B, Beyhan S, Meir J, Yildiz FH. Mol Microbiol. 2006;60:331. doi: 10.1111/j.1365-2958.2006.05106.x. [DOI] [PubMed] [Google Scholar]
- 11.Beyhan S, Yildiz FH. Mol Microbiol. 2007;63:995. doi: 10.1111/j.1365-2958.2006.05568.x. [DOI] [PubMed] [Google Scholar]
- 12.Sudarsan N, et al. Science. 2008;321:411. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beyhan S, Odell LS, Yildiz FH. J Bacteriol. 2008;190:7392. doi: 10.1128/JB.00564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kader A, Simm R, Gerstel U, Morr M, Romling U. Mol Microbiol. 2006;60:602. doi: 10.1111/j.1365-2958.2006.05123.x. [DOI] [PubMed] [Google Scholar]
- 15.Weber H, Pesavento C, Possling A, Tischendorf G, Hengge R. Mol Microbiol. 2006;62:1014. doi: 10.1111/j.1365-2958.2006.05440.x. [DOI] [PubMed] [Google Scholar]
- 16.Hickman JW, Harwood CS. Mol Microbiol. 2008;69:376. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.We are grateful to Stevan Hubbard and Bill Horne for providing access to light scattering and calorimetry, respectively, and to the staff at the National Synchrotron Light Source (NSLS; Brookhaven National Labs) for assistance with synchrotron data collection. The NSLS is supported by the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and by the National Center for Research Resources of the NIH. This work was supported by the NIH (1R01GM081373 to H.S. and RO1AI055987 to F.H.Y.), and by a PEW Scholar award (to H.S.). Atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank under ID code 3KLN and 3KLO. The microarray data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE19479.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.