Abstract
Many ligands that affect nervous system development are members of gene families that function together to coordinate the assembly of complex neural circuits. cpg15/neuritin encodes an extracellular ligand that promotes neurite growth, neuronal survival, and synaptic maturation. Here we identify cpg15-2 as the only paralogue of cpg15 in the mouse and human genome. Both genes are expressed predominantly in the nervous system, where their expression is regulated by activity. cpg15-2 expression increases by more than twofold in response to kainate-induced seizures and nearly fourfold in the visual cortex in response to 24 hours of light exposure following dark adaptation. cpg15 and cpg15-2 diverge in their spatial and temporal expression profiles. cpg15-2 mRNA is most abundant in the retina and the olfactory bulb, as opposed to the cerebral cortex and the hippocampus for cpg15. In the retina, they differ in their cell-type specificity. cpg15 is expressed in retinal ganglion cells, whereas cpg15-2 is predominantly in bipolar cells. Developmentally, onset of cpg15-2 expression is delayed compared with cpg15 expression. CPG15-2 is glycosylphosphatidylinositol (GPI) anchored to the cell membrane and, like CPG15, can be released in a soluble-secreted form, but with lower efficiency. CPG15 and CPG15-2 were found to form homodimers and heterodimers with each other. In hippocampal explants and dissociated cultures, CPG15 and CPG15-2 promote neurite growth and neuronal survival with similar efficacy. Our findings suggest that CPG15 and CPG15-2 perform similar cellular functions but may play distinct roles in vivo through their cell-type- and tissue-specific transcriptional regulation.
Indexing terms: neuritin, glycosylphosphatidylinositol anchor, retina, bipolar cell
A key question in nervous system development is how the brain codes for the emergence of highly patterned structures with intricate yet exquisitely specific connections. A common coding mechanism for both invertebrate and vertebrate species is the use of multigene families that represent proteins with similar physical properties yet distinct in vivo functions. Many growth factors and extracellular signaling cues used for tissue patterning during development are affiliates of broad families whose individual members convey unique, yet sometimes overlapping, information. Although the variations on a theme displayed by these different gene families can derive from nuanced functional differences in the proteins themselves, they frequently arise from differential transcriptional control, which produces unique spatial and temporal expression patterns with tissue and sometimes cell-type-specific resolution. A classic example of differential expression of family members acting to control fate of specific cell types can be seen in the action of the neurotrophin family in the peripheral nervous system (PNS; Reichardt and Farinas, 1997; Bibel and Barde, 2000). All four neurotrophins promote neuronal survival, but each family member controls the survival of specific sensory neuron types through differential expression of the ligands and their receptors. Neurotrophins also show distinct but overlapping temporal and spatial expression in the central nervous system (Maisonpierre et al., 1990), which, though apparently more redundant than in the PNS, still produce effects specific to certain neuronal populations (Xu et al., 2000).
An interesting example of differential expression of family members guiding precise topographic connections is demonstrated by the ephrin family of guidance cues (McLaughlin et al., 2003). A-type ephrins are expressed in a gradient along the anterior-posterior axis of the optic tectum, determining topographic mapping of retinal projections along this axis. In contrast, B-type ephrins show graded expression in the orthogonal orientation and determine mapping along the lateral-medial axis. Thus, the combined action of different family members can result in a specific pattern of cell types or connections depending on the contribution of each family member at a given time and place.
cpg15/neuritin is an activity-regulated gene encoding a small extracellular protein with multiple functions (Nedivi et al., 1996, 1998; Naeve et al., 1997). During early embryonic development, cpg15 is expressed in multiple brain regions and acts as a survival factor for neural progenitors and differentiated neurons (Putz et al., 2005). Later in development, CPG15 promotes growth and stabilization of axonal and dendritic arbors along with synapse formation and maturation (Nedivi et al., 1998; Cantallops et al., 2000; Javaherian and Cline, 2005). cpg15 continues to be expressed in the adult brain, where its expression is correlated with activity-dependent functional plasticity (Nedivi et al., 1996; Lee and Nedivi, 2002; Harwell et al., 2005). The effects of CPG15 are noncell autonomous, suggesting that it acts as a ligand (Nedivi et al., 1998).
To identify cpg15 family members, we searched the human and mouse genome database and found a single potential paralogue that we termed cpg15-2. We compared cpg15 and cpg15-2 gene expression, and the biochemical and functional properties of the proteins they encode. We found that both cpg15 and cpg15-2 are regulated by neuronal activity and that the proteins they encode share many biochemical and functional properties. However, cpg15 and cpg15-2 are differentially expressed, and the proteins undergo different posttranslational processing. Our results suggest that CPG15 and CPG15-2 play distinct roles in the nervous system because of differences in their expression patterns and dispersion properties within the tissue.
Materials and Methods
Isolation and sequence analysis of cpg15-2
A sequence search for genes that encode proteins similar to human CPG15/neuritin (GenBank accession No. AF136631) was done using the T-BLAST-N program (http://www.ncbi.nlm.nih.gov/blast/) with default settings. The search yielded a human predicted mRNA MRCC2446 (GenBank accession No. NM_198443) that we termed human cpg15-2 and a mouse cDNA clone, G630049C14 (Gen-Bank accession No. AK090312) that we termed mouse cpg15-2. The mouse cpg15-2 cDNA was then isolated from adult mouse brain RNA by reverse transcription-polymerase chain reaction (RT-PCR). Poly-(A)+ RNA was reverse transcribed using the SuperScript first-strand synthesis system for RT-PCR (Invitrogen, Carlsbad, CA), and the coding region of the cpg15-2 cDNA was amplified by PCR with primers 15-2-s1 and 15-2-as1 (sequences in Suppl. Table 1) designed according to the mouse cDNA sequence. PCR was carried out using Pfu polymerase (Stratagene, La Jolla, CA) at 95°C 45 seconds, 55°C 45 seconds, and 72°C 3 minutes per cycle for 25 cycles. Amplified DNA was digested with XhoI and SacII and cloned into a modified pcDNA3 vector (Invitrogen) in which the XbaI site was changed to an SacII site. Sequencing of the cloned PCR fragment confirmed its identity with the mouse cDNA G630049C14. The nucleotide sequence of PCR amplified cDNA clone is provided in Supplemental Figure 1.
Protein sequence comparisons were carried out with the Genetyx-Mac program (Software Development, Tokyo, Japan). The SignalP program (http://www.cbs.dtu.dk/services/SignalP/) was used to identify the signal peptide sequence, and big-PI Predictor (http://mendel.imp.univie.ac.at/sat/gpi/gpi_server.html) and NetOGlyc/NetNGlyc (http://www.cbs.dtu.dk/services/) were used to identify the glycosylphosphatidylinositol (GPI)-anchoring sequence and glycosylation sites, respectively. Exon-intron structures of the cpg15 and cpg15-2 genes were deduced from comparison of the cDNA (BC035531, AK090312) and genomic DNA sequences (NT_039579 and NT_078586).
Animals
All animal work was approved by the Massachusetts Institute of Technology Committee on Animal Care and conforms to NIH guidelines for the use and care of vertebrate animals. RNA samples for Northern blots and real-time RT-PCR analysis, protein samples from the retina and the olfactory bulb, and retinal tissue for in situ hybridization and immunostaining were from 8–12-week-old male C57BL/6 mice (Charles River Laboratories, Wilmington, MA), unless otherwise stated. Embryos for hippocampal explant assay and dissociated cultures were from Sprague-Dawley rats (Charles River Laboratories).
Northern blot hybridization and real-time quantitative RT-PCR
For kainate-induced seizures, mice were injected intra-peritoneally with kainate (25 mg/kg). Six hours after injection, cerebral cortices of mice that showed whole-body continuous clonic seizures were harvested. For light-induced expression, six 8-week-old males were dark adapted for 4 weeks. Three mice were then exposed to light for 24 hours while the remaining three were kept in the dark. The retina and the visual cortices were harvested. For the developmental expression profile, whole brains were isolated on embryonic days (E) 12.5, E15.5, and E17.5 and postnatal days (P) 1, 7, 14, 28, and 11 weeks.
