Abstract
During 1998–2005, we analyzed stool samples from 289 children in Rio de Janeiro to detect and genotype norovirus strains. Previous tests showed all samples to be negative for rotavirus and adenovirus. Of 42 (14.5%) norovirus-positive specimens, 20 (47.6%) were identified as genogroup GI and 22 (52.3%) as GII.
Keywords: Norovirus, viral gastroenteritis, viral diagnostics, real-time RT-PCR, dispatch
Noroviruses, a genus within the family Caliciviridae, have emerged as an important cause of epidemic and sporadic diarrheal disease in humans of all ages worldwide (1–3). The norovirus genome consists of a single strand of positive-sense RNA organized into 3 open reading frames (ORFs). ORF1 encodes nonstructural proteins such as RNA-dependent RNA polymerase, ORF2 encodes viral capsid protein 1, and ORF3 encodes a small capsid protein (viral capsid protein 2) associated with stability of viral capsid protein 1 (4–6). According to nucleotide sequence analysis of the polymerase and capsid regions, noroviruses are classified into 5 genogroups, GI to GV; each genogroup can be further divided into several clusters or genotypes. Genogroups GI, GII, and GIV have been found in humans, though GII seems to be the predominant strain around the world (4,7–11). To detect and genotype norovirus in stool samples from Brazilian children <10 years of age who had acute diarrhea, we used real-time Light Cycler reverse transcription–PCR (RT-PCR) and conventional RT-PCR assays.
The Study
From January 1998 through May 2005, a total of 2,421 fecal specimens were collected from children <10 years of age (median age 2.3 years) with acute diarrhea in Rio de Janeiro, Brazil. Of these, 478 (19.7%) specimens were collected from hospitalized children (inpatients) and 1,943 from outpatient children (341 [14.1%] from the emergency department and 1,602 [66.2%] from the walk-in clinic). Of these samples, 14.3% were positive for rotavirus (9.8%) or adenovirus (4.5%). The median age was 12.5 months for rotavirus-positive patients and 12.2 months for adenovirus-positive patients. Overall, of the hospitalized, emergency department, and walk-in clinic patients, 11.7%, 6.2%, and 10.0%, respectively, had samples positive for rotavirus, and 4.8%, 4.1%, and 4.4%, respectively, had samples positive for adenovirus. Enteropathogenic bacteria such as Escherichia coli, Salmonella spp., Yersinia enterocolitica, Campylobacter spp., and Shigella spp. were found in 8% of the specimens. Seven mixed infections were detected (2 adenovirus and Salmonella spp., 2 adenovirus and E. coli, 1 adenovirus and Campylobacter spp., 1 rotavirus and Salmonella spp., and 1 rotavirus and E. coli).
We selected 289 specimens that represented a random subset of samples that had prior negative results for rotavirus and enteric adenovirus. Of these 289, 117 were collected from inpatients and 172 from outpatients (89 emergency department and 83 walk-in clinic). The mean and the median age of the tested patients was 3.1 years. Suspensions of stool (10%) were prepared in diethylpyrocarbonate-treated water and Vertrel XF (Miller-Stephenson, Sylmar, CA, USA) and clarified by centrifugation at 2,100× g for 10 min. We used 200 μL of suspension for RNA extraction with the NucliSens extraction kit (Organon Tekninka, Durham, NC, USA) according to the manufacturer’s instructions. The RNA was eluted in 50 μL of elution buffer and stored at –70°C until use.
A total of 240 samples were tested for norovirus RNA by Light Cycler PCR that used primers and probes for ORF1/ORF2 junction region specific for norovirus GI and GII, as described (3,12), and by the Light Cycler RNA Amplification Kit Hybridization Probes (Roche, Basel, Switzerland). Samples that showed a positive threshold at <38 cycles were considered positive. The 49 remaining samples were tested only by conventional RT-PCR, as described (5). Conventional RT-PCR was performed with the QIAGEN OneStep RT-PCR Kit (QIAGEN, Valencia, CA, USA). The RNA samples were subjected to 1 cycle of reverse transcription (42°C, 10 min) followed by 5 min at 95°C. PCR was performed for 40 cycles, each consisting of 1 min at 94°C, 1 min at 40°C (for GI detection) or 1 min at 44°C for (for GII detection), 1 min at 72°C, and a final extension cycle of 10 min at 72°C.
