Abstract
Summary: BamView is an interactive Java application for visualizing the large amounts of data stored for sequence reads which are aligned against a reference genome sequence. It supports the BAM (Binary Alignment/Map) format. It can be used in a number of contexts including SNP calling and structural annotation. BamView has also been integrated into Artemis so that the reads can be viewed in the context of the nucleotide sequence and genomic features.
Availability: BamView and Artemis are freely available (under a GPL licence) for download (for MacOSX, UNIX and Windows) at: http://bamview.sourceforge.net/
Contact: artemis@sanger.ac.uk
1 INTRODUCTION
Second-generation sequencing produces large volumes of short-read sequence data. In common applications of the technology, such as resequencing or transcriptome sequencing, reads are mapped against a reference genome. In many cases, bases in a reference are covered with alignment depths varying by orders of magnitude and therefore present a challenge for visualization.
SAM (Sequence Alignment/Map) and BAM (Binary Alignment/Map) formats are emerging as a standard representation for read alignments. It is therefore important to have visualization software for this format. BAM files contain the same information as SAM. As BAM format is compressed it provides an efficient means to store the data and enables fast retrieval of regions and so this format has been adopted here.
SAMTools (Li et al., 2009) includes a very simple text alignment viewer using the GNU ncurses library giving a detailed view at the nucleotide resolution level. Lookseq (Manske and Kwiatkowski, 2009) is a perl-cgi application used to display, in a web browser, reads mapped against a reference. It can read the data either directly from BAM files or from a relational database to display reads and plots paired read positions against their inferred size.
Alignment tools can be used to produce BAM format files. For instance, SSAHA (Ning et al., 2001) now supports SAM format (Long et al., 2009) and Maq output (Li et al., 2008) can be converted to SAM/BAM using SAMTools.
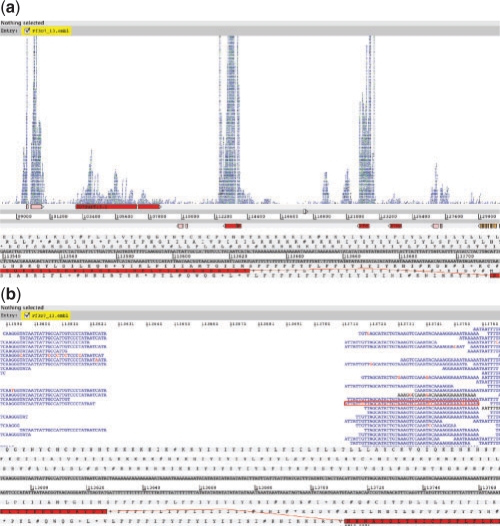
BamView can be used as a stand-alone Java application or displayed in Artemis (Carver et al., 2008; Rutherford et al., 2000, Fig. 1) in conjunction with the reference sequence and annotation. Artemis is a freely available genome browser and annotation tool. BamView in Artemis provides the annotator with an extra level of information that can inform them about structural annotation.
Fig. 1.
(a) Showing the BAM stack view in the Artemis genome browser. This is a region in chromosome 13 of Plasmodium falciparum 3D7. The BAM view is displaying RNA-Seq Solexa reads from the early ring stage in the life cycle of P.falciparum. (b) This is a zoomed in view showing the boundaries of adjacent exons. These boundaries are confirmed by the reads in BamView. A paired read is selected and marked by a red rectangle. SNPs are coloured red. The black reads are single reads.
BamView not only displays the Lookseq type of view (plotting against the paired reads inferred size) but also has other types of views that are described below. It does not require a web server and reads the data from an indexed BAM file.
2 IMPLEMENTATION
The BAM file needs to be sorted and indexed using the SAMTools command tool. This creates the BAM index file that the viewer uses to access the region to display in a fast way. BamView uses picard (picard.sourceforge.net), which is a Java API to read from the BAM file the reads in the region of sequence being displayed. Picard requires Java 1.6+ and so this is the minimum requirement for BamView.
The alignment data can be displayed at different levels of resolution (Fig. 1). Zooming out displays more of the read alignments across the reference sequence. The stand-alone BamView has ‘+’ and ‘−’ buttons to zoom in and out. In Artemis, the resolution of BamView matches the top feature display (Fig. 1). The resolution is controlled using the Artemis zoom scrollbar (on the right-hand side of the feature display). All other BamView options and functionality are in both the standalone and integrated versions.