Total RNA was extracted with the Trizol reagent (Invitrogen) for the kainate experiment, tissue RNA, and developmental series older than E12.5. For light-induced expression, central nervous system subregions, and E12.5 brains, total RNA was extracted using the RNeasy Mini-Kit (Qiagen, Valencia, CA).
For Northern blot analysis, poly-(A)+ RNA was enriched from total RNA by using Oligotex (Qiagen). Ten micrograms of poly-(A)+-enriched RNA was analyzed by Northern blot hybridization as described elsewhere (Fujino et al., 2003). For probe synthesis, a 1.6-kb mouse cpg15 cDNA fragment (nucleotides 12–1581 of GenBank accession No. BC035531), a 0.5-kb mouse cpg15-2 cDNA fragment (nucleotides 1–489 of GenBank accession No. DQ176852), and a 0.3-kb mouse gapdh cDNA fragment (nucleotides 345–660 of GenBank accession No. M32599) excised from pTRI-GAPDH-mouse (Ambion, Austin, TX) were used.
For real-time quantitative RT-PCR, cDNA was synthesized from 0.2–1.0 μg of total or poly-(A)+ RNA using the SuperScript III first-strand synthesis supermix for qRT-PCR (Invitrogen) according to the manufacturer’s instructions. Real-time PCR was run on an Applied Biosystems 7500 system (Foster City, CA). Each reaction contained 1× Power Sybr Green PCR master mix (Applied Biosystems), 1 μl of the cDNA reaction, and 50 nM of each primer. Primer sets were 15-rt1 and 15-rt2 for cgp15, 15-2-rt1 and 15-2-rt2 for cpg15-2, and gapdh-rt1 and gapdh-rt2 for gapdh. Primer sequences are listed in Supplemental Table 1. PCR conditions were 50°C for 2 minutes, 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute per cycle. Plasmid DNA carrying cpg15 or cpg15-2 cDNA or PCR product of gapdh of known concentrations were used as standards. Copy numbers for cpg15 and cpg15-2 were normalized to the copy number of gapdh for each sample. Independent samples from three mice were analyzed unless otherwise stated.
In situ hybridization
Eyes were removed from C57BL/6 mice, embedded in OCT, then frozen on dry ice. Ten-micrometer cryostat sections were thaw mounted on charged slides and processed for in situ hybridization as described by Lee and Nedivi (2002). For probe synthesis, a 0.8-kb mouse cpg15 fragment (nucleotides 733–1581 of GenBank accession No. BC035531) was used. Sections were counterstained with toluidine blue.
Expression constructs
For CPG15 and CPG15-2 expression, we generated constructs in which a FLAG or poly-His tag was inserted after the signal peptide sequence of CPG15 and CPG15-2 to avoid tag removal with signal peptide cleavage to form the mature protein (Suppl. Fig. 2A). To this purpose, we used PCR to amplify two fragments from the full-length cDNA: the N-terminal fragment carrying the signal peptide and the C-terminal fragment carrying the remainder of the cDNA. The latter amplification was done using an upstream primer containing either the FLAG or the His tag. Primers used to amplify each of the fragments are listed in Supplemental Figure 2B, and primer sequences are provided in Supplemental Table 1. The N-terminal fragment and C-terminal fragments were then digested by restriction enzymes as listed in Supplemental Figure 2C and ligated into the pIRES2-EGFP vector (Clontech, Mountain View, CA). Nucleotide sequence of the PCR-amplified clones is provided in Supplemental Figure 1. For viral expression, the FLAG-tagged cpg15 or cpg15-2 cDNAs were cloned into the BamHI site of the FUIGW lentiviral vector (Lois et al., 2002). Replication-incompetent lentiviruses expressing tagged cpg15 and cpg15-2 were then generated as described by Lois et al. (2002).
Cell culture and transfection
Human embryonic kidney 293 (HEK293) and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Cambrex, Walkersville, MD) with 10% fetal bovine serum (Cambrex). For transient transfection, HEK293T cells in 10- or 15-cm dishes were transfected with 12 or 24 μg DNA using Lipofectamine 2000 (Invitrogen) and harvested 3 days later. For generation of stable cell lines expressing FLAG-tagged CPG15 or CPG15-2, HEK293 cells were infected with a lentiviral vector coexpressing enhanced green fluorescent protein (EGFP) and CPG15, EGFP and CPG15-2, or EGFP alone. Single-cell clones expressing EGFP were isolated and expanded.
Antibody production and immunohistochemistry
A rabbit anti-CPG15-2 polyclonal antibody was generated by Open Biosystems (Huntsville, AL) against a synthetic peptide (RERIAGPETNQETLR) corresponding to amino acids 119–133 of mouse CPG15-2 conjugated to keyhole limpet hemocyanin. Sera from two rabbits were affinity purified. The mouse antiprotein kinase Cα (PKCα) monoclonal antibody obtained from BD Biosciences (San Jose, CA; No. 610107, lot No. 01899) was raised against amino acids 270–427 of human PKCα. This anti-PKCα antibody stains a single band of 82 kDa molecular weight on Western blots (manufacturer’s technical information).
C57BL/6 mice were perfused with phosphate-buffered saline (PBS; pH 7.4) and 4% paraformaldehyde, and their eyes were removed and postfixed in 4% paraformaldehyde for 2 hours. After removal of the cornea and lens, the eyes were cryoprotected in 30% sucrose in PBS, embedded in OCT, frozen on dry ice, and sectioned by cryostat at 40 μm. Sections were treated with blocking solution (5% goat serum, 0.3% Triton X-100 in PBS), then primary antibodies [rabbit anti-CPG15-2 (1:100) and mouse anti-PKCα (1:100)] overnight at 4°C, followed by secondary antibodies [Alexa 555-conjugated goat anti-rabbit IgG (Molecular Probes, Eugene, OR; No. A21429) and Alexa 488-conjugated goat anti-mouse IgG (Molecular Probes; No. A11001)] for 1 hour at room temperature. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma, St. Louis, MO). For peptide competition, anti-CPG15-2 antibody was preincubated with 10 μg/ml CPG15 (GLDDKTNIKTVCTYWE) or CPG15-2 (RERIAGPETNQETLR) peptide for 1 hour prior to incubation with sections. Images were acquired with an epifluorescent microscope (Nikon, Tokyo, Japan) equipped with a Spot camera (Spot2; Diagnostic Instruments, Sterling Heights, MI) or with a Nikon Eclipse E600Fn confocal microscope. Adobe Photoshop was used to obtain merged images and to adjust brightness and contrast.
Protein analysis
Cell lysates were prepared by incubating cells with RIPA114 buffer [50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 5 mM EDTA, 1% Triton X-114, 0.2% SDS, protease inhibitor cocktail (1:100; Sigma)] for 1 hour on ice, then centrifuging at 14,000 rpm for 15 minutes at 4°C to remove cell debris.
For protein purification, culture supernatant or cell lysates were immunoprecipitated with anti-FLAG M2 affinity gel (Sigma), washed five times with PBS, then eluted with elution buffer [0.1 mg/ml 3× FLAG peptide (Sigma) in PBS]. Protein concentration was determined by silver staining (Pierce, Rockford, IL).
For coimmunoprecipitation experiments, culture supernatant or cell lysates were incubated with anti-FLAG M2 affinity gel (Sigma) or mouse anti-His monoclonal antibody (Sigma) overnight at 4°C. Protein A agarose (Sigma) was added to samples with anti-His antibody and further incubated for 1 hour. Immunoprecipitates were washed with RIPA114 buffer and PBS, then boiled for 5 minutes in SDS sample buffer. For supernatant mixture experiments, culture supernatants from independently transfected dishes were mixed and incubated at 4°C for 24 hours prior to immunoprecipitation.
Hydrophobic and hydrophilic fractions from the retina and the olfactory bulb of mice were prepared by using a Mem-PER kit (Pierce) and a PAGEprep advance kit (Pierce). Western blots were performed as described elsewhere (Sambrook et al., 1989) using 15% SDS-PAGE gels. Antibodies used were mouse anti-FLAG monoclonal antibody (1:1,000; Sigma; No. F3165, clone M2) recognizing the peptide DYKDDDK, mouse anti-His monoclonal (1: 1,000; Sigma; No. H1029, clone HIS-1) recognizing the peptide HHHHHH, rabbit anti-CPG15-2 (1:100), HRP-conjugated anti-mouse IgG (1:5,000; Jackson Immunoresearch, West Grove, PA), and HRP-conjugated anti-rabbit IgG (1:50,000; Jackson Immunoresearch). Bands were detected by ECL (Amersham, Piscataway, NJ).
