Abstract
Plants are useful in studies of metal toxicity, because their physiological responses to different metals are correlated with the metal exposure dose and chemical state. Moreover a network of proteins and biochemical cascades that may lead to a controlled homeostasis of metals has been identified in many plant species. This paper focuses on the global protein variations that occur in a Populus nigra spp. clone (Poli) that has an exceptional tolerance to the presence of cadmium. Protein separation was based on a two-dimensional liquid chromatography technique. A subset of 20 out of 126 peaks were identified as being regulated differently under cadmium stress and were fingerprinted by MALDI-TOF. Proteins that were more abundant in the treated samples were located in the chloroplast and in the mitochondrion, suggesting the importance of these organelles in the response and adaptation to metal stress.
1. Introduction
Cadmium (Cd) is a widespread element in the environment, mainly from anthropogenic sources. At low concentrations, it is toxic to microorganisms, plants, and animals. In humans, Cd intake is normally by ingestion or inhalation, with the majority of ingested Cd coming from contaminated foods (meat and plants) and water. Thus the transfer of Cd from the soil into plants and from Cd deposited from the troposphere onto edible plants accounts for the majority of human Cd intake [1].
In agriculture, Cd pollution is an increasing problem due to soil amendments and to the intensive use of phosphate fertilizers. In natural environments, Cd is present at concentrations of 0.1–0.5 mg kg−1, but it may reach concentrations as high as 150 mg kg−1 at contaminated sites. The high solubility of Cd causes rapid distribution in the environment, where it is immediate availability to plants and from these primary producers can move up through the entire food chain [2]. On the other hand, the ability of some plants to absorb and accumulate significant quantities of metals in roots, shoots, and leaves offers a unique opportunity to remove inorganic pollutants from contaminated media [3].
Laboratory and field trials have found that many clones from the genus Populus seem promising for the removal of Cd from soil [3, 4]. Poplar plants are ecologically and economically attractive due to their fast growth, high biomass production, and easy propagation through vegetative cuttings. Moreover poplars are used for different industrial applications, including renewable energy production.
The use of plants to remediate metal pollution [5] relies on an understanding of the molecular mechanisms involved in the uptake, translocation, and sequestration of metals by plants. Thus, research focusing on genomic and transcriptomic analyses in plants has been conducted after metal exposure in field trials [6].
A better knowledge of the plant proteome, as a way of understanding phenotypic plasticity and adaptability in plants, is required to effectively exploit plant biological resources [7]. Up to a few years ago, the study of proteomic variability was restricted to a few model plants, such as Arabidopsis thaliana, under a wide range of stresses. Recently, comparative analysis of proteome profiles in plants following metal exposure has attracted greater interest [8, 9].
Comparative proteomics rests on two main approaches: (i) the first is a “gel-based” method, which uses a two-dimensional electrophoresis separation of proteins (2D-PAGE) [10, 11] and (ii) the second is a “gel-less” method, in which proteins are separated by two-dimensional liquid chromatography (2D-LC), a technique which promises to extend the range of protein separation [12, 13]. Using comparative proteomics, protein variations of the Populus nigra clone “Poli” grown without Cd were compared to clones grown with a 50 μM CdSO4 treatment, which was sublethal for this clone [14]. The protein patterns obtained from plant samples of the two experimental conditions were compared using a semiquantitative 2D-LC technique. Three sets of proteins were characterized: (i) more abundant in the treated sample in respect to the control, (ii) less abundant in the treated sample in respect to the control, and (iii) equally abundant in both treated and control samples [13]. Proteins with altered expression ((i) and (ii)) were digested with trypsin and identified by Matrix-assisted LASER desorption / ionization time of flight mass spectrometry (MALDI-TOF/MS). The combination of a software that could quantify protein differences after 2D-LC and identify proteins, with MS technique, gave both qualitative and semiquantitative evidence of some of the proteome changes due to Cd treatment in this particular poplar clone.
2. Materials and Methods
2.1. Plant Material
Stem cuttings of Populus nigra clone “Poli” were grown hydroponically for three weeks in Hoagland's nutrient solution (one-third strength, pH 6.5 (mol l−1)) [15]. Cuttings were placed in a controlled climate chamber equipped with metal halide lamps (Powerstar HQI-TS; Osram, Munich, Germany), which provided a photon flux density of 300 μmol m−2 s−1 for 14 h at 25°C. During the 10 h dark period the temperature was 20°C. The relative humidity was 70%–80%. After three weeks, half of the plants were treated with 50 μM CdSO4 for three more weeks. The hydroponic solution was controlled and maintained at a steady level with periodic supplements to compensate for the high transpiration rates of the plants. At the end of the experimental period, leaves from treated and untreated plants were sampled according to Zacchini et al. [14], frozen in liquid nitrogen, and stored at −80°C for protein extractions. This procedure was repeated three times using three cuttings from each treatment. Leaves were chosen for proteomic analysis because of their relative abundance in the total plant biomass and because their physiological behaviors (e.g., functions, photosynthesis, respiration, transpiration, etc), correlate strongly with the overall health of the whole plant. Detection of Cd uptake and translocation in poplars was previously described by Zacchini et al. [14].
2.2. D-LC Analysis
Five grams of frozen leaves from each treatment were finely ground with a mortar and pestle and then thawed in 4 ml of MgSO4-based extraction buffer as described by Pirondini et al. [13]. Leaf samples from each treatment were extracted in triplicate for crude total proteins mixtures, as described below.
