Abstract
Neutrophils respond to chemotactic stimuli by increasing the nucleation and polymerization of actin filaments, but the location and regulation of these processes are not well understood. Here, using a permeabilized-cell assay, we show that chemotactic stimuli cause neutrophils to organize many discrete sites of actin polymerization, the distribution of which is biased by external chemotactic gradients. Furthermore, the Arp2/3 complex, which can nucleate actin polymerization, dynamically redistributes to the region of living neutrophils that receives maximal chemotactic stimulation, and the least-extractable pool of the Arp2/3 complex co-localizes with sites of actin polymerization. Our observations indicate that chemoattractant-stimulated neutrophils may establish discrete foci of actin polymerization that are similar to those generated at the posterior surface of the intracellular bacterium Listeria monocytogenes. We propose that asymmetrical establishment and/or maintenance of sites of actin polymerization produces directional migration of neutrophils in response to chemotactic gradients.
Neutrophils, cells of the innate immune system, hunt and kill bacteria, which they find by reading chemotactic gradients of formylated peptides released from the bacteria. Neutrophils respond to chemotactic stimuli by increasing the nucleation and polymerization of actin filaments1. They respond to a gradient of chemoattractant by extending actin-rich pseudopodia preferentially in the direction of the highest concentration of chemotactic molecules2. Although actin polymerization is necessary for this morphological polarity and for migration of the neutrophils in response to chemotactic gradients3,4, the spatial distribution of actin polymerization in response to chemotactic gradients is not well understood. A knowledge of this distribution will be crucial in understanding how neutrophils and other chemoattactant-responsive cells spatially rearrange their actin cytoskeletons during chemotaxis.
The Arp2/3 complex, a strong candidate for the regulation of actin polymerization in chemotaxis, has not been studied during chemotaxis. This complex stimulates the nucleation of actin filaments5,6 in a regulatable fashion6,7, and conditional Arp2 and Arp3 mutations in yeast produce several defects in actin function8–10.
Neutrophils would seem to represent an ideal model system for the study of the spatial control of actin polymerization during chemotaxis. However, standard techniques for determining the subcellular localization of actin polymerization, through incorporation of fluorescently labelled actin into microinjected11,12 or permeabilized12,13 cells, have proven difficult or impossible to use when studying neutrophils. It would be useful to express GFP-tagged proteins in neutrophils to analyze protein dynamics in living cells during chemotaxis. However, it has not been possible to express recombinant proteins in neutrophils because they are short-lived terminally differentiated cells.
We have now overcome these difficulties, and report that chemoattractant-stimulated neutrophils establish discrete sites of actin polymerization whose distribution is biased towards the cell surface that is directed towards the highest concentration of chemoattractant. The least-extractable pool of the Arp2/3 complex co-localizes with these sites of actin polymerization, and this complex dynamically redistributes to the region of living neutrophils that receives maximal chemotactic stimulation. We propose that asymmetrical establishment and/or maintenance of these sites of actin polymerization mediates the directional migration of neutrophils in response to gradients of chemoattractant.
Results
Actin distribution and polymerization
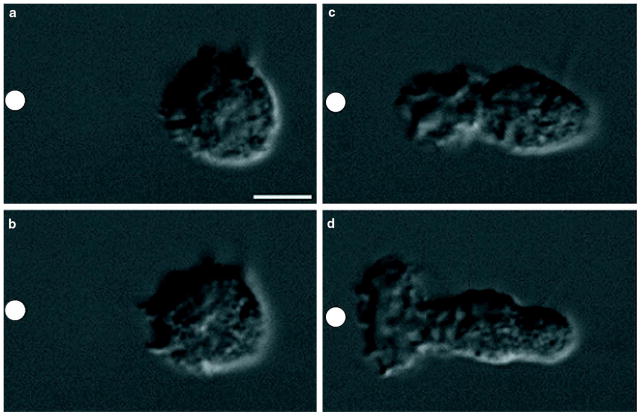
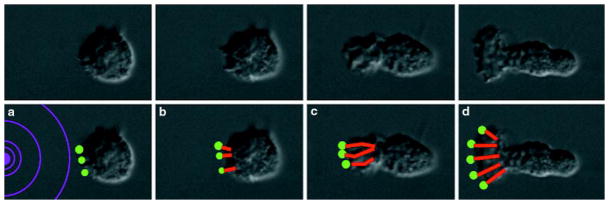
Before stimulation by chemoattractant, neutrophils lack obvious polarity. Between 5 s (Fig. 1a) and 30 s (Fig. 1b) of exposure to a point source of N-formyl-methionyl-leucyl-phenylalanine (FMLP), supplied through a micropipette (Fig. 1a–d, white circle), neutrophils begin to extend their surface toward the chemotactic pipette. Only the neutrophil surface directed up the chemotactic gradient ruffles and extends as neutrophils become polarized in the direction of the micropipette (Fig. 1b–d; see Supplementary Information). Microspikes constantly project from the leading edge towards the micropipette (Fig. 1b–d; see Supplementary Information), some continuing to extend for more than 1 minute if the direction of chemotactic stimulation remains constant. How do neutrophils control actin polymerization to generate these complex, polarized morphologies in response to chemoattractant?