Glycosylation was examined with an enzymatic deglycosylation kit (Prozyme, San Leandro, CA). Thirty nanograms FLAG-tagged CPG15 or CPG15-2 purified from culture supernatant of stably transfected HEK293 cells or 36 μg olfactory lysates were denatured and digested with N-linked glycosylation-specific N-glycanase, then with O-linked glycosylation-specific sialidase, β(1–4)-galactosidase, and β-N-acetylglucosaminidase according to the manufacturer’s instructions. CPG15 and CPG15-2 proteins were detected by Western blot analysis using the anti-FLAG antibody or the anti-CPG15-2 antibody.
Phospholipase C treatment and immunocytochemistry
HEK293 cells stably transfected with FLAG-tagged cpg15 or cpg15-2 were plated on poly-L-lysine-coated coverslips. Phosphatidylinositol-specific phospholipase C (Sigma) was added to the media at 1 U/ml and incubated for 4 hours. Mouse anti-FLAG monoclonal antibody (M2; 1:1,000; Sigma) was added directly to the media and incubated for 30 minutes at 37°C. After three rinses with PBS, cells were fixed with 4% formaldehyde for 15 minutes at 4°C, blocked for 1 hour with 10% goat serum in PBS, then incubated with an Alexa 555-conjugated goat anti-mouse IgG antibody (1:500; Jackson Immunoresearch) for 30 minutes. Images were acquired with an epifluorescent microscope equipped with a Spot camera. Phospholipase C-treated and nontreated cells were imaged under identical conditions. No anti-FLAG antibody staining was observed in HEK293 cells transfected with a control EGFP expression vector (data not shown). Adobe Photoshop was used to obtain merged images and to adjust brightness and contrast.
Hippocampal explant assay
Brains were removed from P3–5 Sprague-Dawley rats. Hippocampi were isolated, then further trimmed into 100–300-μm pieces using a fine tungsten needle knife. Isolated explants were embedded in a 4:3:1 mixture of rat tail collagen, matrigel (Collaborative Research, Bedford, MA), and DMEM, together with HEK293 cell aggregates expressing FLAG-tagged constructs placed at a distance ranging from 100–400 μm (Zhu et al., 1999). Cell aggregates were prepared by the hanging-drop method (Fan and Tessier-Lavigne, 1994). After collagen matrices were solidified, they were cultured in DMEM with 10% fetal bovine serum and 100 μg/ml of penicillin and streptomycin at 37°C in an incubator with 5% CO2 for 60–72 hours. Explants were then fixed and stained for immunocytochemistry with an antineurofilament M rabbit polyclonal antibody (Chemicon, Temecula, CA) to visualize neuronal processes (Li et al., 1999).
Explants were imaged with a Nikon Eclipse E600Fn confocal microscopy system. Images were acquired with Simple PCI software (version 3.5.0.1309; Compix Inc. Image System, Cranberry Township, PA) and analyzed blind to experimental conditions using Object-Image software (http://simon.bio.uva.nl/object-image.html) for process tracing with Morphometry Macros (Ruthazer and Cline, 2002). Number of tips per primary neurite was calculated as total number of tips per explant divided by total primary neurites per explant. Average neurite length was calculated as total neurite length per explant divided by total number of primary neurites. Mean and SEM were calculated from the average value of five to eight explants per condition. Statistical significance was determined by ANOVA and Student-Newman-Keuls post hoc analysis in StatView software (SAS Institute, Cary, NC).
Neurite outgrowth and survival assays
Neurite outgrowth assays were performed as described elsewhere (Lemmon et al., 1989; Nakashiba et al., 2002). Purified protein from culture supernatant of stable lines (18 μl) of indicated concentration was applied in a 50-mm2 circle on nitrocellulose-coated six-well dishes and incubated for 1 hour. Comparison of the protein concentration before and after coating indicated that approximately 80% of the protein adhered to the dish under these conditions. Dishes were then blocked with 10 mg/ml bovine serum albumin (BSA; Sigma) for 30 minutes. Dissociated cortical neurons were prepared as described by Fujino et al. (2003) from cerebral cortices of E19 Sprague-Dawley rats and plated at 1.5 × 105 cells per well. Cells were imaged after 24 hours by phase-contrast microscopy. Neurite length was measured as the direct distance between the center of the soma and the tip of its longest neurite. Counting was blind to experimental conditions. Only large cells with smooth and healthy morphology that were not in contact with neighboring cells were counted. Mean and SEM were calculated from results of three independent experiments. For the survival assay, the same cultures were stained at 24 hours after plating with a Live/Dead Viability/Cytotoxicity Kit (Molecular Probes), and the number of live and dead cells was counted. Increase in survival rate over BSA control levels from three independent experiments was analyzed. Statistical significance was determined by ANOVA and Student-Newman-Keuls post hoc analysis in StatView software (SAS Institute).
Results
cpg15-2 is a novel cpg15 paralogue
To identify cpg15 paralogues, we searched the Genbank database for genes encoding similar proteins. One mouse cDNA clone, G630049C14 (GenBank accession No. AK090312), shared 28% identity and 50% similarity at the amino acid level with mouse CPG15 (Fig. 1A). Henceforth, we refer to this gene as cpg15-2 (GenBank accession No. DQ176852). No other mouse sequence with significant similarity was found, suggesting that cpg15 and cpg15-2 are the only members of this gene family in mouse.
Fig. 1.
cpg15-2 is a new cpg15 paralogue. A: Alignment of the mouse CPG15 and CPG15-2 predicted amino acid sequences. Identical residues are marked by asterisks, and similar residues are marked by dots. Conserved cysteines are boxed. Predicted signal peptides at the N-termini and glycosylphosphatidylinositol (GPI) anchoring signals at the C-termini are underscored. Exon/intron boundaries are marked by arrows. B: Comparison of mouse cpg15 and mouse cpg15-2 genomic structures. Exons, indicated by solid boxes, are numbered in sequence. C: Phylogenetic analysis of the cpg15 gene family of selected species. All the family members in various species belong to either the cpg15 branch or the cpg15-2 branch. Accession numbers are indicated in parentheses. See Supplemental Table 2 for a complete list of known cpg15 gene family members.
The predicted CPG15-2 protein is 162 amino acids long, 20 amino acids longer than CPG15, largely because of an insertion near its C-terminus (Fig. 1A). The two proteins share multiple structural features, including an N-terminal signal sequence and a C-terminal GPI anchor sequence. All six cysteines of CPG15 are conserved in CPG15-2, suggesting they may be crucial for function.
cpg15 and cpg15-2 are also similar in their genomic structure (Fig. 1B). Both genes contain three exons with exon/intron boundaries at equivalent positions in the corresponding protein sequence (Fig. 1A). This is despite the divergence of their nucleotide sequence to a point where no similarity could be detected using the BLASTN program, including both protein coding and promoter regions. The cpg15-2 gene is more compact than cpg15, with shorter introns.