The crude mixture was sonicated for 10 min, and then the solution was centrifuged at 16 000× g for 5 min at 4°C. The pellet containing the larger cellular residues was discharged, and the supernatant was centrifuged at 16 000× g for 30 min at 4°C. The lighter phase contained the purified solubilised proteins. The purified proteins solution was loaded onto a desalted PD-10 column (GE-Healthcare, Uppsala, Sweden). Proteins were quantified determining the O.D. with a spectrophotometer (Beckman Coulter, Fullerton, CA) at 562 nm utilizing a BCA Protein Assay Kit (Novagen, Merck KGaA, Darmstadt, Germany). Protein separation was carried out using a 2D-LC technique (ProteomeLab PF 2D, Beckman Coulter, Fullerton, CA) following a procedure described by Pirondini et al. [13]. Equal amounts (1.3 mg) of total protein extract from leaves of each treatment (untreated or treated for 3 weeks with 50 μM CdSO4) were loaded into the 1st dimension high-performance chromatofocusing (HPCF) column, where protein separation occurred by isoelectric point (pI). The resulting pH fractions were then injected into the 2nd dimension high-performance reversed-phase chromatography (HPRP) column, where separation occurred by protein hydrophobicity. The eluent from the 2nd dimension column was analyzed by U.V. detector for O.D. at 214 nm. This method provided a more consistent and sensitive detection of proteins via peptide bonds. Each protein was then represented by a Gaussian-shaped peak.
2.3. MALDI-TOF Analysis
Eluted proteins were manually collected from the column in single fractions every 0.2 min, starting from the beginning of the peak area. Half of each eluted peak was evaporated and suspended in an SDS-PAGE loading buffer and then loaded onto a 12% 1D-SDS-PAGE gel to check for the number of protein species (data not shown). Only peaks with a single band in the 1D-SDS-PAGE gel were evaporated by Speed Vac into a final volume of 10 μL for further analysis. Protein reduction was performed by incubating each fraction with 25 mM NH4HCO3 and 2 mM DTT in a water bath at 60°C for 1 h. The alkylation of the reduced sulfhydryl groups was carried out by adding 1 mM iodoacetamide (IAA), at 25°C, for 30 minutes in the dark. 1.5 μL of trypsin (125 μg mL−1) in 50 mM NH4HCO3 was subsequently added to each fraction and the digestion was carried out at 37°C for 24 h. The digested samples were then purified with a ZipTipC18 (Millipore, Billerica, MA, USA) using the procedure recommended by the manufacturer. 1 μL of each purified peptide mixture was spotted directly onto a stainless steel MALDI target plate with 1 μL of a saturated solution of α-cyano-4-hydroxycinnamic acid in 0.1% TFA : ACN (2 : 1, v/v). The solution was dried at room temperature and a spot was produced. Positively charged ions were analyzed in reflectron mode. External calibration was performed using a ProteomMass Peptide and protein MALDI/MS calibration kit (Sigma St. Louis, MO, USA). The three detectable peptide fragments were angiotensin II (human) m/z 1046.542, ACTH fragment 18–39 (human) m/z 2465.198, and insulin-oxidized B chain (bovine) m/z 3494.651. The spectra were obtained through random scanning of the sample surface with an ablation LASER. An average of 100 LASER shots was used to improve the signal-to-noise ratio. Positively charged ions were analyzed with a MALDI-LR in TOF/MS mode instrument (Micromass Waters Corporation, Milford Massachusetts, USA). Three technical replicates from each spectrum were analyzed by MS, and only peptides common to all of the resolved spectra were considered for protein identification.
2.4. Bioinformatics and Statistical Analyses
Chromatogram peaks for each pH fraction (hydrophobicity and pI) were converted into band intensity maps using ProteoVue software (Eprogen, Darien, IL, USA). The color intensity of each map was proportional to the relative intensity of each chromatographic peak. Chromatograms obtained from Cd-treated and untreated samples were compared using DeltaVue software (Eprogen, Darien, IL, USA). Protein abundance in treated and untreated samples was semi-quantified by subtracting the difference in peak areas for proteins with the same pI and hydrophobicity. A paired t-test was used to identify significant differences in abundance of proteins between the control and Cd-treated samples. Only peaks with a ratio >3 or <0.33 between untreated and treated samples were included in the analysis. These threshold levels were chosen as being more conservative than those used in [9].
Peptide mass fingerprinting was carried out using the Mascot program (http://www.matrixscience.com). Proteins were identified by searching through the SWISS-PROT and NCBI nonredundant databases (limited to Arabidopsis thaliana and other plant species), as well as against the poplar EST database (http://www.populus.db.umu.se/). The following parameters were used in browsing the protein databases: (i) mass accuracy below 100 ppm, (ii) maximum of one missed cleavage by trypsin, (iii) carbamidomethylation of cysteine, and (iv) oxidation of methionine, where (iii) and (iv) were considered to be fixed modifications. The search was based on the monoisotopic masses of the peptides. For a positive peptide identification, at least five different predicted peptides had to match the observed masses, and the matching peptides had to account for at least 20% of the known sequence.
3. Results and Discussion
3.1. Protein Separation in Leaves
An important aspect of evaluating plants for the phytoremediation of metals is to determine the amount and type of damage to the leaves. An efficient and metal-tolerant photosynthetic system allows plants to maintain high transpiration efficiency, which creates a water flux that can drive metals from the roots and into the stems and leaves where the metal can be compartmentalized. Poplar clone “Poli” was previously selected among 10 Italian poplar clones as having the greatest ability to accumulate Cd in leaves and stem tissues [14].
Several experiments have previously been conducted to characterize the Cd tolerance level of the Populus nigra clone “Poli”. In one study, leaf samples taken from plants after 21 days of growth in the presence of 50 μM CdSO4 showed mild or almost no symptoms of chlorosis and reduction in total leaf area. In comparison, a dose of 50 μM CdSO4 halted radical growth and caused symptoms of leaves chlorosis in other clones in this experiment [14]. Other parameters that distinguished “Poli” clones from the other poplar clones included Cd leaf accumulation after 21 days (450 ± 30 mg/kg d.w.) and a high translocation factor. These parameters were previously quantified by atomic absorption spectrophotometer by Zacchini et al. [14].
Given the interesting physiological features of the clone “Poli”, proteomic characterization was conducted to further understand its response to Cd. This paper describes how a 2D-LC protein separation system, coupled with MS identification, can be used for comparative plant proteomic analysis. This technique has recently been reported as improving semiquantitative comparisons of protein mixtures [12]. Though it has been widely used in human and bacterial comparative proteomic studies [12, 16, 17], few papers have described its application for plants [13, 18]. One of the advantages of this technique is that a larger amount of protein can be analyzed at once (up to 10 mg) in comparison to the conventional 2D SDS-PAGE (0.1–0.3 mg). One benefit of this system is that it works with crude total soluble extracts. This increases the reproducibility of the protein patterns and avoids the loss of proteins due to sequential purification (e.g., the frequent acid precipitation steps). These aspects are crucial in comparative proteomic studies because they also allow the detection of less abundant proteins.
