Abstract
The flowering time of plants is tightly regulated by both promotive and repressive factors. Molecular genetic studies using Arabidopsis have identified several epigenetic repressors that regulate flowering time. TERMINAL FLOWER2 (TFL2), which encodes a homolog of HETEROCHROMATIN PROTEIN1, represses FLOWERING LOCUS T (FT) expression, which is induced by the activator CONSTANS (CO) in response to the long-day signal. Here, we show that TFL2, CO, and FT are expressed together in leaf vascular tissues and that TFL2 represses FT expression continuously throughout development. Mutations in TFL2 derepress FT expression within the vascular tissues of leaves, resulting in daylength-independent early flowering. TFL2 can reduce FT expression even when CO is overexpressed. However, FT expression reaches a level sufficient for floral induction even in the presence of TFL2, suggesting that TFL2 does not maintain FT in a silent state or inhibit it completely; rather, it counteracts the effect of CO on FT activation.
INTRODUCTION
The flowering of plants is regulated by many environmental stimuli and endogenous factors. The flowering of Arabidopsis is promoted by long days, gibberellins, and vernalization, but it can occur eventually even in the absence of environmental cues (autonomous promotion) (reviewed by Araki, 2001; Mouradov et al., 2002; Simpson and Dean, 2002). Recent findings suggest that the ambient temperature also affects the flowering of Arabidopsis (Blazquez et al., 2003).Thus, repressive factors as well as promotive factors are important in the regulation of flowering.
Molecular genetic screens of late and early flowering mutants of Arabidopsis have identified several genes involved in flowering and have found that external and internal floral promotion signals ultimately increase the expression levels of genes called floral pathway integrators, such as LEAFY, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1)/AGAMOUS-LIKE20, and FLOWERING LOCUS T (FT) (Blazquez and Weigel, 2000; Lee et al., 2000; Samach et al., 2000). Among these flowering pathway integrator genes, FT seems to have great importance, because several flowering pathways, including the long-day, vernalization, autonomous promotion, and temperature-dependent pathways, are integrated into the regulation of FT expression. Moreover, loss-of-function ft mutations cause a severe late-flowering phenotype, and overexpression of FT causes an early-flowering phenotype that is independent of daylength and temperature (Kardailsky et al., 1999; Kobayashi et al., 1999; Blazquez et al., 2003).
In the long-day pathway, CONSTANS (CO), which is a B-box–type zinc finger protein that shares identity with GATA transcription factors, directly activates FT expression in response to long-day signals (Samach et al., 2000; Suarez-Lopez et al., 2001; Yanovsky and Kay, 2002), and in the vernalization and autonomous pathways, FLOWERING LOCUS C (FLC) negatively regulates FT expression (Hepworth et al., 2002). Classic experiments suggest that the long-day pathway encompasses several processes that occur in different plant tissues: long-day signals are received by leaves, and signaling molecules generated in the leaves are transmitted to the shoot apical meristem (SAM) to induce flowering. Because of their low expression levels, however, the spatial expression patterns of CO and FT are poorly understood, making it difficult to predict the processes in which these genes are involved.
Recent findings from Arabidopsis research suggest that plants use a chromatin-mediated gene repression system to regulate flowering time. For example, VERNALIZATION2 (VRN2), a SU(Z)12-like Polycomb-group protein, is involved in the vernalization response through the stable silencing of FLC expression: vrn2 mutations abolish the vernalization response and cause the derepression of FLC (Gendall et al., 2001). Mutations in two other Polycomb-group proteins, EMBRYONIC FLOWER2 [SU(Z)12-like] and FERTILIZATION-INDEPENDENT ENDOSPERM (EXTRA SEX COMB-like), also cause the precocious upregulation of several flowering genes, resulting in early-flowering phenotypes (Kinoshita et al., 2001; Yoshida et al., 2001; Moon et al., 2003). TERMINAL FLOWER2 (TFL2), also called LIKE HETEROCHROMATIN PROTEIN1, encodes a protein homologous with an epigenetic repressor, HETEROCHROMATIN PROTEIN1 (HP1), which is involved in heterochromatin formation and the repression of some euchromatic genes in animals and fission yeast (Eissenberg and Elgin, 2000; Gaudin et al., 2001; Li et al., 2002; Kotake et al., 2003). Loss-of-function tfl2 mutants show a daylength-independent early-flowering phenotype (Larsson et al., 1998; Kotake et al., 2003). Upregulation of FT in tfl2 is the main cause of early flowering, because among known flowering genes only FT is upregulated in tfl2 mutants and ft mutations completely suppress the early flowering of tfl2 (Kotake et al., 2003).