To examine the cross-species conservation of the cpg15 gene family and to identify additional family members, we searched for orthologues in other species. The human, rat, and cow genomes were found to contain one copy each of cpg15 and cpg15-2 (Fig. 1C, Suppl. Table 2). In human, an additional cDNA clone, DKFZp761P1315 (GenBank accession No. AL390160), showed significant homology to CPG15, but the homologous region was not part of an open reading frame, suggesting that it may be a pseudogene. In pufferfish, Fugu rubripes, five candidate genes had significant similarity to CPG15 (Fig. 1C), with three more similar to CPG15 and two similar to CPG15-2. Zebrafish had four candidate genes similar to CPG15 or CPG15-2 (Suppl. Table 2). This suggests that, although some of the fish genes may be pseudogenes, fish may have additional CPG15 and CPG15-2 family members. CPG15 is more highly conserved across species, with a homology of 97% between human and mouse, as opposed to 79% for CPG15-2. No gene with significant homology was found in nonvertebrate species sequenced to date including C. elegans and Drosophila. Conservation of cpg15 and cpg15-2 orthologues across species and their similar genomic structure suggest that the two genes arose through a gene duplication event early in vertebrate evolution.
cpg15-2 is an activity-regulated gene with regional and developmental expression profiles distinct from those of cpg15
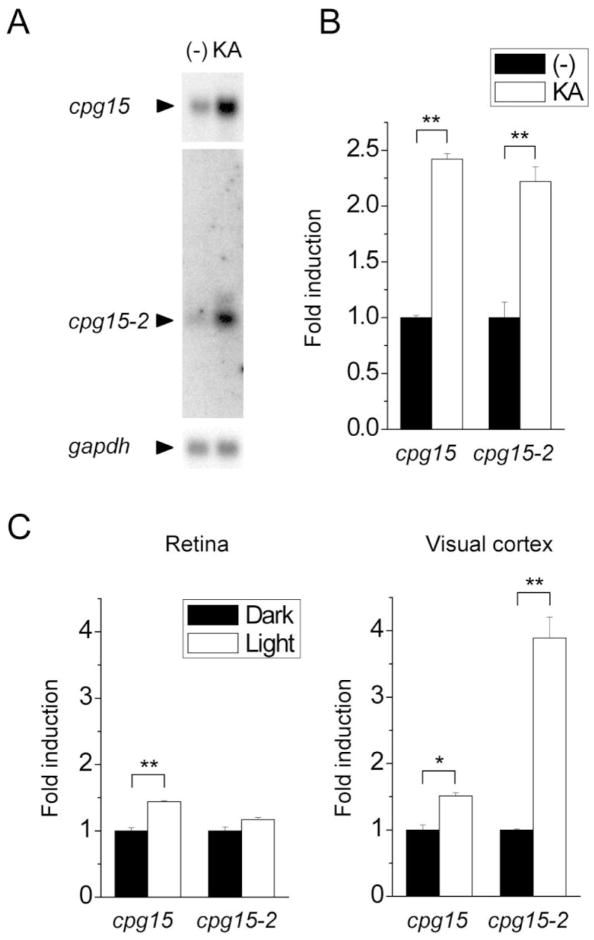
To examine the extent to which cpg15 and cpg15-2 expression was similarly regulated, we first compared their activity-dependent transcriptional regulation. On a Northern blot, the cpg15-2 transcript could be detected as a faint 0.9-kb band corresponding in size to the predicted mRNA sequence (Fig. 2A). Because cpg15 was first characterized as an activity-regulated gene (Nedivi et al., 1993), we tested whether cpg15-2 expression is also regulated by neural activity. We injected mice with kainate to induce strong synchronized neural activity, then compared cpg15 or cpg15-2 expression in brains of kainate-injected and uninjected control mice. The intensity of the band corresponding to cpg15 mRNA increased in response to kainate stimulation (Fig. 2A; Fujino et al., 2003). cpg15-2 mRNA showed a similar increase in response to kainate (Fig. 2A), suggesting that cpg15-2 is also an activity-regulated gene. Quantitative measurement using real-time RT-PCR indicated that both cpg15 and cpg15-2 mRNA increased by more than twofold relative to their basal expression levels (Fig. 2B). The cpg15-2 promoter showed no significant homology to the cpg15 promoter by BLAST-N searches but contained potential binding sites for CREB, USF, and NeuroD (Suppl. Fig. 3), transcription factors implicated in activity-dependent transcription (Shieh et al., 1998; Tao et al., 1998; Chen et al., 2003; Aizawa et al., 2004; Gaudilliere et al., 2004).
Fig. 2.
cpg15 and cpg15-2 are both activity-regulated genes. A: cpg15 and cpg15-2 expression in the brain is induced by kainate. cpg15 and cpg15-2 expression in cerebral cortices of uninjected (—) and kainate-injected (KA) mice was examined by Northern blot analysis. The blot was reprobed with a gapdh probe as a loading control. B: Quantitative measurement of kainate-induced cpg15 and cpg15-2 expression by real-time quantitative reverse transcription polymerase chain reaction (RT-PCR). Expression level of each gene was normalized to gapdh expression levels. Fold induction of each gene relative to its basal expression level in uninjected controls is plotted. Data are represented as mean ± standard error of mean. **P < 0.01, n = 5 for KA, n = 3 for controls. C: Induction by light of cpg15 and cpg15-2 in visual structures. Mice were placed in the dark for 4 weeks, then either exposed to light for 24 hours or kept in the dark. cpg15 and cpg15-2 expression in the retina and the visual cortex was measured by real-time RT-PCR and plotted as in B. *P < 0.05, **P < 0.01, n = 3.
To see whether cpg15-2 could be induced by physiological stimuli, we examined its expression in response to 24 hours of light after 4 weeks of dark adaptation. Compared with dark-adapted mice, mice after 24 hours of light exposure showed 3.9-fold the induction of cpg15-2 and 1.5-fold the induction of cpg15 in the visual cortex (Fig. 2C). Changes in retinal expression were smaller, 1.2-fold and 1.4-fold induction for cpg15-2 and cpg15, respectively. These results indicate that expression of both cpg15-2 and cpg15 is responsive to physiological levels of sensory stimulation, but induction levels differ between the two genes and are region dependent.
In comparing their tissue distributions, we found that cpg15 mRNA was most abundant in the brain and liver, whereas cpg15-2 mRNA was most abundant in the eye and brain, with lower levels in the testis (Fig. 3A). Within the central nervous system, cpg15 and cpg15-2 mRNAs displayed distinct distribution patterns (Fig. 3B). cpg15-2 was expressed most abundantly in the retina, followed by the olfactory bulb and striatum, whereas cpg15 mRNA was most abundant in the cerebral cortex, followed by the hippocampus and thalamus. cpg15 was expressed at higher levels than cpg15-2 in most regions, except for the retina and striatum, where cpg15-2 expression was higher.
Fig. 3.

cpg15 and cpg15-2 show distinct and partially overlapping regional and developmental expression profiles. Tissue distribution (A), distribution in various central nervous system regions (B), and developmental expression (C) of each gene as examined by real-time RT-PCR. Copy number of each message per 1 ng total RNA is shown in A, and expression levels relative to gapdh are shown in B,C. Means ± standard error of mean are plotted (n = 3). Br, brain; Ey, eye; Hr, heart; Lu, lung; Li, liver; Ki, kidney; Sp, spleen; In, intestine; Mu, muscle; Te, testis; Rt, retina; Ol, olfactory bulb; Cx, cerebral cortex; Hp, hippocampus; Cb, cerebellum; St, striatum; Th, thalamus; Mi, midbrain; Me, medulla; E, embryonic day; P, postnatal day.
We next examined cpg15 and cpg15-2 developmental expression profiles in the brain. Expression of both paralogues increased during development and was highest in 11-week-old adults (Fig. 3C). However, onset of expression was different between the two genes. cpg15 was already expressed at about 30% of adult levels at E12.5, whereas cpg15-2 expression remained low during embryonic development and increased around birth between E17.5 and P1. Interestingly, both genes showed a large increase postnatally between 2 and 4 weeks, suggesting a role in late developmental events, such as activity-dependent refinement. In summary, although both paralogues are expressed primarily in the nervous system and are regulated by neural activity, they are distinguished by distinct spatial and temporal expression profiles.
CPG15-2 protein expression in the retina and the olfactory bulb
To characterize CPG15-2 protein expression, we raised antibodies against a C-terminal peptide corresponding to the insert region of CPG15-2 not present in CPG15. We first tested the specificity of the antibody on Western blots of FLAG-tagged CPG15 and CPG15-2 protein expressed and purified from HEK293 cells. The anti-CPG15-2 antibody recognized CPG15-2 but not CPG15 (Fig. 4A). The antibody signal from CPG15-2 could be competed with by a CPG15-2 peptide, further demonstrating the specificity of this antibody to CPG15-2.
Fig. 4.