Figure 1. Polarization of a neutrophil in response to a gradient of chemoattractant.
a–d, Nomarski images of an unpolarized neutrophil responding to a micropipette containing 10 μM FMLP (white circle) at a, 5 s, b, 30 s, c, 81 s, and d, 129 s.
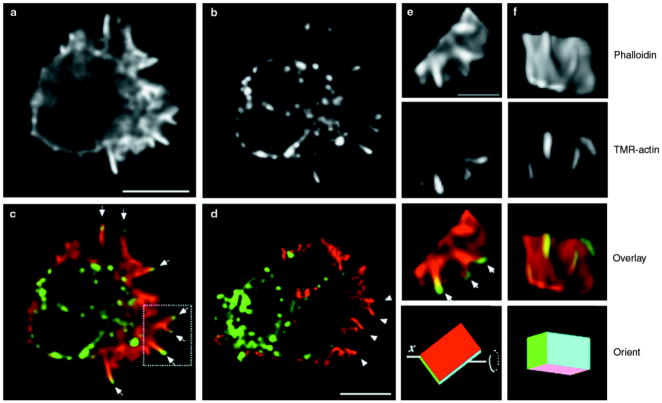
To identify sites of actin polymerization accurately in permeabilized neutrophils, we used fluorescently labelled actin (tetramethylrhodamine–actin (TMR–actin)), but first we had to minimize potential damage to the native cytoskeleton caused by the many neutral proteases of neutrophils. We first treated neutrophils with the membrane-permeable serine-protease inhibitor diisopropyl fluorophosphate (DFP), exposed them to chemoattractant, and then permeabilized them in the presence TMR–actin (procedure modified from refs 12,14). DFP does not alter the cytoskeletal morphology of fixed neutrophils or the ability of neutrophils to undergo chemotaxis. In permeabilized neutrophils polarized by exposure to uniform chemoattractant (that is, chemoattractant was present in the buffer surrounding the cells, rather than in a micro-pipette), exogenous actin incorporates at distinct sites on the pseudopodial surface and in a perinuclear fashion. The pseudopodial incorporation reflects actin polymerization, but the perinuclear incorporation does not, as shown by the latter’s persistence when actin polymerization is inhibited (Fig. 2). Thus we can analyse actin incorporation in regions of the cell away from the nucleus, but cannot determine the total distribution of actin polymerization in polarized cells.
Figure 2. Spatial distribution of incorporation of TMR–actin in a chemoattractant-stimulated permeabilized neutrophil.
a–c, Neutrophil exposed to a uniform concentration (20 nM) of FMLP for 60 s. Scale bar represents 5 μm. a, Phalloidin stain, representing pre-existing filaments and those that incorporated actin during the assay. Note that because phalloidin is excluded by some actin-binding proteins, such as cofilin, phalloidin staining does not necessarily represent all actin filaments. b, TMR–actin stain, representing newly incorporated TMR–actin only. c, Colour overlay, showing phalloidin stain in red and TMR–actin stain in green. Arrows in c indicate sites of new actin incorporation at the tips of finger-like actin bundles. d, A neutrophil stimulated and permeabilized as in a–c but in the presence of 0.2 μm cytochalasin D. Colour scheme is as in c. Arrowheads indicate the absence of TMR–actin incorporation at the tips of finger-like actin bundles. Perinuclear actin incorporation parallels the subcellular distribution of granules (data not shown). Because bright perinuclear but not pseudopodial TMR–actin incorporation is observed when cytochalasin D is present during the permeabilization reaction, we conclude that incorporation at the pseudopodial surface represents new actin polymerization and that perinuclear incorporation results from G-actin-binding proteins or structures, as has been reported for permeabilized fibroblasts12. e, f, Three-dimensional reconstruction of the boxed region of the pseudopodium shown in c. The bottom two panels indicate the relative orientation of the region of the pseudopodium from c as it is rotated along its x-axis. The scale bar in e represents 2 μm. Scale bar in d represents 5 μm.
In neutrophils stimulated with uniform chemoattractant for 60 s, new actin incorporation occurs predominantly at the front surface of the pseudopodium. In horizontal cross-sections, new actin incorporation is concentrated at the tips of radially projecting actin bundles (Fig. 2a–c, arrows). If cells are permeabilized in the presence of cytochalasin D, which inhibits actin polymerization, only perinuclear actin incorporation is observed (Fig. 2d), with no incorporation at the tips of radial actin bundles (arrowheads).
To obtain a more complete picture of the organization of the actin polymerization in the neutrophil pseudopodium, we generated three-dimensional reconstructions of neutrophil pseudopodia (Fig. 3e,f). These reconstructions reveal that what appear to be finger-like actin bundles in horizontal cross-section correspond to radially projecting actin ruffles, that the crescent-like projecting tips of these actin ruffles represent the sites of maximal actin polymerization, and that the sites of actin polymerization are not contiguous with one another. Thus, although sites of actin polymerization are present in many locations throughout the leading edge of stimulated neutrophils, the leading surface of the cell does not polymerize actin uniformly. Instead, the complex ruffled pseudopodium is composed of many distinct sites of actin polymerization organized at the tips of radially distributed actin projections. To our knowledge, this is the first three-dimensional analysis of actin incorporation in a motile cell.