As leaves from poplar trees have very little protein and an abundance of insoluble substances such as waxes and lignocelluloses that may hamper protein solubilisation, applying a 2D-LC separation technique to plant tissues extracts is an effective method of detecting plant proteins [19]. A standardized procedure for this method has previously been developed by Pirondini et al. [13] to optimize protein solubilization while limiting instrumental interference. Recently, a proteomic approach based on a 2D SDS-PAGE separation system was also applied to poplar leaf extracts to identify several proteins related to the Cd stress response [9]. However, as there are as many as 45 555 putative proteins that could potentially be sequenced from the poplar genome [20] and the proteomic approach has only worked with several hundreds of these proteins to date, the use of different complementary techniques could help to decode the entire proteome of a poplar species. Thus the 2D-LC protein separation technique is a suitable complementary tool that can identify proteins that have been upregulated or downregulated in response to Cd stress.
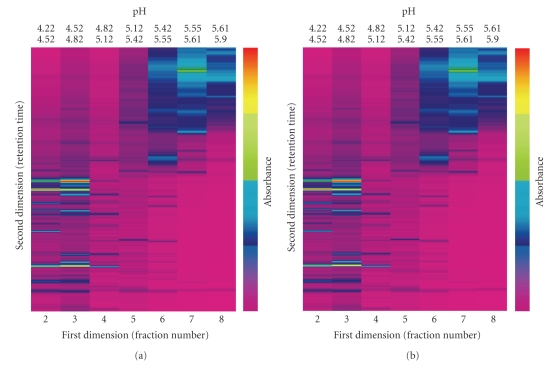
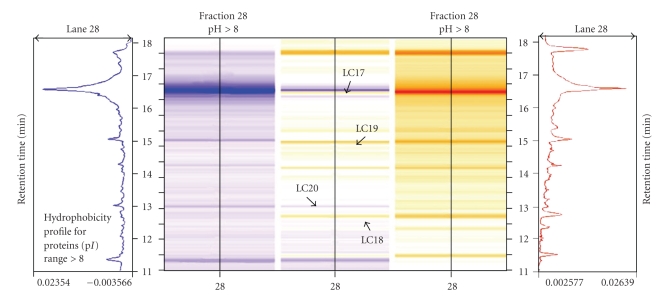
Using the 2D-LC separation technique, about 600 proteins were resolved from treated (50 μM CdSO4) and untreated (0 μM of CdSO4) poplar cuttings from Populus nigra clone “Poli”. Data were displayed on six different virtual gels (three technical replicates for each biological replicate of the control and the treated samples) using ProteoVue software. A comparison of the protein abundance in different samples was carried out using DeltaVue software. Figure 1 shows the reproducibility of the protein profiles from two technical replicates of the control sample. Thus differences between the treated and untreated samples can be determined by matching the corresponding peaks between samples. This concept is further demonstrated in Figure 2, by highlighting differences in protein abundance in Fraction 28.
Figure 1.
Virtual 2D maps created by the software “ProteoVue” for two independent untreated samples. Proteins were extracted as described in the Materials and Methods Section. Equal amounts of protein (1.3 mg) were loaded into the 1st dimension separation column (HPRP). This figure shows the isoelectric point (pI) fractions (from 4.22 to 5.9) for two independent biological replicates.
Figure 2.
Magnified area of a virtual gel obtained after 2nd dimension separation (HPRC), for the 28th pH fraction, and reproduced by “DeltaVue” software. The protein pattern of untreated plants (0 μM CdSO4) is shown in shades of blue on the left and the protein pattern of treated plants (50 μM CdSO4) is shown in shades of yellow on the right. The center column demonstrates the differences in protein abundance between the control and the treated protein samples, as indicated by arrows at the peaks/ bands for proteins of different abundances.
Of the peaks that were separated from the two treatments, 126 peaks (13.6%) were three times more or less abundant in either the treated or untreated samples. 50 % of the 126 peaks had an isoelectric point (pI) between 8 and 4, 33% had a pI > 8 (basic), and 17% had a pI < 4 (acidic). Almost half of the 126 identified peaks had an absorbance less than 0.05 at 214 nm, which is under the limit of detection recommended by the manufacturer for MS analysis. Another limitation of all proteins separation techniques is the possibility that there are multiple protein species in a single spot for 2D SDS-PAGE gels, or in a single peak for 2D-LC. Thus each peak eluted out in a single fraction was analyzed by 1D SDS-PAGE, for protein singularity (data not shown). Twenty peaks were clearly identified as single bands on the gel and were further characterized by MS. As the remaining peaks could not clearly be distinguished as individual bands, they were not analyzed any further.
3.2. Protein Identification by Mass Spectrometry
Proteins were identified by eluting out the same peaks from both untreated and Cd-treated samples and then analysing them by MALDI-TOF mass spectrometry. The resulting tryptic peptides were entered into the SWISS-PROT and/or NCBI nonredundant databases. In addition, a BLAST search was performed on the EST poplar database (http://www.populus.db.umu.se/) to confirm the reliability of the results. Results indicated that there was a consistent correspondence between the experimental and the calculated pI values for most of the analyzed proteins. The few exceptions were likely due to post-transductional modifications of these proteins. Similar inconsistencies have been reported by other authors and have been attributed to the influence of ion concentrations in both the liquid and in the adsorbed phase because a small section of the charged protein appears to bind to the ion-exchange resin [13].
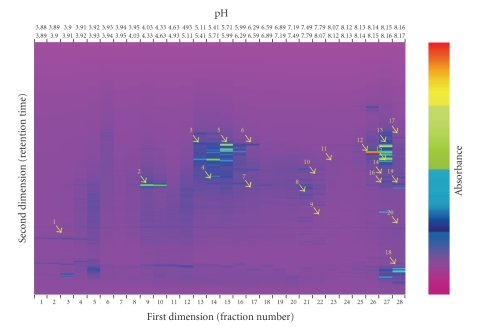
Comparison with the Arabidopsis thaliana protein database gave the highest score for almost all twenty proteins. This result confirms the consistency in synteny between Arabidopsis and Populus for both their genomes and also their proteomes [21]. The proteins identified by MS and their putative function/location are shown in Figure 3 and in Tables 1 and 2.