CPG15-2 is expressed in the retina and the olfactory bulb. A: The anti-CPG15-2 antibody is specific to CPG15-2. Ten nanograms of FLAG-tagged CPG15-2 or CPG15 purified from culture supernatants of stably transfected HEK293 cells was probed on a Western blot with an anti-CPG15-2 antibody. The anti-CPG15-2 antibody recognizes FLAG-tagged CPG15-2 bands but does not cross-react with FLAG-tagged CPG15. CPG15-2 bands are significantly reduced by the addition of CPG15-2 peptide. Probing the blot with anti-FLAG antibody shows similar loading of FLAG-tagged CPG15 and CPG15-2. B: Forty micrograms of hydrophobic and hydrophilic protein fractions from the retina and olfactory bulb were probed with the anti-CPG15-2 antibody. Right blot shows competition by the CPG15-2 peptide. O, hydrophobic fraction; I, hydrophilic fraction; Olf, olfactory.
We then used the anti-CPG15-2 antibody to examine expression of the endogenous protein in the retina and olfactory bulb, the two CNS regions showing the highest cpg15-2 mRNA levels. Because of CPG15-2’s potential membrane association, we separated lysates into hydrophilic and hydrophobic fractions by using a detergent-based method. Western blot analysis with the anti-CPG15-2 antibody revealed the same bands in the hydrophobic fractions derived from both regions, the most intense band being of 13 kDa (Fig. 4B). This is close to the predicted size of 11.3 kDa for CPG15-2 after the cleavage of signal peptide and the GPI anchoring signal, assuming that the mobility may be slightly slowed by addition of the GPI moiety. The CPG15-2 bands could be eliminated by competition with the CPG15-2 peptide. The presence of most CPG15-2 in the hydrophobic fraction suggests that in vivo CPG15-2 is predominantly membrane associated, likely residing in the plasma membrane (but possibly also in cytoplasmic organelles, such as the endoplasmic reticulum or the Golgi apparatus).
FLAG-tagged CPG15-2 expressed in HEK293 cells migrated at a size larger than endogenous CPG15-2 in the retina and olfactory bulb (see Fig. 4A,B). We tested whether this larger size could be accounted for by CPG15-2 glycosylation, by treating CPG15-2 with a glycosidase and assaying for a size change on Western blots. Glycosidase treatment of CPG15-2 expressed in HEK293 cells resulted in a marked shift in band size to 13 and 15 kDa (Suppl. Fig. 4A). In contrast, glycosidase had no effect on the olfactory bulb protein (Suppl. Fig. 4B), suggesting that CPG15-2 undergoes aberrant glycosylation in HEK293 cells.
Differential expression of cpg15 and cpg15-2 in the retina
Other members of multigene families with differential expression patterns are sometimes preferentially expressed in different cell types within the same tissue. To examine CPG15-2 tissue localization further, we stained the retina with anti-CPG15-2 antibody. We found CPG15-2 immunofluorescence in the outer plexiform layer, and a fiber-type staining could be seen spanning the inner nuclear layer and inner plexiform layer (Fig. 5A,B). Anti-CPG15-2 immunoreactivity could be competed away by the CPG15-2 peptide against which the antibody was generated, but not by a CPG15 peptide (Fig. 5A), indicating that the staining is CPG15-2 specific.
Fig. 5.
CPG15 and CPG15-2 are expressed by different cell types in the retina. A: CPG15-2 immunohistochemistry. Retinal sections were immunostained with the anti-CPG15-2 antibody (magenta), then counterstained with 4′,6-diamidino-2-phenylindole (DAPI; green). CPG15-2 is localized to the outer plexiform layer, inner nuclear layer, and inner plexiform layer. Anti-CPG15-2 staining was competed out by the addition of a CPG15-2 peptide but not by a CPG15 peptide. B: PKCα-positive rod bipolar cells express CPG15-2. Dual staining of the retina with anti-CPG15-2 (magenta) and anti-PKCα (green) antibodies shows colocalization of CPG15-2 with PKCα (white) in the axons of rod bipolar cells. Not all CPG15-2-stained fibers are PKCα positive, as indicated by the arrows. Maximum intensity projections of five confocal images taken at 1-μm intervals are shown. C: In situ hybridization on retinal sections probed for cpg15. cpg15 signal can be seen in the ganglion cell layer. Signal in the pigment epithelium is due to nonspecific light scattering. PE, pigment epithelium; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. Scale bars = 50μm.
Because the fiber-like CPG15-2 staining resembled bipolar cell axons, we double stained the retina with antibodies against CPG15-2 and PKCα, a rod bipolar cell marker (Grunert and Martin, 1991). Staining with anti-PKCα antibody showed an immunoreactivity pattern similar to that previously described, with the classic morphology of bipolar cells outlined in the outer plexiform, inner nuclear, and inner plexiform layers (Grunert and Martin, 1991). Approximately 90% of the fiber-like CPG15-2 staining colocalized with PKCα-positive fibers (Fig. 5B), indicating that rod bipolar cells express CPG15-2. Anti-CPG15-2 stained fibers extending in a similar orientation and negative for PKCα (Fig. 5B) indicate that other cell types, most likely cone bipolar cells, also express CPG15-2. Anti-CPG15-2 staining was low or undetectable in the ganglion cell layer and the outer nuclear layer, suggesting that it is not expressed by ganglion or photoreceptor cells. These results show that CPG15-2 expression in the retina is restricted to cells in the inner nuclear layer, including rod bipolar cells, where it is preferentially localized to the axons.
We were unable to generate or obtain an anti-CPG15 antibody suitable for immunohistochemistry, so we performed in situ hybridization to assay for cpg15 retinal expression. cpg15 mRNA was found predominantly in the ganglion cell layer (Fig. 5C), suggesting that ganglion cells or displaced amacrine cells are the major retinal cell types expressing cpg15. Thus, in the retina, the two genes are expressed by distinct cell types, cpg15-2 in bipolar cells and cpg15 in ganglion or amacrine cells.
Extracellular localization and posttranslational modification of CPG15-2
Localization of CPG15-2 in hydrophobic fractions of brain extracts suggests that it may be membrane linked via a GPI anchor similar to CPG15. To test for this possibility, we immunostained HEK293 cells expressing FLAG-tagged CPG15 or CPG15-2 with an anti-FLAG antibody. Under nonpermeabilizing staining conditions, the two proteins showed a similar punctate membrane staining (Fig. 6A), indicating that they are present on the outer cell surface. Addition of phospholipase C, which cleaves the GPI anchor, significantly reduced the surface staining of CPG15 and CPG15-2 (Fig. 6A), showing that CPG15-2 is GPI anchored on the cell surface.
Fig. 6.
CPG15-2 is a GPI anchored protein expressed in membrane-bound and soluble-secreted forms. A: CPG15-2 is a GPI-anchored membrane protein. HEK293 cells expressing FLAG-tagged CPG15 or CPG15-2 were immunostained with an anti-FLAG antibody (magenta) under nonpermeabilizing conditions. EGFP (green) coexpressed from the same vector marks the transfected cells. Both CPG15 and CPG15-2 show a punctate staining on the cell surface that is significantly reduced by phospholipase C treatment prior to immunostaining. B: Coexisting membrane-bound and soluble forms of CPG15 and CPG15-2. Cell lysate and culture supernatant from HEK293 cells stably transfected with FLAG-tagged cpg15, cpg15-2, or empty vector (—) were immunoprecipitated with an anti-FLAG antibody, then probed on a Western blot with an anti-FLAG antibody. PLC, phospholipase C; cell, cell lysate fraction; sup, culture supernatant fraction. Scale bar = 20 μm.
CPG15 is a GPI anchored protein that is also secreted in a soluble form (Putz et al., 2005). To see whether the same is true for CPG15-2, we immunoprecipitated FLAG-tagged proteins from the cell lysate and culture supernatant of transfected HEK293 cells using an anti-FLAG antibody and visualized them on a Western blot using the same antibody. As with CPG15, CPG15-2 was present in both cell lysate and culture supernatant fractions (Fig. 6B), indicating that CPG15-2 is secreted. The soluble CPG15 was similar in size to the CPG15 in the cell lysate fraction, whereas the soluble CPG15-2 was about 1 kDa smaller than the CPG15-2 in the cells. In addition, the relative abundance of secreted protein was lower for CPG15-2 than for CPG15. These findings suggest that, although both proteins are secreted, CPG15-2 may undergo different types of processing to generate its soluble forms.
