Abstract
Summary: The Microbial Proteomic Resource (MPR) is a repository service that contains non-redundant protein databases of related bacterial strains, which were generated through an in-house developed software called Multi-Strain Mass Spectrometry Prokaryotic DataBase Builder (MSMSpdbb). MSMSpdbb merges and clusters protein sequences inferred from genomic sequences, and provide a protein list in FASTA format that covers for divergence in gene annotation, translational start site choice and presence of single nucleotide polymorphisms and other mutations.
Availability: MSMSpdbb was developed in C++ using the Qt libraries (Nokia) and licensed under the GNU General Public License version 2. MSMSpdbb is freely available, and its installation files, instructions for use and additional documentation can be found at the MPR web site http://org.uib.no/prokaryotedb/ can also be found at Proteomecommons.org (see Supplementary Methods for Hash number).
Contact: Gustavo.Souza@biomed.uib.no
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
There are currently >900 completed bacterial genomic sequences available and over 3000 ongoing genome projects (Genomes Online Database GOLD) (Kyrpides, 1999). For proteomic purposes, there are often several relevant annotated genomic sequences available, and it may be difficult to select the best option for characterization of the experimental data. Different bioinformatic approaches for annotation of genes are currently used and this affects the quality of annotated protein lists used in proteomics (de Souza et al., 2008; de Souza et al., 2009). Characterization of clinical bacterial isolates with many specific genetic variations is suboptimal if they are not represented in the selected database. Recent data suggest that microbial virulence can be related to single nucleotide polymorphisms (SNPs) (Garcia Pelayo et al., 2009), and information from more than one sequenced genome can be relevant. Therefore, it is crucial to provide protein databases that can compensate for annotation errors and cover genetic variations among closely related organisms, and that provide readily identifiable unique observations. This limitation is acknowledged by the efforts of investigators to provide non-redundant protein databases with high genomic coverage, as for example the International Protein Index (Kersey et al., 2004).
The software Multi-Strain Mass Spectrometry Prokaryotic DataBase Builder (MSMSpdbb) can merge protein databases from several sources and be applied on any prokaryotic organism. The clustering is performed so that sequences sharing high similarity are merged and reported only once, and unique peptide sequences observed in polymorphisms or with different translational start site (TSS) choices are appended to the primary sequence in a MS-friendly manner using non-assigned characters as previously described by Schandorff et al. (2007). In short, N-terminal tryptic peptides from divergent predictions of the TSS are appended to the main sequence and separated by the code ‘O’, and polymorphic peptides are appended with the code ‘J’. Currently, this can be applied only using an in-house Mascot server, since the engine search tool need to be set up to recognize those two letters as a theoretical amino acid of mass zero, and also a new enzyme rule for trypsin can be created, which will recognize J and O as cleavage sites. Details of the database entry structure are described in Supplementary Methods.
LTQ-Orbitrap tandem mass spectrometry data from Helicobacter pylori (data not shown) or Mycobacterium tuberculosis H37Rv whole-cell lysate samples were used to test the new databases. The data of M.tuberculosis H37Rv were analyzed using the clustered database of annotated proteins from eight genomic sequences of the M.tuberculosis complex (MTC) (see Supplementary Methods). The original annotation of M.tuberculosis H37Rv (Tuberculist) was used as a reference. We have previously shown that for analysis of M.tuberculosis H37Rv proteins, the Tuberculist annotation performed better than a posterior annotation provided by the TIGR Institute (not used in this work), but some peptides only entered in the secondary annotation were also found (de Souza et al., 2008). The question is how much extra peptide information can be obtained from M.tuberculosis H37Rv using all genomic sequences with annotations from MTC organisms. Supplementary Figure 1 shows that the MTC database had 14% more protein entries and 24% more theoretical unique peptides (7–50 amino acids long) as compared with the Tuberculist database (Supplementary File 1 lists all proteins and peptides from both databases). A larger MTC database did not significantly compromise the statistical validation of the data with Mascot. By using MaxQuant (Cox and Mann, 2008) for validation, the number of identified MS/MS events were very similar at 1% false discovery rate. The MTC database identified 0.34% more peptides than the Tuberculist database, observed in high-quality spectra.