Figure 3.
A “ProteoVue” 2D map of a leaf protein extract from Populus nigra clone “Poli”, grown for 3 weeks in nutrient solution supplemented with 50 μM CdSO4. The x-axis is in isoelectric point (pI) units from 4.0 to 8.0. The y-axis displays increasing hydrophobicity. The color scale of the bands represents the relative intensity of each band by UV detection at 214 nm. The proteins detected by MALDI/TOF analysis are numbered and highlighted with yellow arrows.
Table 1.
Proteins regulated by Cd treatment in leaves of Populus nigra clone “Poli”, separated by 2D-LC.
| Nos.a | Homologous proteinb | Accession numberb | Measured pI intervalc | Predicted pI/Mr (Da)c | Score / coverage (%)d | Ratioe | ESTs poplarf |
|---|---|---|---|---|---|---|---|
| LC1 | Aquaporin | NIP12_ARATH | <4.00 | 8.63/31249 | 56/40 | +5.6 | P047H06 |
| LC2 | Dehydration-responsive protein | gi|18405331 | 4.03–4.33 | 8.60/67472 | 60/35 | +3.4 | P082D06 |
| LC3 | Predicted protein | gi|162695261 | 5.11–5.41 | 6.20/45886 | 69/32 | −7.2 | T020D11 |
| LC4 | Acetyl coenzime A carboxylase | gi|74272305 | 5.41–5.71 | 5.60/120177 | 69/35 | −3.5 | Q001C04 |
| LC5 | NADPH isocitrate dehydrogenase | gi|169989 | 5.71–5.99 | 6.13/49508 | 51/28 | +4.4 | N006H08 |
| LC6 | Thylakoidal processing peptidase 2 | TPP2_ARATH | 6.29–6.59 | 6.54/40581 | 56/21 | +4.8 | T040C06 |
| LC7 | Hypothetical protein | gi|125603717 | 6.29–6.59 | 6.67/47148 | 71/26 | −3.0 | Y004E07 |
| LC8 | Predicted protein | gi|168009327 | 7.49–7.79 | 7.22/42542 | 69/40 | +5.3 | V017A06 |
| LC9 | Chaperone ClpB | CLPB_RHOBA | >8.00 | 5.06/97498 | 110/56 | +8.4 | P058E06 |
| LC10 | Rubisco large subunit | gi|33636375 | >8.00 | 5.13/6207 | 69/80 | −8.5 | No match |
| LC11 | Aldo/keto reductase protein | gi|15232354 | >8.00 | 6.46/35185 | 64/35 | +4.5 | V056F06 |
| LC12 | Cytochrome P450 | C71BT_ARATH | >8.00 | 8.06/56318 | 105/64 | +7.3 | M122H12 |
| LC13 | ATP-dependent Clp protein | CLPR1_ARATH | >8.00 | 8.81/42942 | 51/35 | +4.7 | UM61TE04 |
| LC14 | Cysteine proteinase inhibitor 2 | CYT2_ORYSJ | >8.00 | 8.82/16550 | 56/25 | +3.2 | FL_GENBANK_63 |
| LC15 | Hydroxyacylglutathione hydrolase 1 | GLO2M_ARATH | >8.00 | 8.83/36933 | 51/40 | −4.6 | S065B02 |
| LC16 | Hypothetical protein | gi|38175624 | >8.00 | 9.10/27188 | 69/36 | −4.5 | N054D02 |
| LC17 | GTP binding protein | gi|145359760 | >8.00 | 9.46/20079 | 60/40 | −3.2 | K027P68 |
| LC18 | Protein kinase APK1B | APK1B_ARATH | >8.00 | 9.37/45946 | 56/37 | +5.3 | N041C12 |
| LC19 | CRS2A chloroplastic intron splicing facilitator | CRS2A_ARATH | >8.00 | 9.40/27356 | 69/60 | +3.5 | K028P24 |
| LC20 | Nucleotide translocator | gi|444790 | >8.00 | 9.78/43995 | 92/43 | −7.4 | T027D03 |
aThe nos. refer to the spot numbers as given in Figure 3. bPutative protein identification and accession number for the closest match in the SWISS PROT and NCBI databases. cpI values as estimated from the 2D-LC together with the predicted pI and Mr of the closest match in the databases. dScore and percentage of coverage of the matching peptide sequence tags. eRatio between values of the peak areas of the proteins from treated leaves (50 μM CdSO4) and untreated leaves (0 μM CdSO4) obtained from the Deltavue software. fMatches with the poplar EST database.
Table 2.
Functional classes and putative locations of the proteins identified by MS.
| Protein class | Protein name | Localization | Predicted function or domains |
|---|---|---|---|
| Uptake | Aquaporin NIP-2 | Plasma membrane | Channel/ pore transporter |
| Proteolysis | Thylakoidal processing peptidase | Chloroplast membrane | Peptidase activity |
| Stress proteins | Cysteine proteinase inhibitor | Cytosol | Inhibition of exogenous proteases such as those present in digestive tracks of insects and nematodes |
| Aldo/keto reductase | Cytosol | Oxidoreductase activity involved in response to cadmium ions | |
| Cytochrome P450 | Endoplasmic reticulum | Ferulate 5-hydrolase activity, monoxygenase activity | |
| Dehydration responsive protein | Golgi apparatus | Biological process unknown | |
| ClpB, chloroplastic | Chloroplast | ATP binding, ATPase activity | |
| ATP-dependent Clp | Chloroplast | Protease activity | |
| Metabolism | NADPH isocitrate dehydrogenase | Mitochondrion | TCA cycle |
| CRS2A, chloroplastic intron splicing facilitator | Chloroplast | Aminoacyl-tRNA hydrolase activity | |
| Ribulose 1,5-bisphosphate carboxylase/oxygenase large subunit | Chloroplast | Calvin cycle | |
| Nucleotide translocator | Mitochondrial inner membrane | ATP : ADP antiporter activity | |
| Acetyl-CoA carboxylase | Mitochondrion | Fatty acid metabolism | |
| Predicted protein | Mitochondrion | Succinyl-CoA synthetase subunit beta | |
| Regulatory protein | APK1B kinase | Chloroplast | Serine/ threonine activity |
| GTP binding protein | Mitochondrion | Protein targeting, sorting and translocation | |
| Predicted protein | — | Serine/ threonine kinase domain | |
| Glutathione pathway | Hydroxyacylglutathione hydrolase 1 | Mitochondrion | Thiolesterase that catalyzes the hydrolysis of S-D-lactoyl glutathione to form glutathione and D-lactic acid |
Predictions were obtained by checking putative protein acquisition with the annotation in SWISS-PROT, NCBI, and databases.