We selected 6 samples that were positive by real-time Light Cycler PCR (2 GI and 4 GII) for analysis by conventional RT-PCR with specific primers in capsid region D of norovirus GI and GII, as described above. The amplified cDNA samples were purified from the gel by using QIAquick gel extraction kit (QIAGEN), and the sequences were determined with the BigDye terminator cycle sequencing kit and the ABI PRISM 3100 automated DNA sequencer (Applied Biosystems, Foster City, CA, USA) by using the same primers as used for the conventional RT-PCR. The nucleotide sequences of the amplicons were aligned with corresponding sequences of selected norovirus strains available in the GenBank database and analyzed by using the CLUSTAL V algorithm of the MegAlign program in the DNASTAR software package (Madison, WI, USA). The nucleotide sequences obtained in this study were deposited in GenBank under accession nos. DQ496212, DQ496213, DQ496214, DQ496215, DQ496216, and DQ496217.
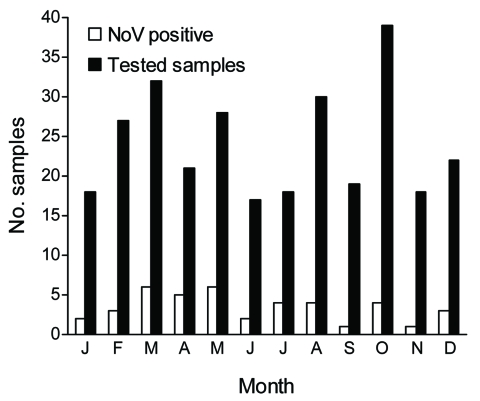
Of the 289 fecal specimens tested, 42 (14.5%) were positive for norovirus: 36 (15%; n = 240) by real time Light Cycler PCR and 6 (12.2%; n = 49) by conventional RT-PCR. These percentages correspond only to single infections because we did not test samples already known to be positive for other pathogens such as rotavirus and adenovirus. Positive samples and genogroups varied by year with no obvious yearly pattern (Table 1). Although norovirus is often referred to as “the winter vomiting disease,” we detected infection throughout the year, with no seasonal pattern (Figure). Norovirus infections were equally common among outpatients and inpatients. Among 117 inpatients, 18 (15.4%) had positive norovirus test results compared with 24 (14%) of 172 outpatients (11 emergency department and 13 walk-in clinic). Although the disease caused by norovirus is described as mild (diarrhea, vomiting, abdominal pain, and fever) and generally does not lead to hospitalization (13,14), of 42 norovirus-infected children, 29 (69%) were either hospitalized or received medical care in the emergency department, which suggests that they had a more severe illness. Only 13 (31%) of the 42 norovirus-infected children attended the walk-in clinic, which suggests that they had mild disease (Table 2). Other than diarrhea, fever was the most common symptom among the 42 norovirus-positive patients in this study, reported for 11 (26.2%) patients. Vomiting only was described for 8 (19.0%); vomiting and fever was described for 6 (14.3%). No mixed infection with bacteria was observed.