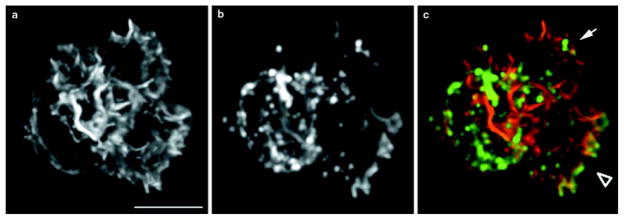
Figure 3. Spatial distribution of TMR–actin incorporation in a neutrophil with two pseudopodia.
Images represent maximum-intensity projections of three-dimensional immunofluorescence data. a, Phalloidin staining, representing total actin. b, TMR–actin staining, representing newly incorporated actin. c, Colour overlay, with phalloidin staining in red and newly incorporated actin in green. Arrow indicates a pseudopodium with minimal new actin incorporation. Arrowhead indicates a pseudopodium that is predominant in terms of new actin incorporation. The intense perinuclear TMR–actin stain does not represent new actin polymerization (Fig. 2). Scale bar represents 5 μm.
Neutrophils only transiently contain more than one pseudopodium; in neutrophils with two pseudopodia, one pseudopodium gains dominance and actively extends while the other pseudopodium is retracted (D.R. Soll, personal communication; O.D.W., unpublished observations). Of 25 randomly chosen neutrophils with multiple pseudopodia, 17 exhibited much more dramatic actin incorporation for one pseudopodial projection than the other(s), as shown for a single neutrophil in Fig. 3 (compare arrowhead and arrow in Fig. 3c). This indicates that actin polymerization does not depend solely on the pre-existing actin distribution and that two morphologically similar regions of the cell can differ in their abilities to polymerize F-actin.
To determine whether external chemotactic gradients bias the spatial distribution of sites of actin polymerization, we exposed neutrophils to a chemotactic micropipette and then permeabilized them in the presence of fluorescently labelled actin. Actin fingers showing polymerization at their tips are observed only on the up-gradient face of cells exposed to a point source of chemoattractant (data not shown).
Distribution and dynamics of the Arp2/3 complex
To study the dynamics of the Arp2/3 complex during neutrophil chemotaxis, it would be useful to follow the distribution of a green fluorescent protein (GFP)-tagged component of the complex. To overcome the difficulties of expressing a recombinant protein in short-lived neutrophils, we took advantage of the human promyelocytic cell line PLB-985. These cells can be cultured indefinitely, transfected using retroviruses, and then differentiated into cells that resemble human neutrophils in their signalling properties and response to chemoattractant.
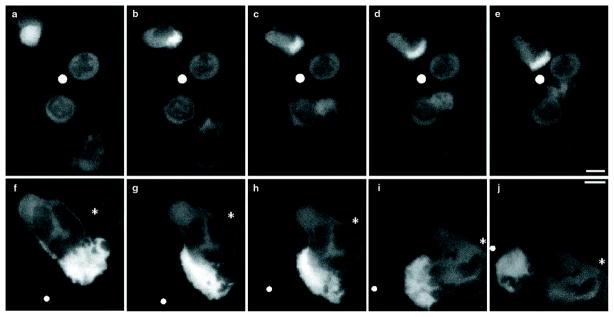
Immediately after exposure to the chemotactic micropipette (Fig. 4, white circle), neutrophils either lack polarity and show a uniform distribution of Arp3–GFP (Fig. 4a, top left neutrophil), or exhibit a slight polarity with Arp3–GFP uniformly distributed throughout the cytosol and excluded from the nucleus (Fig. 4a, bottom right cell). Within about a minute of exposure to the chemotactic micropipette, Arp3–GFP concentrates in the region of neutrophils that is beginning to exhibit cell polarity (that is, the region of the cell that is facing up the gradient of chemoattractant; Fig. 4b, top left cell), or in the pseudopod of polarized cells that are starting to migrate towards the chemotactic micropipette (Fig. 4b, lower right cell; the two cells shown in the centre were unresponsive during this experiment). Arp3–GFP remains strongly concentrated in the pseudopod as the cells migrate towards the chemotactic micropipette (Fig. 4c–e; see Supplementary Information). In contrast, GFP is present throughout the cytosol of cells expressing GFP alone (data not shown). When the chemotactic micropipette (Fig. 4f–j) moves, Arp3–GFP dynamically redistributes with the moving pseudopod to concentrate on the surface of the cell nearest the chemoattractant (see Supplementary Information). To our knowledge, this is the first analysis of the dynamics of the Arp2/3 complex in a living cell in response to a directional chemoattractant.
Figure 4. Response of neutrophils expressing Arp3–GFP to a stationary or moving chemotactic micropipette.
a–e, Response to a stationary micropipette. f–j, Response to a motile micropipette. Images are single optical sections from near the bottom of a cell. a, Image taken immediately after exposing neutrophils to a chemotactic micropipette (white circle). b–e, Same group of neutrophils at b, 72 s, c, 166 s, d, 196 s, and e, 240 s of exposure to the chemotactic micropipette. f, A polarized neutrophil responding to a moving chemotactic micropipette (white circle). The white asterisk represents a fixed reference point. g–h, Same cell as that shown in f at g, 78 s, h, 109 s, l, 193 s, and j, 305 s of exposure to the micropipette. Scale bar represents 5 μm.
