Abstract
Summary: CD-HIT is a widely used program for clustering and comparing large biological sequence datasets. In order to further assist the CD-HIT users, we significantly improved this program with more functions and better accuracy, scalability and flexibility. Most importantly, we developed a new web server, CD-HIT Suite, for clustering a user-uploaded sequence dataset or comparing it to another dataset at different identity levels. Users can now interactively explore the clusters within web browsers. We also provide downloadable clusters for several public databases (NCBI NR, Swissprot and PDB) at different identity levels.
Availability: Free access at http://cd-hit.org
Contact: liwz@sdsc.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The size of the biological sequence databases is rapidly growing due to large-scale genome projects and the emerging field of metagenomics (Yooseph et al., 2007). New sequencing technologies are now producing sequence data at a very high rate, and this has created a greater need for bioinformatics tools to effectively organize and analyze the data. Fortunately, biological sequences are related and may share homology, and thus clustering these sequences into groups and finding a representative or a consensus for each group are practical ways to solve the sequence analysis problems.
Our previous works (Li and Godzik, 2006; Li et al., 2001; Li et al., 2002) introduced CD-HIT based on short word filtering and a greedy incremental clustering algorithm to cluster and compare large biological sequence datasets. One advantage of CD-HIT is its ultrahigh speed and the ability to handle large datasets. Since its release, CD-HIT has been widely used by many groups in various fields, including UniRef (Suzek et al., 2007), SMART (Letunic et al., 2009) and metagenome data analyses (Turnbaugh et al., 2009; Yooseph et al., 2008).
In the last few years, we have been continuously improving this program with more functions and better accuracy, scalability and flexibility. We also implemented a new web server to allow users to cluster or compare sequences without installing and executing the command-line version of CD-HIT locally. The server provides interactive interface and additional visualization tools. It also provides precalculated and regularly updated sequence clusters for several widely used databases, including NCBI NR, Swissprot and PDB.
2 METHODS AND IMPLEMENTATION
The detailed algorithms and benchmark results for CD-HIT can be found from our previous works (Li and Godzik, 2006; Li et al., 2001; Li et al., 2002). Here, we highlight the novel features and functions.
2.1 Improved clustering algorithm
The original CD-HIT uses a fast greedy incremental clustering process. Briefly, sequences are first sorted by decreasing length. The longest one becomes the representative of the first cluster. Then, each remaining sequence is compared with the existing representatives. If the identity with any representative is above a given threshold, it is grouped into that cluster without comparing it to other representatives. Otherwise, it becomes the representative of a new cluster. In the updated CD-HIT, we added a refined greedy incremental clustering process that produces more accurate clusters. In this process, a sequence is grouped into the most similar cluster instead of the first similar cluster. The refined process does not change the representative sequences.
CD-HIT uses a short word filter to avoid unnecessary alignments. In short, the minimum number of identical short words (k-mers) shared by two sequences depends on their sequence identity and can be calculated analytically or statistically. Without an actual alignment, we can still determine that the identity of two sequences is below a given threshold by counting short words. A short word filter performs much better with a higher identity threshold. Clustering in the refined process is implemented with a dynamic short word filter. For each sequence to be clustered, the initial filter matches the user-defined identity threshold. But during the clustering procedure, if this sequence hits any cluster with better identity, the filter is reset to match this better identity to increase the performance of the filter. With the dynamic short word filter, although the refined clustering process needs to evaluate the similarities of a sequence and all the existing representatives, it only requires about 1.5–3× CPU time of the original process.
2.2 Improved clustering control
The original CD-HIT uses global sequence identities. The improved CD-HIT also works with local identities. Users can finely control the clustering behavior by including more criteria besides sequence identity cutoffs. We include alignment length, unaligned length and alignment coverage for both aligned sequences as new clustering parameters into the current CD-HIT. For example, users can make clusters of sequences of similar length by specifying that the alignments must cover both sequences at similar coverage.
2.3 Clustering at low identity thresholds
The performance of the native short word filter drops significantly with a lower identity threshold; therefore, the original CD-HIT does not provide protein clustering under 40% identity. However, clustering at low identities has been frequently requested by CD-HIT users. We implemented a script, called PSI-CD-HIT, to perform protein sequence clustering at a low identity threshold such as 30%. It uses the similar greedy incremental clustering strategy, but it uses BLAST to calculate the similarities. So users can also specify an expect-value cutoff. PSI-CD-HIT runs on a stand-alone computer or a LINUX cluster. It can cluster a PDB-sized dataset in ∼20 min.