Table 1. Distribution of norovirus-positive samples detected in Rio de Janeiro, Brazil, 1998–2005*.
| Year | Real-time reverse transcription–PCR |
Conventional reverse transcription–PCR |
Total no. positive samples/genogroup | |||
|---|---|---|---|---|---|---|
| No. samples tested | No. positive samples/genogroup | No. samples tested | No. positive samples/genogroup | |||
| 1998 | 23 | 4/3GI + 1GII | 0 | NA | 4/3GI + 1GII | |
| 1999 | 29 | 3/GII | 5 | 1/GI | 4/1GI + 3GII | |
| 2000 | 31 | 4/2GI + 2GII | 7 | 0 | 4/2GI + 2GII | |
| 2001 | 26 | 4/GII | 0 | NA | 4/GII | |
| 2002 | 31 | 5/4GI + 1GII | 10 | 0 | 5/4GI + 1GII | |
| 2003 | 32 | 8/5GI + 3GII | 9 | 1/GI | 9/6GI + 3GII | |
| 2004 | 39 | 4/1GI + 3GII | 16 | 3/GI | 7/4GI + 3GII | |
| 2005 |
29 |
4/GII |
|
2 |
1/GII |
5/5GII |
| Total | 240 | 36/15GI + 21GII | 49 | 6/5GI + 1GII | 42/20GI + 22GII | |
*NA, not applicable.
Figure.
Seasonal distribution of norovirus (NoV) infections in Rio de Janeiro, Brazil, 1998–2005.
Table 2. Distribution of all tested samples by age groups and patient status, Rio de Janeiro, Brazil, 1998–2005.
| Age, y | Outpatients |
Inpatients |
|||||
|---|---|---|---|---|---|---|---|
| No. tested | PCR-positive, no. (%) | PCR-negative, no. (%) | No. tested | PCR-positive, no. (%) | PCR-negative, no. (%) | ||
| <1 | 29 | 4 (13.8) | 25 (86.2) | 45 | 5 (11.1) | 40 (88.9) | |
| 1–5 | 64 | 9 (14.0) | 55 (86.0) | 88 | 15 (17.0) | 73 (83.0) | |
| 6–10 | 24 | 5 (20.8) | 19 (79.2) | 39 | 4 (10.3) | 35 (89.7) | |
Although norovirus belonging to genogroup GII is considered the most prevalent strain worldwide (7–9,11,15), we found no important difference in the prevalence of the 2 genogroups detected in our study. Overall, 20 (48%) of the 42 samples were identified as genogroup GI and 22 (52%) as GII (Table 1). No statistically significant difference in the prevalence of GI and GII was observed between inpatients and outpatients.
Conclusions
Our study documents that noroviruses are a common cause of acute gastroenteritis in children who are inpatients or outpatients in Brazil and are likely second only to rotavirus as a cause of severe childhood diarrhea. Our study was exploratory and has limitations. Nonetheless, it documents how common norovirus infections may be and indicates that further study will be necessary to assess their role among Brazilian children, to understand the epidemiology of the disease, and to seek evidence of immunity in children, which might encourage development of a vaccine.
Acknowledgments
This article represents a portion of a thesis submitted by C.C.S. to the Universidade Federal do Rio de Janeiro, Brazil, as partial fulfillment of the requirements for a Doctor of Science degree.
This study was supported in part by Conselho Nacional de Desenvolvimento Científico e Tecnológico, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro, Brazil; and an American Society for Microbiology International Fellowship for Latin America.
Biography
Dr Soares is assistant researcher at Fiocruz–Oswaldo Cruz Institute, Rio de Janeiro, Brazil. Her research interests include diagnosis and epidemiology of enteric and respiratory viruses.