To determine the subcellular distribution of the endogenous Arp2/3 complex in human neutrophils, we tested several affinity-purified rabbit polyclonal antibodies, raised against the p21–Arc, p34–Arc (for Arp complex) and Arp3 components of the human Arp2/3 complex15, on chemoattractant-stimulated human neutrophils. Because all these antibodies showed similar distributions, we describe only the results obtained with the anti-p21–Arc antibody, which consistently produced the brightest staining. In neutrophils stimulated with uniform FMLP for 90 s, fixed with formaldehyde, extracted with methanol, and processed for immunocytochemistry with antibodies to actin (Fig. 5a) and p21–Arc (Fig. 5b), p21–Arc (green) is enriched at the tips of radially projecting actin fingers (red) (Fig. 5c). To determine the relationship between sites of actin polymerization and the Arp2/3 complex, we processed neutrophils permeabilized in the presence of tetramethylrhodamine–actin (TMR–actin) for p21–Arc immunocytochemistry. p21–Arc immunostaining co-localizes with sites of actin polymerization at the tip of actin fingers (Fig. 5d–g). This actin finger extends into the cell periphery, a region in which all exogenous actin incorporation is prevented by cytochalasin D (Fig. 2d).
Figure 5. Immunofluorescence localization of endogenous Arp2/3 complex and actin in human neutrophils and the relationship of Arp2/3 localization to sites of actin polymerization.
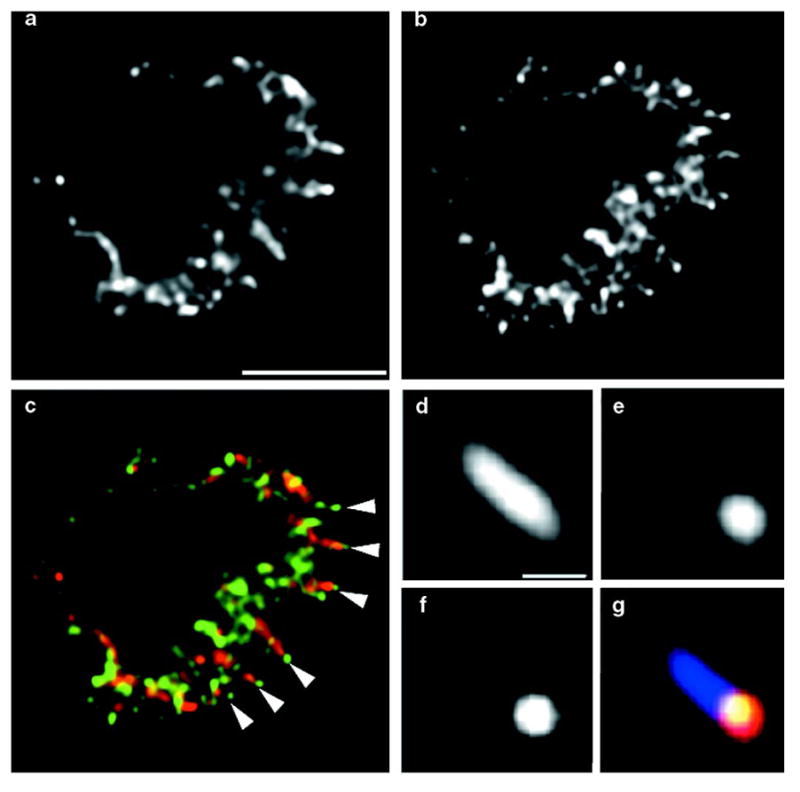
Images are single optical sections from near the midsection of cell. a, Actin immunostaining. Scale bar represents 5 μm. b, p21–Arc immunostaining. c, Colour overlay, showing actin in red and p21–Arc in green. Arrowheads indicate the localization of p21–Arc at the tips of actin fingers. d–g, Detail of an actin finger, from a cell permeabilized in the presence of TMR–actin to detect sites of actin polymerization and then processed for p21–Arc and actin immunostaining. d, Anti-actin staining, showing pre-existing and newly incorporated actin filaments. Scale bar represents 0.5 μm. e, TMR–actin staining, showing newly incorporated actin. f, p21–Arc staining. g, Triple overlay of d–f, showing total actin shown in light blue, newly incorporated actin in red, and p21–Arc in green. The green staining co-localizes with red and appears yellow.
Discussion
In our experiments with permeabilized neutrophils, chemoattractant-induced actin polymerization is restricted to the pseudopodial surface of stimulated cells and is concentrated at the tips of actin bundles that project into the plasma membrane. This pattern of actin incorporation resembles that seen in in vivo studies of the intracellular bacterium Listeria monocytogenes, a situation in which new actin polymerization occurs only at the most proximal portion of the actin tail, immediately adjacent to the posterior surface of the bacterium16,17. This pattern of actin polymerization differs from that seen previously in neutrophils14, where incorporation of exogenous (fluorescently labelled) actin paralleled the distribution of endogenous F-actin. We attribute this discrepancy to the fact that here we protected the endogenous actin cytoskeleton from proteolysis. If neutrophils are not pretreated with DFP before permeabilization, extensive proteolysis of actin and actin-associated proteins is observed18, possibly generating, through the severing and uncapping of pre-existing actin filaments, barbed actin ends, which can be elongated. When we omit DFP treatment before permeabilizing neutrophils, we typically observe incorporation of exogenous actin throughout the distribution of endogenous F-actin (data not shown).