2.4 Hierarchical clustering
In the hierarchical clustering process, the program first performs clustering on the original input dataset at a high identity threshold, and the representatives of each previous clustering step will be the input of the following clustering run at a lower identity threshold. The whole process iteratively joins the similar sequences into families and therefore produces a hierarchical structure. For protein sequences, the last step is performed with PSI-CD-HIT if the final identity threshold is <40%. This strategy can maximize the computational efficiency and the quality of clustering. We have applied such strategy in a protein family analysis of a large metagenomic dataset with 17 million sequences (Li et al., 2008).
2.5 Annotation enrichment of sequence cluster
We provide an option for joint analysis of sequence clustering and annotation information. Users can place annotation terms (Gene Ontology, protein family, etc.) in the definition lines of input FASTA files. For each annotation term A and each cluster C, we use the following numbers:
NAC = number of sequences with A in C;
NC = number of sequences in C;
NAI = number of sequences with A in the input;
NI = number of the input sequences.
A P-value is calculated using the one-tailed Fisher's exact test to assess whether NAC/NC > NAI/NI and annotation term A is enriched in cluster C. Such functionality is very useful to check the cluster quality at different identity levels and also for function assignment of proteins with unknown function.
2.6 Web server
All basic functions of CD-HIT are provided through tab-based interfaces in our web server. We provided CD-HIT (CD-HIT-EST) to cluster a protein (DNA/RNA) dataset. Users can upload a FASTA file and select a desired sequence identity level and other parameters. CD-HIT-2D (CD-HIT-EST-2D) can compare two databases uploaded by users. H-CD-HIT and H-CD-HIT-EST in our server performs hierarchical clustering up to three steps.
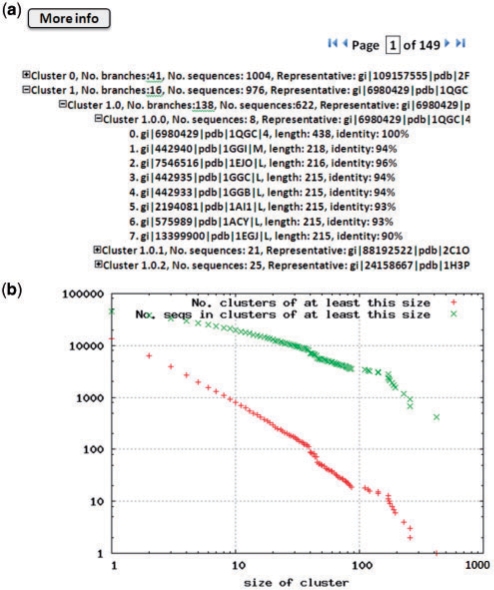
After submitting a clustering or comparison job, a unique identifier will be assigned. A user can use the identifier to track the status of the job. After the job is finished, we provide the raw outputs generated by the command-line CD-HIT. Additionally, we provide tools to visualize the clustering results with cluster explorer and cluster distribution plots. Cluster explorer uses a tree structure to represent the clustering results Figure 1a. Each cluster is represented by a clickable text object on the web page, and users can click on a representative sequence to retrieve information of the sequences belong to the cluster. This option is most useful for investigating the results from hierarchical clustering. In this situation, each sequence could be a representative sequence from the previous clustering step, and users can click it to explore the results from the previous clustering. Cluster distribution plots are scatter plots where the X-axis is the cluster size (number of sequences in a cluster), and then the Y-axis represents the number of clusters of at least this size and the number of corresponding sequences Figure 1b. This tool is very useful to observe the global structure of a sequence database.
Fig. 1.
Screenshots of CD-HIT Suite. (a) Cluster Explorer for investigating clusters. (b) A cluster distribution plot to explore the global structure of a whole dataset.
3 CONCLUSION
CD-HIT has been significantly improved from our previous work. CD-HIT Suite provides users with a friendly web interface to perform biological sequence clustering and comparison with additional visualization tools. It also provides precalculated clusters for several public sequence databases which are regularly updated.
ACKNOWLEDGEMENTS
We thank Mr Michael Chiu for his excellent editorial assistance.
Funding: National Institutes of Health (1R01RR025030) from National Center for Research Resources.
Conflict of Interest: none declared.
REFERENCES
- Letunic I, et al. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Li W, et al. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics. 2001;17:282–283. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- Li W, et al. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics. 2002;18:77–82. doi: 10.1093/bioinformatics/18.1.77. [DOI] [PubMed] [Google Scholar]
- Li W, et al. Probing metagenomics by rapid cluster analysis of very large datasets. PLoS ONE. 2008;3:e3375. doi: 10.1371/journal.pone.0003375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzek BE, et al. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yooseph S, et al. The Sorcerer II global ocean sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yooseph S, et al. Gene identification and protein classification in microbial metametagenomic sequence data via incremental clustering. BMC Bioinformatics. 2008;9:182. doi: 10.1186/1471-2105-9-182. [DOI] [PMC free article] [PubMed] [Google Scholar]