Several of the identified proteins were more abundant in the treated samples than in the untreated samples. For example, Aquaporin NIP-2 is a protein that belongs to the large family of major intrinsic proteins (MIPs) and is present in many eukaryotic organisms from yeast to humans [22, 23]. The physiological role of aquaporins in plants is not well understood, but reports indicate that they may play an important role in transport selectivity and gating (i.e. opening and closure of pores) and can help transport or diffuse physiologically important molecules such as CO2, H2O2, ammonium (NH4+), and its gaseous conjugated base (NH3, ammonia) through the cell membrane. Aquaporins can also aid in the uptake of metalloids such as As and Sb and are involved in carbon fixation mechanisms, cell signaling transduction, stress responses, and cell osmotic and pH stabilization [24]. Another protein identified as being particularly abundant in the Cd-treated samples was the thylakoidal processing peptidase, which is a chloroplast enzyme relevant for protein maturation within organelles [25].
Kinase 1B (APK1B), a protein homolog in Arabidopsis, was also found to be more abundant in leaves of Cd-treated plants. It is involved in phosphorylation of tyrosine, serine, and threonine residues and contributes to signal transduction in the chloroplast [26]. In animal cells, protein tyrosine kinases have been identified in both receptor and cytosolic forms and play a role in cell growth, development and the oxidative stress response [27].
Two proteins involved in the stress response, ClpB and ATP-dependent Clp, were more prevalent in the Cd-treated samples. The heat shock protein ClpB is a member of the caseinolytic protease (Clp) family, whose members act as molecular chaperones in many organisms [28, 29]. ClpB is found in the chloroplastic stroma and helps with thermotolerance. In Arabidopsis thaliana, ClpB-p knockouts mutants did not survive for long as they failed to accumulate chlorophyll and could not properly develop the chloroplasts. Thus, in plants, ClpB proteins are not limited to the heat stress response, but play an essential role in chloroplast development under different stress conditions (i.e., exposure to toxic metals) [30]. The related protein, ATP-dependent Clp, is also involved in chloroplast functions essential for cell survival. The recent characterization of several chloroplast proteases determined that ClpRs are the noncatalytic subunits of the Clp core proteolytic complex [31].
The chloroplast-specific NADPH isocitrate dehydrogenase was also more abundant in the Cd-treated samples, along with CRS2, a putative chloroplast RNA Splicing 2 factor that is required for splicing of group II introns. The importance of RNA splicing, and alternative RNA splicing patterns for plant survival in response to stress conditions, has been deduced using a transient expression system in monocots [32]. In plant chloroplasts and in animal mitochondria, different metabolic reactions are coupled with NADPH, including those involved in thiol group reduction. Mitochondrial NADP+-dependent isocitrate dehydrogenase is an enzyme linked to the above mentioned process, although the precise role of this enzyme is still unknown. It was suggested that this protein may be a regulatory switch in the tricarboxylic acid cycle flux and in the reductive modulation of alternative oxidase (AOX) [33]. Castro-Guerrero et al. [34] have demonstrated that an enhancement in AOX appears to be related to the Cd-resistance mechanism developed in the algae E. gracilis. When their primary respiration pathway is impaired, the algae switch to an alternative oxidative route to maintain a relatively unaltered level of metabolism [34].
Interestingly, a cysteine proteinase inhibitor was more abundant in the Cd–treated leaves. This enzyme can potentially improve resistance to biotic stresses such as pathogens and insects in both animals and plants [35, 36]. An aldo-ketoreductase protein, previously found in animals and recently identified in plants in response to stress, was also more abundant after Cd stress. Oberschall et al. [37] have shown that a novel plant NADPH-dependent aldo-keto reductase functions in a detoxification pathway which can prevent stress damage.
Another protein more abundant in the Cd-treated plants is a member of the cytochrome P450 family, which have a key role in the response to abiotic and biotic stresses both in animals and plants [38, 39]. Information on the mechanism of how metal ions inhibit NADPH-cytochrome P450 reductase is limited in comparison with the effect of organic pollutants on both poplars and Arabidopsis [40]. The cytochrome P450 enzymes also regulate the production of the plant hormone abscisic acid (ABA). As ABA 8′-hydroxylase belongs to a cytochrome P450 family, it implies that this protein plays a major regulatory role in controlling the level of ABA in plants [41]. ABA is involved in apoptosis, senescence, and leaf shedding, and ABA regulation is essential for maintenance of plant fitness in abiotic stress conditions, such as drought and also Cd stress [42].
Eight proteins were identified as being less abundant after the Cd treatment. A RuBisCo large subunit was found to be more abundant in the untreated samples, as the photosynthetic system was likely damaged or less efficient in the Cd-treated plants. In this case, the measured and the theoretical pI values were different, but the “score” and “coverage” parameters of the protein were high enough to confirm its identification. In literature, examples of post-transductional modifications to the large and small subunits of RuBisCo such as acetylations, deformylation, methylation, and phosphorylation support the possibility of different pIs [43].