Our results show that, in living neutrophils, a GFP-tagged Arp3 component of the Arp2/3 complex is uniformly distributed in unpolarized cells before stimulation, rapidly accumulates in pseudopodia following exposure of the neutrophils to chemoattractant, and dynamically redistributes in response to a moving source of chemoattractant. These data indicate that external spatial signals may modulate the behaviour of the Arp2/3 complex. The localization of Arp3–GFP throughout the newly polymerized pseudopod supports the hypothesis that the Arp2/3 complex acts as a nucleus for actin polymerization and is incorporated into growing actin filaments as a pointed-end cap. These data are consistent with the localization of Arp2/3 to the lamellipodia of fixed fibroblasts15,19 and the pseudopodia of fixed Acanthamoeba castellanii20–22. In living, unstimulated neutrophils, the localization of GFP–Arp3 resembles that in living unstimulated fibroblasts23, with diffuse cytoplasmic staining and a weak signal in the lamellipodia, although we do not observe dynamic Arp2/3 dots under any conditions. Upon chemotactic stimulation, neutrophils exhibit massive recruitment of the Arp2/3 complex to the pseudopodia; thus protrusive structures of resting cells contain a small amount of Arp2/3 complex and chemoattractant induces a marked recruitment of this complex to the growing pseudopod.
In unstimulated neutrophils expressing GFP–Arp3 that are fixed with formaldehyde and then permeabilized, the Arp2/3 complex is present throughout the actin cytoskeleton (data not shown). In contrast, after chemoattractant-stimulated neutrophils are permeabilized and then fixed and extracted with methanol, the Arp2/3 complex is predominantly associated with the sites of active actin polymerization. This enrichment of a ‘less-extractable pool’ of the Arp2/3 complex at sites of actin polymerization is observed only for stimulated neutrophils and is most dramatic for samples permeabilized before fixation. We suspect that our fixation and permeabilization conditions preferentially extract Arp2/3 complexes present in actin tails but stabilize Arp2/3 complexes that are associated with factors that induce its recruitment or activation. A reasonable candidate for recruitment of the Arp2/3 complex is the small GTPase Cdc42. Activated Cdc42 can induce actin polymerization in neutrophil, Dictyostelium and Xenopus extracts24,25. The Arp2/3 complex is biochemically downstream of Cdc42-mediated actin polymerization in cell extracts7,26, and activated Cdc42 forms a stable complex with the Arp2/3 complex and other associated proteins7, perhaps accounting for the behaviour of the least-extractable pool of the Arp2/3 complex in our permeabilized neutrophil system.
Chemoattractant-induced sites of actin polymerization in neutrophils are at the tips of actin fingers that extend into the plasma membrane; these sites co-localize with the least-extractable pool of the Arp2/3 complex, and the fingers correspond to radially protruding structures that extend from the neutrophil towards point sources of chemoattractant. On the basis of these data, we present a model for actin polymerization in response to chemotactic stimulation; this model is analogous to models proposed for actin-based motility of the intracellular bacterial pathogen Listeria monocytogenes.
Listeria monocytogenes can move rapidly in the cytoplasm of infected host cells27,28. The bacterially expressed protein ActA stimulates the ability of the host Arp2/3 complex to nucleate actin filaments6 and this localized actin polymerization at the bacterial surface is thought to drive bacterial motility. In well preserved Listeria pseudopodial projections in macrophages, the actin tail consists of long axial filaments and short randomly orientated filaments29, supporting the suggestion29,30 that the actin filaments nucleated at the surface of Listeria continue to polymerize only while next to the bacterial surface; after the filaments have left the zone of actin polymerization adjacent to the bacterial surface, they are capped at their barbed ends and cease growing.
We propose that stimulation of neutrophil chemoattractant receptors leads to the organization of foci of actin polymerization, at or just under the plasma membrane, that are functionally equivalent to the zone of actin polymerization generated by the ActA protein at the posterior surface of Listeria monocytogenes (Fig. 6). In this model, each focus mediates actin polymerization at its surface by activating the nucleating ability of the Arp2/3 complex. Actin filaments continue to polymerize only while in the zone of actin polymerization at the surface of the polymerization focus, and filaments are capped at their barbed ends after they have left this zone. Actin polymerization at the surface of the polymerization focus propels it and the cell membrane forward, forming an actin finger similar to that of a Listeria tail, with polymerization taking place at the tip of the growing finger (Fig. 6).
Figure 6. Model of actin polymerization in response to a chemotactic signal.
Top, Nomarski images of an unpolarized neutrophil exposed to a chemotactic micropipette (just to left of field) for a, 5 s, b, 30 s, c, 81 s, and d, 129 s. Bottom, the model. a, A neutrophil exposed to a gradient of chemoattractant (purple concentric circles) generates an asymmetric distribution of polymerization foci. b–d, The force generated by polymerization of actin (red lines) propels the polymerization focus forward and pushes the membrane outwards. The preferential activation of polymerization foci nearest to the chemoattractant could result in directional migration of neutrophils in response to chemotactic gradients and would be consistent with the behaviour of the pseudopod in response to a changing direction of chemoattractant. Note that this figure represents a single optical section of a neutrophil responding to a chemotactic gradient; the three-dimensional organization of the sites of actin polymerization and actin projections is shown in Fig. 2e, f.