CPG15 and CPG15-2 form homo- and heterodimers
On Western blots, purified CPG15 and CPG15-2 both presented as multiple bands, with the higher molecular weight bands approximately twice the size of the predicted monomer, suggestive of dimer formation (see Fig. 6B). To test whether CPG15 and CPG15-2 form homodimers and perhaps heterodimers, we performed coimmunoprecipitation experiments on extracts from HEK293T cells cotransfected with FLAG- and His-tagged CPG15 or CPG15-2. We found that His-tagged CPG15 or CPG15-2 coimmunoprecipitated with both FLAG-tagged CPG15 and CPG15-2 (Fig. 7A). This demonstrates that CPG15 and CPG15-2 can form both homodimers and heterodimers with each other.
Fig. 7.
CPG15 and CPG15-2 form both homo- and heterodimers. A: Coimmunoprecipitation of CPG15 and CPG15-2. cpg15 and cpg15-2 each tagged with either FLAG or poly-His were cotransfected into HEK293T cells as indicated. Cell lysates of the transfected cells were immunoprecipitated with an anti-FLAG antibody, then probed with an anti-His antibody to detect coprecipitating proteins. CPG15 and CPG15-2 both form homodimers and can also interact with each other. B: Dimerization requires cellular processing. CPG15 or CPG15-2 was immunoprecipitated either from culture supernatant of HEK293T cells cotransfected with FLAG- and His-tagged constructs or from a mixture of two supernatants from cells independently transfected with FLAG- or His-tagged constructs. Immunoprecipitation and Western blots were done using the indicated antibodies. Coimmunoprecipitation was observed from the supernatant of cotransfected cells but not from the supernatant mixture of independently transfected cells, indicating that dimerization does not occur in a cell-free solution. Similar amounts of FLAG- and His-tagged protein were present in both supernatants. Co-tf, cotransfected; mix, supernatants mixed; IP, immunoprecipitation.
Because CPG15 and CPG15-2 are extracellular proteins, we examined whether dimer formation could occur in solution or requires coexpression for correct assembly. We mixed and incubated culture supernatants from two independent HEK293T cultures, one transfected with FLAG-tagged and the other with His-tagged constructs, for 24 hours at 4°C. Neither CPG15 nor CPG15-2 FLAG-and His-tagged proteins were coimmunoprecipitated from mixed supernatants (Fig. 7B). Positive controls of culture supernatants from HEK293T cells cotransfected with both FLAG- and His-tagged constructs did show coimmunoprecipitation under similar conditions with similar amounts of each protein. These results suggest that dimers formed during intracellular processing are stable, with little exchange reaction, and are thus unlikely to occur in cell-free solution.
CPG15 and CPG15-2 promote neurite extension with similar efficacy
Conservation of many biochemical features between CPG15 and CPG15-2 suggests that their functions might also be similar. Overexpression of CPG15 in Xenopus has been shown to promote axonal and dendritic arbor growth (Nedivi et al., 1998; Cantallops et al., 2000), and purified recombinant CPG15 promotes neurite outgrowth and arborization in primary embryonic hippocampal and cortical cultures (Naeve et al., 1997). To compare the effects of CPG15 and CPG15-2 on neurite growth, hippocampal explants were cocultured in a collagen/matrigel matrix with HEK293 cell aggregates expressing CPG15 or CPG15-2. Sixty to seventy-two hours later, the neurites extending from each explant were measured for length and branching. We found that, when explants were cocultured with CPG15 or CPG15-2 expressing cells, their neurites were significantly longer than those from explants cocultured with control HEK293 cells (Fig. 8A,B). Explants cocultured with CPG15- or CPG15-2-expressing HEK293 cells also had significantly more branch tips per neurite compared with control explants (Fig. 8C), indicating that CPG15 and CPG15-2 promote neurite growth and branching.
Fig. 8.
CPG15 and CPG15-2 promote neurite extension and branching in hippocampal explants. A: Maximum intensity projection confocal images of representative explants cocultured with control HEK293, CPG15-expressing, or CPG15-2-expressing cell aggregates for 60–72 hours, then fixed and immunostained for Neurofilament M to visualize neuronal processes. Brightness and contrast were adjusted in Photoshop. B: Quantification of average neurite length measured as tip distance from the explant. Explants cocultured with CPG15- or CPG15-2-expressing HEK293 cells have significantly longer neurites. *P < 0.01. C: Quantification of branching measured as number of branch tips per each neurite growing out of an explant. Explants cocultured with CPG15- or CPG15-2-expressing HEK293 cells have significantly more branch tips (*P < 0.01). Scale bar = 100 μm.
To compare the growth promoting functions of CPG15 and CPG15-2 quantitatively, we also performed the neurite extension assay on dissociated cortical neurons. Dissociated neurons were plated on dishes coated with FLAG-tagged CPG15 or CPG15-2 purified from the culture supernatant, or with BSA, and neurite growth was assayed 24 hours later. Neurons plated on CPG15- or CPG15-2-coated dishes had significantly longer neurites than those plated on BSA-coated dishes (Fig. 9A,B), confirming the growth-promoting effect of these proteins observed in the hippocampal explant assay. When dishes were coated with a control solution purified from the supernatant of empty-vector-transfected HEK293 cells, no effect on neurite growth was observed, demonstrating that the positive growth effect of CPG15 and CPG15-2 is not due to contaminating material from the HEK293 cells or the culture media (data not shown). We compared the efficacy of CPG15 and CPG15-2 by assaying neurite growth in neurons plated on dishes with a range of protein concentrations. Neurons showed a similar dose-response curve for the growth-promoting activity of CPG15 and CPG15-2. For both proteins, neurite growth could be seen from concentrations starting at 20 ng/cm2 and was saturated at approximately 50 ng/cm2 (Fig. 9B). We did not find a statistically significant effect of CPG15 and CPG15-2 on neurite branching, possibly because of the shorter culture period for the dissociated cortical cultures (24 hours) compared with the hippocampal explants (60–72 hours).
Fig. 9.
CPG15 and CPG15-2 have similar neurite growth and survival effects. Dissociated cortical neurons were plated on dishes coated with bovine serum albumin (BSA) or purified CPG15 or CPG15-2, and imaged 24 hours later. A: Representative phase-contrast images of cortical neurons plated on BSA-, CPG15-, or CPG15-2-coated dishes. B: Neurite length of neurons plated on different concentrations of CPG15 (open bars) or CPG15-2 (solid bars; *P < 0.05). Neurite length in BSA controls is indicated by the dotted line. C: Nonadditive effects of CPG15 and CPG15-2 on neurite growth. Neurons were plated on CPG15-, CPG15-2-, or CPG15- and CPG15-2-coated dishes (20 ng/cm2 for each protein). The percentage of neurons (ordinate) with neurites longer than the given length (abscissa) is plotted for each treatment. D: Survival rate of neurons plated on different concentrations of CPG15 (open bars) or CPG15-2 (solid bars; *P < 0.05). Increase in survival rate over baseline levels in BSA controls is plotted. Scale bar = 100 μm.
To examine whether CPG15 and CPG15-2 have additive effects on neurite growth, we plated neurons on dishes coated with both CPG15 and CPG15-2. To assess the changes within the population more precisely, we plotted the distribution of neurons according to their neurite length (Fig. 9C). CPG15 and CPG15-2 each showed a significant shift in the distribution curve compared with BSA controls, confirming their neurite growth effect. Co-application of CPG15 and CPG15-2 produced a distribution curve almost overlapping that of CPG15 and CPG15-2, showing the absence of any further additive effect compared with application of either of the individual proteins. These results suggest that both proteins affect similar subpopulation of neurons within the cerebral cortex.
In summary, CPG15 and CPG15-2 enhance neurite growth with similar efficacy in both dissociated cortical neurons and hippocampal explant cultures. Both proteins also promote neurite branching in the explant culture system.