Four mitochondrial proteins were found to be less abundant after the Cd treatment. (1) Hydroxyacylglutathione hydrolase 1 is a mitochondrial precursor that functions as a thiolesterase and catalyzes the hydrolysis of S-D-lactoyl-glutathione to form glutathione and D-lactic acid. This enzyme has been described in animals and in plants [44] and the ability of glutathione to bind Cd has been well established [45]. (2) Acetyl-CoA carboxylase is an enzyme that catalyzes the first step in fatty acid synthesis, the carboxylation of acetyl-CoA to malonyl-CoA. Two physically distinct types of this enzyme are present in nature, and most plants have both forms. The heteromeric form (four subunits similar to those found in prokaryotes) is found in the plastids, where de novo synthesis of fatty acids occurs. The homomeric form (single large polypeptide similar to that found in eukaryotes) is found in the cytosol. This enzyme may play a role in the oxidative stress response, (i.e., after exposure to metals) [46]. (3) A GTP-binding protein. G proteins bind to heterotrimeric GTP-binding proteins and transduce extracellular signals into intracellular signals by activating effector molecules (i.e., adenylate cyclases), which catalyze cAMP formation. cAMP signaling pathways have been reported as being able to protect cells from ionizing radiation-induced apoptosis. However the mechanism of this protection is still unknown [47]. (4) A nucleotide translocator is protein known to be involved in the alteration of mitochondrial membrane permeability for free radicals and metal ions in animals and plants [48, 49].
Another paper was recently published on proteomic responses in poplar leaves after exposure to a prolonged Cd treatment [9]. The papers differed in (i) plant material: Populus nigra versus Populus tremula, (ii) length of the Cd treatment and concentration: 50 μM Cd for 21 days versus 20 μM Cd for 28 days, (iii) protein extraction, separation and identification procedures: 2D-LC versus 2D-PAGE gel, and (iv) threshold level for the comparative analysis: 3-fold versus 1.5-fold ratios. However, some common features were found with respect to the classes and the cellular location of some of the identified proteins, including stress proteins, carbohydrate and general metabolism-related proteins, and proteolysis proteins: Table 2 and [9]. Kieffer et al. [9] concluded that after the first “alarm phase” there was a stabilization of the stress response and plants remained photosynthetically active. In addition, plants increased their energy supply by activating mitochondrial respiration. In comparison, this paper determined that some regulatory proteins such as homolog of a GTP-binding protein and the protein kinase APK1B that were more abundant after Cd treatment (21 days) were not detected in the Populus tremula clone [9]. It was hypothesized that these proteins may account for the robust response by Poli clone to Cd stress. The synthesis of these proteins could limit the damages and/or optimize the different pathways useful for Cd adaptation and Cd homeostasis (Table 2). To date, no studies have been found regarding the regulation of these proteins under Cd stress, probably due to their low abundance in plant samples, confirming the value of this separation technique.
4. Conclusion
Environmental exposure to toxic elements such as Cd forces both plants and animals to enter into a “tradeoff” situation to minimize the impact of these compounds on the organism. “Tradeoffs” may include the activation of a stress defence response, reduction of metabolic rate, and/or activation of novel metabolic routes. A phenotypic study of the adaptive response is technically more difficult than transcriptomic analysis. For instance, the proteomic analyses described in this study and in Kieffer et al. by [9] focus on only a small percentage of the putative poplar proteomes. However, phenotypic plasticity and environmental adaptations from “tradeoffs” between gene expression and environmental conditions would be difficult to quantify without protein analysis. Comparative proteomics of metal treated and untreated plants (e.g., Poli) is likely to be applicable to multiple areas of phytoremediation in future. In particular, proteins with altered synthesis due to changes in gene expression may be used to design molecular markers for the selection of more advantageous genotypes.
Acknowledgments
The authors wish to thank the International Phytosociety (IPS) coediting program for their assistance in reviewing the manuscript language. We acknowledge the kind assistance of Dr. Aliosha Malcevschi (Department of Environmental Sciences, University of Parma) for protein MS analysis and CIM (Centro Interdipartimentale Misure, Parma, Italy) for access to MALDI-TOF-MS facilities. This research was supported by funding Professor Nelson Marmiroli from FIL of the University of Parma Local Funding for Research and by funding Dr Marta Marmiroli from PRIN “Trees and forest plantations for environmental restoration: physiological and molecular mechanisms in the selection of Salicaceae for phytoremediation of heavy metals and hydrocarbons”. The contributions of Professor Giuseppe Scarascia Mugnozza and his colleagues at IBAF (Istituto di Biologia Agro-Ambientale e Forestale, Monterotondo Scalo, Roma, Italy) in the selection and propagation of poplar clone “Poli” were instrumental in the success of this work.