How might neutrophils regulate the behaviour of these polymerization foci to mediate cell migration up chemotactic gradients? We have shown that neutrophils extend microspikes and radial actin ruffles towards a point source of chemoattractant, that the tips of these actin bundles represent the sites of maximum actin polymerization, and that the spatial distribution of actin polymerization does not depend solely on the distribution of pre-existing actin and can be biased by chemotactic gradients. On the basis of these data, we propose that asymmetric establishment and/or maintenance of sites of actin polymerization produce the cytoskeletal and morphological rearrangements that mediate cell migration up gradients of chemoattractants.
Methods
Neutrophil preparation and stimulation
A drop of healthy human blood, acquired by pinprick, was collected on the centre of a sterile coverslip and neutrophils were isolated as described31. The cells were covered with mHBSS medium (150 mM NaCl, 4 mM KCl, 1.2 mM CaCl2, 1 mM MgCl2, 10 mg ml−1 glucose and 20 mM HEPES, pH 7.2) and incubated at 37 °C for 15 min before chemotactic stimulation. All subsequent steps were carried out at room temperature.
For uniform chemotactic stimulation, cells were incubated in mHBSS containing 20 nM FMLP (Sigma). For delivery of a point source of FMLP, we allowed a solution of 10 μM FMLP to diffuse passively out of the tip of a micropipette of diameter ~0.2 μm whose position relative the coverslip could be controlled with a Narishige micromanipulator (procedure modified from ref. 32).
Preparation of TMR–actin
Actin was prepared33 from frozen rabbit muscle (Pel-Freez Biologicals, Rogers, AR), polymerized into filamentous form, and derivatized with N-hydroxysuccinimidyl-5-carboxytetramethylrhodamine (Molecular Probes) as described34. TMR–actin was stored at a concentration of 100 μM at −80 °C after freezing aliquots in liquid nitrogen. Before use, TMR–actin was rapidly thawed from −80 °C, diluted ten times in G-buffer, sonicated using a Vibracell sonicator (Sonics and Materials, Danbury, CT), and clarified in a microfuge for 20 min at 4 °C.
Permeabilized cells
Before permeabilization, neutrophils were treated for 5 min in mHBSS containing 1 mM DFP (Sigma). Neutrophils were then permeabilized (using a modification of the procedure in refs 12,14). For experiments involving uniform chemoattractant stimulation, neutrophils were stimulated for 60 s in mHBSS containing 20 nM FMLP and then permeabilized for 3 min in cytoskeleton buffer with sucrose (CBS; containing 10 mM MES, pH 6.1, 138 mM KCl, 3mM MgCl2, 2 mM EGTA; refs 12,35) containing 0.2 mg ml−1 saponin or 1% NP40, 1 mM ATP, 20 nM FMLP and 0.35 μM TMR–actin, added immediately before use. For some experiments, 1 μg ml−1 fluorescein isothiocyanate (FITC)–phalloidin was used to stabilize the F-actin cytoskeleton during permeabilization.
Immunocytochemistry
To visualize the actin cytoskeleton, cells were fixed for 20 min in a solution of 3.7% paraformaldehyde in CBS and then incubated with 10 units ml−1 Texas-Red-labelled X-phalloidin (Molecular Probes) for 20 min. For experiments involving affinity-purified rabbit anti-p21–Arc primary antibodies15, neutrophils were fixed for 40 min in 3.7% paraformaldehyde, briefly washed in PBS containing 0.5% Triton-X100, and then incubated in methanol at −20 °C for 3 min before incubation with a 1:50 dilution of anti-p21–Arc antibody for 1 h. Mouse anti-actin antibodies (5 μg ml−1; Boehringer Mannheim) were used to label the actin cytoskeleton for methanol-fixed samples. All secondary antibodies were from Jackson ImmunoResearch Laboratories.
Amphotropic retrovirus generation and PLB985-cell transduction
For experiments involving the GFP-tagged construct, we used the human promyelocytic cell line PLB-985 (ref. 36). CDNAs encoding EGFP (Clontech) and Arp3–GFP15 were cloned into the pLNCX retroviral vector37 under the control of the CMV promoter. Retroviruses were produced and PLB-985 cells were stably transduced as described38.
Microscopy and analysis
All images were acquired with a scientific-grade, cooled, charge-coupled device on a multiwavelength wide-field three-dimensional microscopy system (ref. 39 and references therein) in which the shutters, filter wheels, focus movement, and data collection are all computer driven. Neutrophils were imaged using a ×60 1.4 NA lens (Olympus) and n=1.518 immersion oil (RP Cargille Laboratories, Cedar Grove, NJ). Immunofluorescent samples were imaged in successive 0.25-μm focal planes through the sample, and out-of-focus light was removed with a constrained iterative deconvolution algorithm40,41. Maximum-intensity projections, side views and rotated reconstructions of the three-dimensional data stacks were generated using image-visualization environment software42. Unless otherwise indicated, all images represent single optical sections of immunofluorescence data.