It remains largely unknown how wild-type plants can overcome the epigenetic repression of flowering genes during floral induction. TFL2 represents a good system in which to examine gene activation under such epigenetic repression, because FT is known to be activated directly by CO (Samach et al., 2000). Because TFL2 is expressed in various regions of plant tissues, in this study we initially characterized the spatial and temporal expression patterns of CO and FT in an attempt to understand their regulation. We found that both TFL2 and CO regulate FT expression in the vascular tissues of leaves, which suggests that CO and FT may have leaf-specific functions. With regard to the mechanisms of TFL2, we show that TFL2 can reduce, but cannot completely repress, the expression of FT by counteracting the activity of CO. Our results suggest that this counterbalance of TFL2 and CO activity on the expression of FT ensures the daylength-regulated flowering response of Arabidopsis.
RESULTS
Expression Patterns of TFL2, CO, and FT in Wild-Type Plants
To address whether TFL2 and CO regulate FT in the same tissue regions, we first analyzed the spatial expression patterns of these genes. As a result of their low expression, little is known about the expression domains of either CO or FT. Therefore, we generated transgenic plants using genomic fragments of TFL2, CO, and FT, which were sufficient to complement the respective tfl2, co, and ft mutants, coupled to the β-glucuronidase (GUS) reporter gene.
In CO gene (gCO):GUS transgenic plants, GUS was expressed in a CO genomic context: the start codon of an 8.4-kb genomic fragment spanning the gCO, which was sufficient to complement co-101 (Table 1), was replaced with the GUS coding region (Figure 1A). For the detection of TFL2, we used gTFL2:GUS transgenic plants, in which a functional TFL2:GUS fusion protein was expressed in a TFL2 genomic context (Figure 1A) (Kotake et al., 2003). To detect FT expression, an 8.9-kb region upstream of the FT start codon was fused to the GUS coding region (Figure 1A). An 11.8-kb genomic fragment (gFT) spanning the same 8.9-kb upstream sequence largely recovered the late flowering of ft-101 (Table 1). Also, the expression of FT cDNA under the control of this 8.9-kb fragment was sufficient to rescue the late flowering of ft-1 (Table 1), and this 8.9-kb promoter region seemed to contain regulatory sequences responsive to CO and TFL2 (see below). Moreover, both gCO:co-101 and pFT:FT/ft-1 transgenic plant lines delayed flowering time under short-day conditions (data not shown). Therefore, each construct contained the full regulatory sequence, indicating that the GUS expression pattern would mimic the expression pattern of endogenous mRNA, thereby facilitating highly sensitive whole-mount expression analysis, which is difficult to achieve by in situ hybridization.
Table 1.
Complementation of co, ft, and tfl2 Mutants
| Plant | No. of Rosette Leaves | se | n |
|---|---|---|---|
| Wild type (Col) | 6.9 | 0.1 | 29 |
| tfl2-2 | 4.9 | 0.2 | 9 |
| gTFL2:GUS #17/tfl2-2 | 7 | 0.14 | 26 |
| ft-101 | 43.7 | 1.93 | 6 |
| gFT #1/ft-101 | 8.3 | 0.31 | 8 |
| gFT #7/ft-101 | 8.5 | 0.29 | 4 |
| ft-1 (Col) | 36.1 | 1.49 | 7 |
| pFT:FT #5/ft-1 | 6.5 | 0.27 | 21 |
| pFT:FT #11/ft-1 | 7.8 | 0.2 | 20 |
| co-101 | 16.7 | 0.87 | 7 |
| gCO #7/co-101 | 7.8 | 0.31 | 20 |
| gCO #18/co-101 | 7.3 | 0.35 | 29 |
Plants were grown on Murashige and Skoog (1962) plates containing 2% sucrose at a density of 30 plants/9-cm-diameter plate, except for ft and co-101 mutants, which were grown at a density of 9 plants/plate. Kanamycin-resistant T2 progeny were used for analysis.