CPG15 and CPG15-2 promote survival of cortical neurons
Another known function of CPG15 is promoting survival of developing and mature neurons (Putz et al., 2005). To test whether CPG15-2 also acts as a neuronal survival factor, we differentially stained live and dead cells in the same dissociated cultures used for the neurite growth assay. In control dishes coated with BSA, only about 30% of the cells survive, likely because of the initial damage from dissociation and plating. In dishes coated with CPG15 or CPG15-2, cells showed increased survival compared with the survival rate of cells plated on BSA-coated dishes (Fig. 9D). Significant survival was observed with addition of CPG15 or CPG15-2 at concentrations above 20 ng/cm2, indicating that survival-promoting and neurite growth functions are effective at similar doses.
Discussion
Here we describe for the first time cpg15-2, an activity-regulated gene encoding an extracellular protein that promotes neurite growth and neuronal survival and the only paralogue of the previously characterized cpg15. By examining the similarities and differences between these genes and their proteins, we can begin to understand the conserved features likely to be important for function and divergent properties that may underlie specific in vivo roles. We find that CPG15-2 and CPG15 differ in their expression profiles and posttranslational processing but share basic biochemical properties and cellular function.
Differential expression of cpg15 and cpg15-2
cpg15 and cpg15-2 are members of a novel gene family in vertebrates that likely arose through gene duplication. The paralogues have diverged to a point where no similarity is detectable at the nucleotide level, including coding and regulatory regions, suggesting the evolution of distinct in vivo roles. Functionally, the divergence of transcriptional control regions may relate to the different tissue distribution as well as temporal and regional expression profiles seen for cpg15 and cpg15-2. cpg15 is most abundant in the cerebral cortex, whereas cpg15-2 is most abundant in the retina. Both genes gradually increase their expression during embryogenesis and postnatal development, but onset of cpg15 expression is earlier than that of cpg15-2. In the retina, the two genes are expressed in distinct cell types. This suggests that, even in regions where the two genes are seemingly “coexpressed,” higher resolution examination could reveal mutually exclusive expression of cpg15 or cpg15-2 in specific cell types within the region. Another example of mutually exclusive expression of gene family members in the retina, where they are seemingly coexpressed, can be seen in the cell adhesion molecules called Sidekicks (Yamagata et al., 2002). Two family members are expressed in the same layers of the retina, yet in nonoverlapping subsets of neurons, thereby guiding lamina-specific synaptic connections.
It is interesting that, despite differential expression patterns of CPG15 and CPG15-2, both genes are induced in response to increased activity. There are several potential binding sites for activity-regulated transcription factors, including CREB, in the cpg15-2 promoter, but their number and positions are different from those in the cpg15 promoter. Thus, cpg15-2 may be utilizing a distinct set of transcription factors to achieve activity dependence in different cell types. This may also account for the difference in the magnitude of light-induced expression of the two genes in the visual cortex. The fact that the two promoters evolved to support divergent expression patterns but still retained activity dependence suggests that regulation by activity is a critical part of their in vivo function. Activity-dependent regulation adds another layer of specificity to the expression pattern of cpg15 and cpg15-2. Even within regions and cell types capable of expressing these genes, the expression level in a particular neuron at any time will depend on the level of its synaptic input.
Biochemical and functional properties
Although both coding and noncoding regions are significantly diverged between cpg15 and cpg15-2, their protein structure retains common features, such as the signal peptide, GPI anchoring signal, and conserved cysteines. In addition, CPG15 and CPG15-2 both exist in a GPI anchored membrane-bound form and a soluble-secreted form. A GPI anchor can direct proteins to lipid rafts (Tsui-Pierchala et al., 2002), so it may serve to target CPG15 and CPG15-2 to a subregion of the cell membrane, thus increasing their local concentration. In this way, lower protein levels would be required to pass the threshold for neurite growth and survival (20 ng/cm2 in dissociated cultures), perhaps enhancing the in vivo efficacy of CPG15 and CPG15-2. CPG15-2 is localized preferentially but not exclusively to axons, as is CPG15 (Nedivi et al., 2001), consistent with axonal targeting of GPI-anchored proteins (Dotti et al., 1991). The GPI anchor may also provide a mechanism for generating both membrane-bound and soluble-secreted proteins from the same gene. Indeed, several other GPI anchored proteins are also known to be secreted (Faivre-Sarrailh and Rougon, 1997).
The significance of coexisting soluble-secreted and membrane-bound forms may be in long distance vs. localized effects. The conversion from the membrane-bound to the soluble-secreted form would likely increase the effective range of each protein through dispersion from the expressing cell. In this respect, it is interesting to note the difference in quantity and quality of the CPG15- and CPG15-2-soluble forms. CPG15 is secreted more efficiently than CPG15-2 by HEK293 cells. Comparison of the protein size between cell and supernatant fractions shows no difference for CPG15 but a marked reduction of CPG15-2 size in the supernatant compared with the cell fraction, hinting at different posttranslational processing of the two proteins. The CPG15-2 cleavage site may be farther from the GPI anchor compared with CPG15, thus generating a shorter soluble form. An alternative possibility is that the majority of CPG15 is processed into a soluble form within the cell and stored in vesicles before being presented on the cell surface, so the CPG15 in the cell fraction would be similar in size to that in supernatant fraction. These differences in posttranslational processing would be consistent with more efficient secretion of CPG15.
CPG15 and CPG15-2 form both homodimers and heterodimers, suggesting that dimerization is important for protein function, perhaps by clustering CPG15 and CPG15-2 molecules for effective signaling. Other ligands such as the neurotrophins also exist in a dimeric form (Ibanez, 1998). The ability of CPG15 and CPG15-2 to bind to each other suggests that their tertiary structure at the interaction site is conserved despite only 28% amino acid identity. The presence of dimer bands in Western blots indicates that dimers are stable even after denaturation by boiling, consistent with our finding that dimer formation does not occur passively in solution in the absence of cells. Because both membrane-bound and soluble CPG15 and CPG15-2 exist in dimeric forms, it is likely that dimerization is an intracellular process that occurs soon after addition of the GPI anchor. Intracellular dimerization has several implications. First, CPG15 and CPG15-2 are unlikely to interact homophilically when expressed on the surface of two opposing cells, as seen for some cell adhesion molecules. Second, CPG15 and CPG15-2 are unlikely to serve as receptors for themselves, even if they were capable of bidirectional signaling. Third, if CPG15 and CPG15-2 are expressed in a mutually exclusive manner, as observed in the retina, CPG15/CPG15-2 heterodimers may not exist in vivo, despite the ability of the two proteins to bind to each other in vitro.
In neurite outgrowth and neuronal survival assays, hippocampal explants and cortical neurons responded similarly to CPG15 and CPG15-2, suggesting that the two ligands signal through similar if not identical downstream pathways. The absence of an additive effect of CPG15 and CPG15-2 on neurite growth further supports this notion. Thus, despite the divergence of their primary sequence, CPG15 and CPG15-2 apparently maintained sufficient similarity in tertiary structure to allow binding to a common receptor. Alternatively, their specific receptors co-evolved to support similar cellular functions of each ligand.
In conclusion, CPG15 and CPG15-2 constitute a family of activity-regulated ligands that promote neurite growth and neuronal survival. Most of their basic protein properties are conserved despite low sequence homology, suggesting that properties such as presentation on the cell surface, cleavage in a soluble-secreted form, and ability to dimerize are critical for their function. Divergent features are spatial and temporal expression profiles of CPG15 and CPG15-2 and secretion efficiency. As with other ligand families, differential expression may define their specific in vivo roles. Activity-regulated expression may further restrict their availability in vivo and thus play a role in activity-dependent refinement of neuronal circuits.
Supplementary Material
Acknowledgments
We are grateful for the advice of Dr. Toshiaki Nakashiba on neurite growth assays, Drs. Yi Rao and Guofa Lin on explant assays, and Drs. Connie Cepko and Doug Kim on retinal immunostaining. We thank Dr. Martha Constantine-Paton for use of the confocal microscope, Rachel Schecter for help with RT-PCR experiments, and members of the Nedivi laboratory for critical reading of the manuscript.
Grant sponsor: National Eye Institute; Grant number: EY011894 (to E.N.).
Footnotes
This article includes Supplementary Material available via the Internet at http://www.interscience.wiley.com/jpages/0021-9967/suppmat.