Supplementary Material
Acknowledgments
We thank A. Abo, D. Agard, C. Bargmann, D. Drubin, Z. Kam, C. Kenyon, R. Mullins, J. Taunton, J. Weissman, S. Zigmond and members of the Bourne and Sedat laboratories for discussions; A. Abo for the PLB-985 promyelocytic cell line; and C. Bargmann, C. Kenyon, J.V. Small, and S. Zigmond for critical reading of the manuscript. This work was supported in part by grants from the NIH (to H.R.B., J.W.S. and T.J.M.). M.D.W. is a Leukemia Society of America Special Fellow; G.S. is a Medical Research Council of Canada Postdoctoral Fellow; and O.D.W. is an HHMI Predoctoral Fellow.
Footnotes
Supplementary information is available on Nature Cell Biology’s World-Wide Web site (http://cellbio.nature.com).
References
- 1.Cano ML, Lauffenburger DA, Zigmond SH. Kinetic analysis of F-actin depolymerization in polymorphonuclear leukocyte lysates indicates that chemoattractant stimulation increases actin filament number without altering the filament length distribution. J Cell Biol. 1991;115:677–687. doi: 10.1083/jcb.115.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zigmond SH. Mechanisms of sensing chemical gradients by polymorphonuclear leukocytes. Nature. 1974;249:450–452. doi: 10.1038/249450a0. [DOI] [PubMed] [Google Scholar]
- 3.Zigmond SH, Hirsch JG. Effects of cytochalasin B on polymorphonuclear leucocyte locomotion, phagocytosis and glycolysis. Exp Cell Res. 1972;73:383–393. doi: 10.1016/0014-4827(72)90062-6. [DOI] [PubMed] [Google Scholar]
- 4.Watts RG, Crispens MA, Howard TH. A quantitative study of the role of F-actin in producing neutrophil shape. Cell Motil Cytoskeleton. 1991;19:159–168. doi: 10.1002/cm.970190304. [DOI] [PubMed] [Google Scholar]
- 5.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welch MD, Rosenblatt J, Skoble J, Portnoy DA, Mitchison TJ. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- 7.Ma L, Rohatgi R, Kirschner MW. The Arp2/3 complex mediates actin polymerization induced by the small GTP-binding protein Cdc42. Proc Natl Acad Sci USA. 1998;95:15362–15367. doi: 10.1073/pnas.95.26.15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCollum D, Feoktistova A, Morphew M, Balasubramanian M, Gould KL. The Schizosaccharomyces pombe actin-related protein, Arp3, is a component of the cortical actin cytoskeleton and interacts with profilin. EMBO J. 1996;15:6438–6446. [PMC free article] [PubMed] [Google Scholar]
- 9.Moreau V, Madania A, Martin RP, Winson B. The Saccharomyces cerevisiae actin-related protein Arp2 is involved in the actin cytoskeleton. J Cell Biol. 1996;134:117–132. doi: 10.1083/jcb.134.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winter D, Podtelejnikov AV, Mann M, Li R. The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr Biol. 1997;7:519–529. doi: 10.1016/s0960-9822(06)00223-5. [DOI] [PubMed] [Google Scholar]
- 11.Okabe S, Hirokawa N. Actin dynamics in growth cones. J Neurosci. 1991;11:1918–1929. doi: 10.1523/JNEUROSCI.11-07-01918.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Symons MH, Mitchison TJ. Control of actin polymerization in live and permeabilized fibroblasts. J Cell Biol. 1991;114:503–513. doi: 10.1083/jcb.114.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan AY, et al. EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells. J Cell Sci. 1998;111:199–211. doi: 10.1242/jcs.111.2.199. [DOI] [PubMed] [Google Scholar]
- 14.Redmond T, Zigmond SH. Distribution of F-actin elongation sites in lysed polymorphonuclear leukocytes parallels the distribution of endogenous F-actin. Cell Motil. Cytoskeleton. 1993;26:7–18. doi: 10.1002/cm.970260103. [DOI] [PubMed] [Google Scholar]
- 15.Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol. 1997;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanger JM, Sanger JW, Southwick FS. Host cell actin assembly is necessary and likely to provide the propulsive force for intracellular movement of Listeria monocytogenes. Infect Immun. 1992;60:3609–3619. doi: 10.1128/iai.60.9.3609-3619.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Theriot JA, Mitchison TJ, Tilney LG, Portnoy DA. The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization. Nature. 1992;357:257–260. doi: 10.1038/357257a0. [DOI] [PubMed] [Google Scholar]
- 18.Amrein PC, Stossel TP. Prevention of degradation of human polymorphonuclear leukocyte proteins by diisopropylfluorophosphate. Blood. 1980;56:442–447. [PubMed] [Google Scholar]