References
- 1.Dudka S, Miller WP. Accumulation of potentially toxic elements in plants and their transfer to human food chain. Journal of Environmental Science and Health B. 1999;34(4):681–708. doi: 10.1080/03601239909373221. [DOI] [PubMed] [Google Scholar]
- 2.Schützendübel A, Polle A. Plant responses to abiotic stresses: heavy metal-induced oxidative stress and protection by mycorrhization. Journal of Experimental Botany. 2002;53(372):1351–1365. [PubMed] [Google Scholar]
- 3.Robinson BH, Mills TM, Petit D, Fung LE, Green SR, Clothier BE. Natural and induced cadmium-accumulation in poplar and willow: implications for phytoremediation. Plant and Soil. 2000;227(1-2):301–306. [Google Scholar]
- 4.Unterbrunner R, Puschenreiter M, Sommer P, et al. Heavy metal accumulation in trees growing on contaminated sites in Central Europe. Environmental Pollution. 2007;148(1):107–114. doi: 10.1016/j.envpol.2006.10.035. [DOI] [PubMed] [Google Scholar]
- 5.Marmiroli N, McCutcheon SC. Making phytoremediation a successful technology. In: McCutcheon SC, Schnoor JL, editors. Phytoremediation: Transformation and Control of Contaminants. Hoboken, NJ, USA: Wiley-Interscience; 2003. pp. 75–107. [Google Scholar]
- 6.van de Mortel JE, Schat H, Moerland PD, et al. Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant, Cell and Environment. 2008;31(3):301–324. doi: 10.1111/j.1365-3040.2007.01764.x. [DOI] [PubMed] [Google Scholar]
- 7.Rossignol M, Peltier J-B, Mock H-P, Matros A, Maldonado AM, Jorrín JV. Plant proteome analysis: a 2004–2006 update. Proteomics. 2006;6(20):5529–5548. doi: 10.1002/pmic.200600260. [DOI] [PubMed] [Google Scholar]
- 8.Sarry J-E, Kuhn L, Ducruix C, et al. The early responses of Arabidopsis thaliana cells to cadmium exposure explored by protein and metabolite profiling analyses. Proteomics. 2006;6(7):2180–2198. doi: 10.1002/pmic.200500543. [DOI] [PubMed] [Google Scholar]
- 9.Kieffer P, Schroeder P, Dommes J, Hoffmann L, Renaut J, Hausman J-F. Proteomic and enzymatic response of poplar to cadmium stress. Journal of Proteomics. 2009;72(3):379–396. doi: 10.1016/j.jprot.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 10.Rose JKC, Bashir S, Giovannoni JJ, Jahn MM, Saravanan RS. Tackling the plant proteome: practical approaches, hurdles and experimental tools. The Plant Journal. 2004;39(5):715–733. doi: 10.1111/j.1365-313X.2004.02182.x. [DOI] [PubMed] [Google Scholar]
- 11.Rabilloud T. Two-dimensional gel electrophoresis in proteomics: old, old fashioned, but it still climbs up the mountains. Proteomics. 2002;2(1):3–10. [PubMed] [Google Scholar]
- 12.Lambert J-P, Ethier M, Smith JC, Figeys D. Proteomics: from gel based to gel free. Analytical Chemistry. 2005;77(12):3771–3787. doi: 10.1021/ac050586d. [DOI] [PubMed] [Google Scholar]
- 13.Pirondini A, Visioli G, Malcevschi A, Marmiroli N. A 2-D liquid-phase chromatography for proteomic analysis in plant tissues. Journal of Chromatography B. 2006;833(1):91–100. doi: 10.1016/j.jchromb.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 14.Zacchini M, Pietrini F, Scarascia Mugnozza G, Iori V, Pietrosanti L, Massacci A. Metal tolerance, accumulation and translocation in poplar and willow clones treated with cadmium in hydroponics. Water, Air, and Soil Pollution. 2009;197(1–4):23–34. [Google Scholar]
- 15.Arnon DI, Hoagland RD. Crop production in artificial culture solutions and in soils with special reference to factors influencing yield and absorption of inorganic nutrient. Soil Science. 1940;50:463–483. [Google Scholar]
- 16.Ruelle V, Falisse-Poirrier N, ElMoualij B, et al. An immuno-PF2D-MS/MS proteomic approach for bacterial antigenic characterization: to Bacillus and beyond. Journal of Proteome Research. 2007;6(6):2168–2175. doi: 10.1021/pr060661g. [DOI] [PubMed] [Google Scholar]
- 17.Schlautman JD, Rozek W, Stetler R, Mosley RL, Gendelman HE, Ciborowski P. Multidimensional protein fractionation using ProteomeLab PF 2DTM for profiling amyotrophic lateral sclerosis immunity: a preliminary report. Proteome Science. 2008;6, article 26 doi: 10.1186/1477-5956-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komatsu S, Zang X, Tanaka N. Comparison of two proteomics techniques used to identify proteins regulated say gibberellin in rice. Journal of Proteome Research. 2006;5(2):270–276. doi: 10.1021/pr0502929. [DOI] [PubMed] [Google Scholar]
- 19.Plomion C, Lalanne C. Protein extraction from woody plants. In: Thiellement H, Zivy M, Damerval C, Méchin V, editors. Plant Proteomics: Methods and Protocols. Vol. 335. Totowa, NJ, USA: Humana Press; 2007. [Google Scholar]
- 20.Tuskan GA, DiFazio S, Jansson S, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313(5793):1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 21.Taylor G. Populus: Arabidopsis for forestry. Do we need a model tree? Annals of Botany. 2002;90(6):681–689. doi: 10.1093/aob/mcf255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanghe A, Van Dijck P, Dumortier F, Teunissen A, Hohmann S, Thevelein JM. Aquaporin expression correlates with freeze tolerance in baker’s yeast, and overexpression improves freeze tolerance in industrial strains. Applied and Environmental Microbiology. 2002;68(12):5981–5989. doi: 10.1128/AEM.68.12.5981-5989.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang B. The human aquaporin gene family. Current Genomics. 2000;1(1):91–102. [Google Scholar]
- 24.Bienert GP, Thorsen M, Schüssler MD, et al. A subgroup of plant aquaporins facilitate the bi-directional diffusion of As(OH)3 and Sb(OH)3 across membranes. BMC Biology. 2008;6, article 26 doi: 10.1186/1741-7007-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaal BK, Ishida K-I, Green BR. A thylakoidal processing peptidase from the heterokont alga Heterosigma akashiwo. Plant Molecular Biology. 2003;52(2):463–472. doi: 10.1023/a:1023900100803. [DOI] [PubMed] [Google Scholar]
- 26.Hirayama T, Oka A. Novel protein kinase of Arabidopsis thaliana (APK1) that phosphorylates tyrosine, serine and threonine. Plant Molecular Biology. 1992;20(4):653–662. doi: 10.1007/BF00046450. [DOI] [PubMed] [Google Scholar]