Right clicking on the window gives a pop-up menu with a ‘View’ menu. There are four views provided by BAMView. The first is similar to one devised within Lookseq, whereby paired reads are plotted against their inferred insert size. An option displays single read alignments. There is an option to draw this against a log scale of the inferred size. In situations where the distribution of inferred sizes is broad this has the advantage of showing them closer together. The second is a ‘stack view’ where reads in a region are displayed piled against the reference to reduce the alignment depth. A variation is the ‘paired stack view’, where lines join paired reads within the stacks. When forward and reverse reads are described in the BAM file, the ‘strand stack view’ can be used to display reads above and below a line representing the reference.
The reads are colour coded so that paired reads are blue and those with an inversion are red. Reads that do not have a mapped mate are black and are optionally shown in the inferred insert size view. In the stack view, duplicated reads that span the same region are collapsed into one green line.
In addition to these views, when zoomed in the reads are displayed at the nucleotide level to shows how they are aligned to the reference sequence. Bases can be coloured by their mapping quality score (blue <10; green <20; orange <30; black ≥30) and there is an option to display insertions.
Bases that disagree with the reference consensus (sequencing errors or polymorphism) can also be displayed by selecting an option in the pop-up menu. These are red vertical lines on the reads and displayed as red nucleotides when zoomed in.
From the pop-up menu, BamView has the option to filter the reads displayed. This enables the user to display reads that are of significance and it also has the advantage of reducing the memory required by the programme. Reads can be filtered based on their mapping quality (i.e. the MAPQ field in the BAM file). Read alignments can also be filtered using the FLAG field. This field is used in the SAM/BAM specification to describe predefined properties of each read alignment (e.g. proper pair, mate unmapped, first of pair).
When the user points the mouse cursor over a read, a tool-tip will display information about the read (name, coordinates, insert size and reference sequence name). The reads can be selected which is useful to track the read when zooming in and out. When fully zoomed in, the selected reads are highlighted by a red rectangle. Right clicking on a reads shows a pop-up menu that includes an option to go to the other read of a mate pair.
3 DISCUSSION
Using the different views in BamView and as an integrated window in the Artemis tool means that it has a range of uses. For example, BamView can be used to inspect the confidence of a deep alignment of short reads, by highlighting base discrepancies. As it is integrated into Artemis, the underlying reference consensus sequence can be edited directly based on the aligned evidence. More commonly, the view is used in-house to assess the evidence behind a SNP or other polymorphism call. The alignment depth and base quality can both be easily browsed and inspected. Perhaps the greatest strength of BamView, when embedded in Artemis, is that it allows annotation to be changed based on manually inspecting data from transcriptome sequencing experiments (RNAseq, Otto et al., 2009; Wang et al., 2009). An annotator can zoom into intron–exon boundaries, identified from coverage plots, and see the quality of evidence supporting a prediction or manually adjust exons coordinates to fit the evidence. Simply viewing individually aligned reads cannot resolve alternate splicing patterns, but by clicking through read-pairs, an annotator can reconstruct the phase of exons in different isoforms.
ACKNOWLEDGEMENTS
We would like to thank Gary Dillon, Jacqui McQuillan, Anna Protasio for their suggestions in the development of this application.
Funding: Wellcome Trust through their funding of the Pathogen Genomics group at the Wellcome Trust Sanger Institute (WT 085775/Z/08/Z).
Conflict of Interest: none declared.
REFERENCES
- Carver T, et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, et al. HI: haplotype improver using paired-end short reads. Bioinformatics. 2009;15:2436–2437. doi: 10.1093/bioinformatics/btp412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manske HM, Kwiatkowski DP. LookSeq: a browser-based viewer for deep sequencing data. Genome Res. 2009;19:2125–2132. doi: 10.1101/gr.093443.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Z, et al. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto TD, et al. New insights into the blood stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol. Microbiol. 2009 doi: 10.1111/j.1365-2958.2009.07026.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, et al. Artemis: sequence visualisation and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Wang Z, et al. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]