- 19.Machesky LM, et al. Mammalian actin-related protein 2/3 complex localizes to regions of lamellipodial protrusion and is composed of evolutionarily conserved proteins. Biochem J. 1997;328:105–112. doi: 10.1042/bj3280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J Cell Biol. 1994;127:107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelleher JF, Atkinson SJ, Pollard TD. Sequences, structural models, and cellular localization of the actin-related proteins Arp2 and Arp3 from Acanthamoeba. J Cell Biol. 1995;131:385–397. doi: 10.1083/jcb.131.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mullins RD, Kelleher JF, Xu J, Pollard TD. Arp2/3 complex from Acanthamoeba binds profilin and cross-links actin filaments. Mol Biol Cell. 1998;9:841–852. doi: 10.1091/mbc.9.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schafer DA, et al. Visualization and molecular analysis of actin assembly in living cells. J Cell Biol. 1998;143:1919–1930. doi: 10.1083/jcb.143.7.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zigmond SH, Joyce M, Borleis J, Bokoch GM, Devreotes PN. Regulation of actin polymerization in cell-free systems by GTP-γS and Cdc42. J Cell Biol. 1997;138:363–374. doi: 10.1083/jcb.138.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma L, Cantley LC, Janmey PA, Kirschner MW. Corequirement of specific phosphoinositides and small GTP-binding protein Cdc42 in inducing actin assembly in Xenopus egg extracts. J Cell Biol. 1998;140:1125–1136. doi: 10.1083/jcb.140.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullins RD, Pollard TD. Rho-family G-proteins act through Arp2/3 complex to stimulate actin polymerization in Acanthamoeba extracts. Curr Biol. doi: 10.1016/s0960-9822(99)80187-0. (in the press) [DOI] [PubMed] [Google Scholar]
- 27.Dabiri GA, Sanger JM, Portnoy DA, Southwick FS. Listeria monocytogenes moves rapidly through the host-cell cytoplasm by inducing directional actin assembly. Proc Natl Acad Sci USA. 1990;87:6068–6072. doi: 10.1073/pnas.87.16.6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tilney LG, Tilney MS. The wily ways of a parasite: induction of actin assembly by Listeria. Trends Microbiol. 1993;1:25–31. doi: 10.1016/0966-842x(93)90021-i. [DOI] [PubMed] [Google Scholar]
- 29.Sechi AS, Wehland J, Small JV. The isolated comet tail pseudopodium of Listeria monocytogenes: a tail of two actin filament populations, long and axial and short and random. J Cell Biol. 1997;137:155–167. doi: 10.1083/jcb.137.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchand JB, et al. Actin-based movement of Listeria monocytogenes: actin assembly results from the local maintenance of uncapped filament barbed ends at the bacterium surface. J Cell Biol. 1995;130:331–343. doi: 10.1083/jcb.130.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cassimeris L, McNeill H, Zigmond SH. Chemoattractant-stimulated polymorphonuclear leukocytes contain two populations of actin filaments that differ in their spatial distributions and relative stabilities. J Cell Biol. 1990;110:1067–1075. doi: 10.1083/jcb.110.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerisch G, Keller HU. Chemotactic reorientation of granulocytes stimulated with micropipettes containing fMet-Leu-Phe. J Cell Sci. 1981;52:1–10. doi: 10.1242/jcs.52.1.1. [DOI] [PubMed] [Google Scholar]
- 33.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85B:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 34.Kellogg DR, Mitchison TJ, Alberts BM. Behaviour of microtubules and actin filaments in living Drosophila embryos. Development. 1988;103:675–686. doi: 10.1242/dev.103.4.675. [DOI] [PubMed] [Google Scholar]
- 35.Small JV. Organization of actin in the leading edge of cultured cells: influence of osmium tetroxide and dehydration on the ultrastructure of actin meshworks. J Cell Biol. 1981;91:695–705. doi: 10.1083/jcb.91.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tucker KA, Lilly MB, Heck L, Jr, Rado T. A Characterization of a new human diploid myeloid leukemia cell line (PLB- 985) with granulocytic and monocytic differentiating capacity. Blood. 1987;70:372–378. [PubMed] [Google Scholar]
- 37.Miller AD, Rosman GJ. Improved retroviral vectors for gene transfer and expression. Biotechniques. 1989;7:980–982. 984–986, 989–990. [PMC free article] [PubMed] [Google Scholar]
- 38.Servant G, Weiner OD, Neptune E, Sedat JW, Bourne HR. Dynamics of a chemoattractant receptor in living neutrophils during chemotaxis. Mol Biol Cell. 1999;10:1163–1178. doi: 10.1091/mbc.10.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiraoka Y, Swedlow JR, Paddy MR, Agard DA, Sedat JW. Three-dimensional multiple-wavelength fluorescence microscopy for the structural analysis of biological phenomena. Semin Cell Biol. 1991;2:153–165. [PubMed] [Google Scholar]
- 40.Agard DA, Hiraoka Y, Shaw P, Sedat JW. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. doi: 10.1016/s0091-679x(08)60986-3. [DOI] [PubMed] [Google Scholar]
- 41.Swedlow JR, Sedat JW, Agard DA. In: Deconvolution of Images and Spectra. Jansson PA, editor. Academic; San Diego: 1997. pp. 284–307. [Google Scholar]
- 42.Chen H, Hughes DD, Chan TA, Sedat JW, Agard DA. IVE (Image Visualization Environment): a software platform for all three-dimensional microscopy applications. J Struct Biol. 1996;116:56–60. doi: 10.1006/jsbi.1996.0010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.