- 27.Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241(4861):42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 28.Kitagawa M, Wada C, Yoshioka S, Yura T. Expression of ClpB, an analog of the ATP-dependent protease regulatory subunit in Escherichia coli, is controlled by a heat shock sigma factor (sigma 32) Journal of Bacteriology. 1991;173(14):4247–4253. doi: 10.1128/jb.173.14.4247-4253.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsell DA, Kowal AS, Lindquist S. Saccharomyces cerevisiae Hsp104 protein. Purification and characterization of ATP-induced structural changes. Journal of Biological Chemistry. 1994;269(6):4480–4487. [PubMed] [Google Scholar]
- 30.Lee U, Rioflorido I, Hong S-W, Larkindale J, Waters ER, Vierling E. The Arabidopsis ClpB/Hsp100 family of proteins: chaperones for stress and chloroplast development. The Plant Journal. 2007;49(1):115–127. doi: 10.1111/j.1365-313X.2006.02940.x. [DOI] [PubMed] [Google Scholar]
- 31.Adam Z, Rudella A, van Wijk KJ. Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Current Opinion in Plant Biology. 2006;9(3):234–240. doi: 10.1016/j.pbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Sinibaldi RM, Mettler IJ. Intron splicing and intron-mediated enhanced expression in monocots. Progress in Nucleic Acid Research and Molecular Biology. 1992;42:229–257. doi: 10.1016/s0079-6603(08)60577-2. [DOI] [PubMed] [Google Scholar]
- 33.Gray GR, Villarimo AR, Whitehead CL, McIntosh L. Transgenic tobacco (Nicotiana tabacum L.) plants with increased expression levels of mitochondrial NADP+-dependent isocitrate dehydrogenase: evidence implicating this enzyme in the redox activation of the alternative oxidase. Plant and Cell Physiology. 2004;45(10):1413–1425. doi: 10.1093/pcp/pch162. [DOI] [PubMed] [Google Scholar]
- 34.Castro-Guerrero NA, Rodríguez-Zavala JS, Marín-Hernández A, Rodríguez-Enríquez S, Moreno-Sánchez R. Enhanced alternative oxidase and antioxidant enzymes under Cd2+ stress in Euglena. Journal of Bioenergetics and Biomembranes. 2008;40(3):227–235. doi: 10.1007/s10863-007-9098-6. [DOI] [PubMed] [Google Scholar]
- 35.Masoud SA, Johnson LB, White FF, Reeck GR. Expression of a cysteine proteinase inhibitor (oryzacystatin-I) in transgenic tobacco plants. Plant Molecular Biology. 1993;21(4):655–663. doi: 10.1007/BF00014548. [DOI] [PubMed] [Google Scholar]
- 36.Ettari R, Micale N, Schirmeister T, et al. Novel peptidomimetics containing a vinyl ester moiety as highly potent and selective falcipain-2 inhibitors. Journal of Medicinal Chemistry. 2009;52(7):2157–2160. doi: 10.1021/jm900047j. [DOI] [PubMed] [Google Scholar]
- 37.Oberschall A, Deak M, Torok K, et al. A novel aldose/aldehyde reductase protects transgenic plants against lipid peroxidation under chemical and drought stresses. The Plant Journal. 2000;24(4):437–446. doi: 10.1046/j.1365-313x.2000.00885.x. [DOI] [PubMed] [Google Scholar]
- 38.Lee WM. Drug-induced hepatotoxicity. The New England Journal of Medicine. 2003;349(5):474–485. doi: 10.1056/NEJMra021844. [DOI] [PubMed] [Google Scholar]
- 39.Narusaka Y, Narusaka M, Seki M, et al. Crosstalk in the responses to abiotic and biotic stresses in Arabidopsis: analysis of gene expression in cytochrome P450 gene superfamily by cDNA microarray. Plant Molecular Biology. 2004;55(3):327–342. doi: 10.1007/s11103-004-0685-1. [DOI] [PubMed] [Google Scholar]
- 40.Doty SL, James CA, Moore AL, et al. Enhanced phytoremediation of volatile environmental pollutants with transgenic trees. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(43):16816–16821. doi: 10.1073/pnas.0703276104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quarrie SA, Gulli M, Calestani C, Steed A, Marmiroli N. Location of a gene regulating drought-induced abscisic acid production on the long arm of chromosome 5A of wheat. Theoretical and Applied Genetics. 1994;89(6):p. 794. doi: 10.1007/BF00223721. [DOI] [PubMed] [Google Scholar]
- 42.Hsu YT, Kao CH. Role of abscisic acid in cadmium tolerance of rice (Oryza sativa L.) seedlings. Plant, Cell and Environment. 2003;26(6):867–874. doi: 10.1046/j.1365-3040.2003.01018.x. [DOI] [PubMed] [Google Scholar]
- 43.Houtz RL, Magnani R, Nayak NR, Dirk LMA. Co- and post-translational modifications in Rubisco: unanswered questions. Journal of Experimental Botany. 2008;59(7):1635–1645. doi: 10.1093/jxb/erm360. [DOI] [PubMed] [Google Scholar]
- 44.Hutchinson TH, Mahshid Y, Jönsson R, Björklund C, Kenne K. Proteomic analysis of phospholipidosis in citalopram treated U937 cells—support for the cholesterol biosynthesis hypothesis. Toxicology in Vitro. 2008;22(5):1198–1204. doi: 10.1016/j.tiv.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 45.Mendoza-Cózatl DG, Butko E, Springer F, et al. Identification of high levels of phytochelatins, glutathione and cadmium in the phloem sap of Brassica napus. A role for thiol-peptides in the long-distance transport of cadmium and the effect of cadmium on iron translocation. The Plant Journal. 2008;54(2):249–259. doi: 10.1111/j.1365-313X.2008.03410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sasaki Y, Nagano Y. Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Bioscience, Biotechnology and Biochemistry. 2004;68(6):1175–1184. doi: 10.1271/bbb.68.1175. [DOI] [PubMed] [Google Scholar]
- 47.Kim S-Y, Seo M, Oh J-M, Cho E-A, Juhnn Y-S. Inhibition of gamma ray-induced apoptosis by stimulatory heterotrimeric GTP binding protein involves Bcl-xL down-regulation in SH-SY5Y human neuroblastoma cells. Experimental and Molecular Medicine. 2007;39(5):583–593. doi: 10.1038/emm.2007.64. [DOI] [PubMed] [Google Scholar]
- 48.Chen JJ, Bertrand H, Yu BP. Inhibition of adenine nucleotide translocator by lipid peroxidation products. Free Radical Biology and Medicine. 1995;19(5):583–590. doi: 10.1016/0891-5849(95)00066-7. [DOI] [PubMed] [Google Scholar]
- 49.Hashimoto H, Nishi R, Umeda M, Uchimiya H, Kato A. Isolation and characterization of a rice cDNA clone encoding ATP/ADP translocator. Plant Molecular Biology. 1993;22(1):163–164. doi: 10.1007/BF00039005. [DOI] [PubMed] [Google Scholar]