Footnotes
Suggested citation for this article: Soares CC, Santos N, Beard RS, Albuquerque MCM, Maranhão AG, Rocha LN, et al. Norovirus detection and genotyping for children with gastroenteritis, Brazil. Emerg Infect Dis [serial on the Internet]. 2007 Aug [date cited]. Available from http://www.cdc.gov/eid/content/13/8/1244.htm
References
- 1.Kageyama T, Shinohara M, Uchida K, Fukushi S, Hoshino F, Kojima S, et al. Coexistence of multiple genotypes, including newly identified genotypes, in outbreaks of gastroenteritis due to norovirus in Japan. J Clin Microbiol. 2004;42:2988–95. 10.1128/JCM.42.7.2988-2995.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pang X, Lee B, Chui L, Preiksaitis J, Monroe SS. Evaluation and validation of real-time reverse transcription-PCR assay using the LightCycler System for detection and quantification of norovirus. J Clin Microbiol. 2004;42:4679–85. 10.1128/JCM.42.10.4679-4685.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pang XL, Preiksaitis JK, Lee B. Multiplex real time RT-PCR for the detection and quantification of norovirus genogroups I and II in patients with acute gastroenteritis. J Clin Virol. 2005;33:168–71. 10.1016/j.jcv.2004.12.014 [DOI] [PubMed] [Google Scholar]
- 4.Bull RA, Hansman GS, Clancy LE, Tanaka MM, Rawlinson WD, White PA. Norovirus recombination in ORF1/ORF2 overlap. Emerg Infect Dis. 2005;11:1079–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vinjé J, Hamidjaja R, Sobsey M. Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. J Virol Methods. 2004;116:109–17. 10.1016/j.jviromet.2003.11.001 [DOI] [PubMed] [Google Scholar]
- 6.Wu FT, Oka T, Katayama K, Wu HS, Jiang DS, Miyamura T, et al. Genetic diversity of noroviruses in Taiwan between November 2004 and March 2005. Arch Virol. 2006;151:1319–27. 10.1007/s00705-005-0717-4 [DOI] [PubMed] [Google Scholar]
- 7.Hirakata Y, Arisawa K, Nishio O, Nakagomi O. Multiprefectural spread of gastroenteritis outbreaks attributable to a single genogroup II norovirus strain from a tourist restaurant in Nagasaki, Japan. J Clin Microbiol. 2005;43:1093–8. 10.1128/JCM.43.3.1093-1098.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vidal R, Solari V, Mamani N, Jiang X, Vollaire J, Roessler P, et al. Caliciviruses and foodborne gastroenteritis, Chile. Emerg Infect Dis. 2005;11:1134–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zintz C, Bok K, Parada E, Barnes-Eley M, Berkre T, Staat M, et al. Prevalence and genetic characterization of caliciviruses among children hospitalized for acute gastroenteritis in the United States. Infect Genet Evol. 2005;5:281–90. 10.1016/j.meegid.2004.06.010 [DOI] [PubMed] [Google Scholar]
- 10.Lindell AT, Grillner L, Svensson L, Wirgart BZ. Molecular epidemiology of norovirus infections in Stockholm, Sweden, during the years 2000 to 2003: association of the GGIIb genetic cluster with infection in children. J Clin Microbiol. 2005;43:1086–92. 10.1128/JCM.43.3.1086-1092.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vainio K, Myrmel M. Molecular epidemiology of norovirus outbreaks in Norway during 2000 to 2005 and comparison of four norovirus real-time reverse transcriptase PCR assays. J Clin Microbiol. 2006;44:3695–702. 10.1128/JCM.00023-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trujillo AA, McCaustland KA, Zheng DP, Hadley LA. Vaughn G, Adams SM, et al. Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of norovirus. J Clin Microbiol. 2006;44:1405–12. 10.1128/JCM.44.4.1405-1412.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolin R, Blacklow N, DuPont H, Formal S, Buscho R, Kasel J, et al. Transmission of acute infectious nonbacterial gastroenteritis to volunteers by oral administration of stool filtrates. J Infect Dis. 1971;123:307–12. [DOI] [PubMed] [Google Scholar]
- 14.Green K. Caliciviridae: the noroviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, et al., editors. Fields virology, 5th ed. Philadelphia: Lippincott Williams & Wilkins. p. 949–79. [Google Scholar]
- 15.Hansman GS, Katayama K, Maneekarn N, Peerakone S, Khamrin P, Tonusin S, et al. Genetic diversity of norovirus and sapovirus in hospitalized infants with sporadic cases of acute gastroenteritis in Chiang Mai, Thailand. J Clin Microbiol. 2004;42:1305–7. 10.1128/JCM.42.3.1305-1307.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]