LITERATURE CITED
- Aizawa H, Hu SC, Bobb K, Balakrishnan K, Ince G, Gurevich I, Cowan M, Ghosh A. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science. 2004;303:197–202. doi: 10.1126/science.1089845. [DOI] [PubMed] [Google Scholar]
- Bibel M, Barde YA. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000;14:2919–2937. doi: 10.1101/gad.841400. [DOI] [PubMed] [Google Scholar]
- Cantallops I, Haas K, Cline HT. Postsynaptic CPG15 promotes synaptic maturation and presynaptic axon arbor elaboration in vivo. Nat Neurosci. 2000;3:1004–1011. doi: 10.1038/79823. [DOI] [PubMed] [Google Scholar]
- Chen WG, West AE, Tao X, Corfas G, Szentirmay MN, Sawadogo M, Vinson C, Greenberg ME. Upstream stimulatory factors are mediators of Ca2+-responsive transcription in neurons. J Neurosci. 2003;23:2572–2581. doi: 10.1523/JNEUROSCI.23-07-02572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti CG, Parton RG, Simons K. Polarized sorting of glypiated proteins in hippocampal neurons. Nature. 1991;349:158–161. doi: 10.1038/349158a0. [DOI] [PubMed] [Google Scholar]
- Faivre-Sarrailh C, Rougon G. Axonal molecules of the immunoglobulin superfamily bearing a GPI anchor: their role in controlling neurite outgrowth. Mol Cell Neurosci. 1997;9:109–115. doi: 10.1006/mcne.1997.0609. [DOI] [PubMed] [Google Scholar]
- Fan CM, Tessier-Lavigne M. Patterning of mammalian somites by surface ectoderm and notochord: evidence for sclerotome induction by a hedgehog homolog. Cell. 1994;79:1175–1186. doi: 10.1016/0092-8674(94)90009-4. [DOI] [PubMed] [Google Scholar]
- Fujino T, Lee WC, Nedivi E. Regulation of cpg15 by signaling pathways that mediate synaptic plasticity. Mol Cell Neurosci. 2003;24:538–554. doi: 10.1016/s1044-7431(03)00230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudilliere B, Konishi Y, de la Iglesia N, Yao G, Bonni A. A CaMKII-NeuroD signaling pathway specifies dendritic morphogenesis. Neuron. 2004;41:229–241. doi: 10.1016/s0896-6273(03)00841-9. [DOI] [PubMed] [Google Scholar]
- Grunert U, Martin PR. Rod bipolar cells in the macaque monkey retina: immunoreactivity and connectivity. J Neurosci. 1991;11:2742–2758. doi: 10.1523/JNEUROSCI.11-09-02742.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwell C, Burbach B, Svoboda K, Nedivi E. Regulation of cpg15 expression during single whisker experience in the barrel cortex of adult mice. J Neurobiol. 2005;65:85–96. doi: 10.1002/neu.20176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez CF. Emerging themes in structural biology of neurotrophic factors. Trends Neurosci. 1998;21:438–444. doi: 10.1016/s0166-2236(98)01266-1. [DOI] [PubMed] [Google Scholar]
- Javaherian A, Cline HT. Coordinated motor neuron axon growth and neuromuscular synaptogenesis are promoted by CPG15 in vivo. Neuron. 2005;45:505–512. doi: 10.1016/j.neuron.2004.12.051. [DOI] [PubMed] [Google Scholar]
- Lee WCA, Nedivi E. Extended plasticity of visual cortex in dark-reared animals may result from prolonged expression of genes like cpg15. J Neurosci. 2002;22:1807–1815. doi: 10.1523/JNEUROSCI.22-05-01807.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon V, Farr KL, Lagenaur C. L1-mediated axon outgrowth occurs via a homophilic binding mechanism. Neuron. 1989;2:1597–1603. doi: 10.1016/0896-6273(89)90048-2. [DOI] [PubMed] [Google Scholar]
- Li HS, Chen JH, Wu W, Fagaly T, Zhou L, Yuan W, Dupuis S, Jiang ZH, Nash W, Gick C, Ornitz DM, Wu JY, Rao Y. Vertebrate slit, a secreted ligand for the transmembrane protein roundabout, is a repellent for olfactory bulb axons. Cell. 1999;96:807–818. doi: 10.1016/s0092-8674(00)80591-7. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Maisonpierre PC, Belluscio L, Friedman B, Alderson RF, Wiegand SJ, Furth ME, Lindsay RM, Yancopoulos GD. NT-3, BDNF, and NGF in the developing rat nervous system: parallel as well as reciprocal patterns of expression. Neuron. 1990;5:501–509. doi: 10.1016/0896-6273(90)90089-x. [DOI] [PubMed] [Google Scholar]
- McLaughlin T, Hindges R, O’Leary DD. Regulation of axial patterning of the retina and its topographic mapping in the brain. Curr Opin Neurobiol. 2003;13:57–69. doi: 10.1016/s0959-4388(03)00014-x. [DOI] [PubMed] [Google Scholar]
- Naeve GS, Ramakrishnan M, Rainer K, Hevroni D, Citri Y, Theill LE. Neuritin: a gene induced by neural activity and neurotrophins that promotes neuritogenesis. Proc Natl Acad Sci U S A. 1997;94:2648–2653. doi: 10.1073/pnas.94.6.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashiba T, Nishimura S, Ikeda T, Itohara S. Complementary expression and neurite outgrowth activity of netrin-G subfamily members. Mech Dev. 2002;111:47–60. doi: 10.1016/s0925-4773(01)00600-1. [DOI] [PubMed] [Google Scholar]
- Nedivi E, Hevroni D, Naot D, Israeli D, Citri Y. Numerous candidate plasticity-related genes revealed by differential cDNA cloning. Nature. 1993;363:718–722. doi: 10.1038/363718a0. [DOI] [PubMed] [Google Scholar]
- Nedivi E, Fieldust S, Theill LE, Hevron D. A set of genes expressed in response to light in the adult cerebral cortex and regulated during development. Proc Natl Acad Sci U S A. 1996;93:2048–2053. doi: 10.1073/pnas.93.5.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedivi E, Wu GY, Cline HT. Promotion of dendritic growth by CPG15, an activity-induced signaling molecule. Science. 1998;281:1863–1866. doi: 10.1126/science.281.5384.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedivi E, Javaherian A, Cantallops I, Cline HT. Developmental regulation of CPG15 expression in Xenopus. J Comp Neurol. 2001;435:464–473. doi: 10.1002/cne.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putz U, Harwell C, Nedivi E. Soluble CPG15 expressed during early development rescues cortical progenitors from apoptosis. Nat Neurosci. 2005;8:322–331. doi: 10.1038/nn1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt LF, Farinas I. Neurotrophic factors and their receptors. In: Cowan WM, Jessell TM, Zipursky SL, editors. Molecular and cellular approaches to neural development. New York: Oxford University Press; 1997. pp. 220–263. [Google Scholar]
- Ruthazer ES, Cline H. Multiphoton imaging of neurons in living tissue: acquisition and analysis of time-lapse morphological data. Real-Time Imaging. 2002;8:175–188. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shieh PB, Hu SC, Bobb K, Timmusk T, Ghosh A. Identification of a signaling pathway involved in calcium regulation of BDNF expression. Neuron. 1998;20:727–740. doi: 10.1016/s0896-6273(00)81011-9. [DOI] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Tsui-Pierchala BA, Encinas M, Milbrandt J, Johnson EM., Jr Lipid rafts in neuronal signaling and function. Trends Neurosci. 2002;25:412–417. doi: 10.1016/s0166-2236(02)02215-4. [DOI] [PubMed] [Google Scholar]
- Xu B, Zang K, Ruff NL, Zhang YA, McConnell SK, Stryker MP, Reichardt LF. Cortical degeneration in the absence of neurotrophin signaling: dendritic retraction and neuronal loss after removal of the receptor TrkB. Neuron. 2000;26:233–245. doi: 10.1016/s0896-6273(00)81153-8. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Weiner JA, Sanes JR. Sidekicks: synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell. 2002;110:649–660. doi: 10.1016/s0092-8674(02)00910-8. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Li H, Zhou L, Wu JY, Rao Y. Cellular and molecular guidance of GABAergic neuronal migration from an extracortical origin to the neocortex. Neuron. 1999;23:473–485. doi: 10.1016/s0896-6273(00)80801-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.