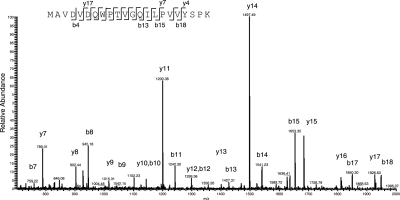
The higher number of peptides identified by the MTC database was expected to be caused by differences in the interpretation of coding sequences. For example, Figure 1 illustrate an MS/MS fragmentation pattern of ion m/z 1221.641, identified as peptide MAVDVDQWPTVGQILPVVYSPK from entry MT3222 (from the M.tuberculosis CDC1551 annotation) with a Mascot Score of 108 (score 24 indicated P < 0.01), 0.32 p.p.m. mass accuracy and a MaxQuant posterior error probability of 8.67E-36. This protein entry is only annotated in the M.tuberculosis CDC1551, H37Ra and KZN-1435 genomes, and it was identified with two peptides covering 23.6% of the sequence. However, the gene coding region is present in all eight genomes showing that the gene was simply not annotated in five of the genomes. Similar observations of previously unrecognized gene products in M.tuberculosis H37Rv are between 50 and 130 amino acids long (data not shown). This shows that gene prediction tools may fail to correctly annotate small genes.
Fig. 1.
Identification of MT3222 in M.tuberculosis H37Rv. A representative MS/MS spectrum present in the M.tuberculosis H37Rv whole-cell lysate sample. The fragmentation profile matched a peptide (inset) with a Mascot score of 108 derived from MT3222, a hypothetical protein annotated only in the CDC1551, H37Ra and KZN-1435 genomes.
Additionally, peptides with minor sequence variations were also successfully identified using the MTC database. For example, the protein Rv0412c contains a peptide, INSDISVGNYR, with a SNP that is specific to the M.tuberculosis H37Rv and H37Ra genomes. Supplementary Figure 2 shows the MS/MS fragmentation pattern of the peptide with a Mascot score of 72. In the remaining genomes, the aspartic acid is replaced by tyrosine, and we also identified this peptide in clinical isolate samples of M.tuberculosis (data not shown). Peptides containing SNPs were also observed in the H.pylori sample (data not shown).
In conclusion, multi-strain proteomic databases allow for identification of sequence variations between strains such as SNPs and divergent TSSs, in addition to cover limitations of gene annotations without compromising peptide redundancy and peptide identification rates. The application of this type of databases for proteomic characterization of relevant clinical strains which have not been sequenced is of interest.
MSMSpdbb and its source code are freely available at the Microbial Proteomic Resource at http://org.uib.no/prokaryotedb/ and can be modified to be used for other bacterial species (instructions about software usage are given in Supplementary Methods). We have applied MSMSpdbb for many different species of clinical and industrial interest, generating both full translations and annotated-only clustered databases. The full translation option from MSMSpdbb can also be used on individual genomes in order to be able to identify unannotated proteins. While full translation databases of merged genomes are often very large and could compromise statistical validation of the data, a full translation of an individual genome can be used at almost no false positive cost (de Souza et al., 2009). So far, we have available databases of 32 species, and updates are done on a weekly basis, with further species being added. Already available databases are also updated when new genome sequences are released. On request, MSMSpdbb users will also be allowed to upload their databases.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Matthias Mann (Max Planck Institute, Martinsried, Germany) and Dr Matthias Selbach (Max-Delbruck Centre, Berlin, Germany) for providing H.pylori data which were also used to validate the MSMSpdbb software.
Funding: Norwegian Research Council (175141) (FUGE).
Conflict of Interest: none declared.
REFERENCES
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- de Souza GA, et al. High accuracy mass spectrometry analysis as a tool to verify and improve gene annotation using Mycobacterium tuberculosis as an example. BMC Genomics. 2008;9:316. doi: 10.1186/1471-2164-9-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza GA, et al. Validating divergent ORF annotation of the Mycobacterium leprae genome through a full translation data set and peptide identification by tandem mass spectrometry. Proteomics. 2009;9:3233–3243. doi: 10.1002/pmic.200800955. [DOI] [PubMed] [Google Scholar]
- Garcia Pelayo MC, et al. A comprehensive survey of single nucleotide polymorphisms (SNPs) across Mycobacterium bovis strains and M. bovis BCG vaccine strains refines the genealogy and defines a minimal set of SNPs that separate virulent M. bovis strains and M. bovis BCG strains. Infect. Immun. 2009;77:2230–2238. doi: 10.1128/IAI.01099-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ, et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- Kyrpides NC. Genomes OnLine Database (GOLD 1.0): a monitor of complete and ongoing genome projects world-wide. Bioinformatics. 1999;15:773–774. doi: 10.1093/bioinformatics/15.9.773. [DOI] [PubMed] [Google Scholar]
- Schandorff S, et al. A mass spectrometry-friendly database for cSNP identification. Nat. Methods. 2007;4:465–466. doi: 10.1038/nmeth0607-465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.