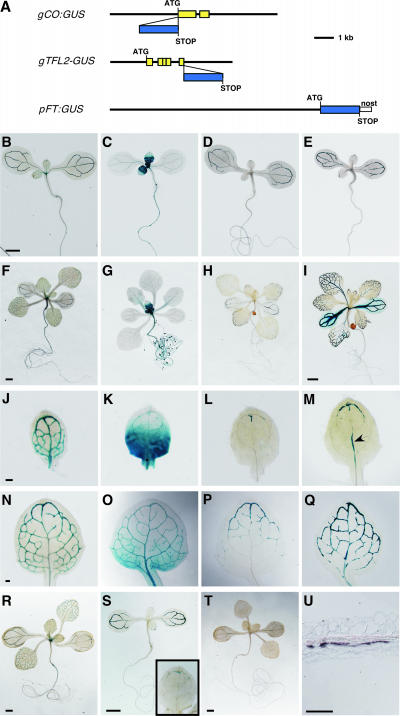
Figure 1.
Whole-Mount Expression Analyses of CO, FT, and TFL2.
(A) Constructs used in expression analyses (see Methods). Yellow boxes indicate open reading frames. Blue boxes indicate the uidA open reading frame (GUS gene). ATG and STOP indicate what actually function, so that only gTFL2:GUS encodes a fusion protein. nost, the nopaline synthase terminator.
(B) to (Q) GUS expression patterns of gCO:GUS ([B], [F], [J], and [N]), gTFL2:GUS ([C], [G], [K], and [O]), and pFT:GUS ([D], [H], [L], and [P]) in ecotype Columbia (Col) and pFT:GUS in tfl2-2 ([E], [I], [M], and [Q]) in whole-mount staining of 6-day-old seedlings ([B] to [E]), 12-day-old seedlings ([F] to [I]), the first true leaves of 6-day-old seedlings ([J] to [M]), and the first true leaves of 8-day-old seedlings ([N] to [Q]).
(R) to (T) GUS expression in short-day conditions of 12-day-old gCO:GUS (R), 6-day-old pFT:GUS (S), and 12-day-old pFT:GUS (T) plants. The inset in (S) shows a higher magnification of the first true leaf.
(U) In situ hybridization against FT mRNA in the cotyledon of tfl2-2 (longitudinal section).
The arrowhead indicates GUS expression in the primary vein in (M). Bars = 1 mm in (B) for (B) to (E), 1 mm in (F) for (F) to (H), 1 mm in (I), 0.1 mm in (J) for (J) to (M), 0.1 mm in (N) for (N) to (Q), 1 mm in (R) to (T), and 0.1 mm in (U).
In wild-type plants, the expression of pFT:GUS (i.e., an FT promoter fused to GUS) was seen first in the vascular tissues of cotyledons (Figure 1D) and was detected later in the vascular tissues of the apical part of the leaves (Figure 1P). GUS expression was not obvious in the primary veins or in the basal parts of the leaves, even in mature seedlings (Figures 1H and 1P). pFT:GUS expression was not continuous in vascular tissues and was interrupted (Figure 1P). pFT:GUS expression was not detected in leaf primordia, SAMs, hypocotyls, or roots (Figures 1D, 1H, and 2C). In inflorescences, pFT:GUS was expressed in the vascular tissues of inflorescence stems, pedicels, and floral organs but not in the inflorescence meristem (data not shown). Under short-day conditions, the expression pattern of pFT:GUS was similar to that under long-day conditions in 6-day-old seedlings (cf. Figure 1S with Figures 1D and 1L); however, after 8 days, pFT:GUS expression in true leaves diminished (Figure 1T). This photoperiod-dependent pFT:GUS expression also guarantees that this construct has full regulatory elements.
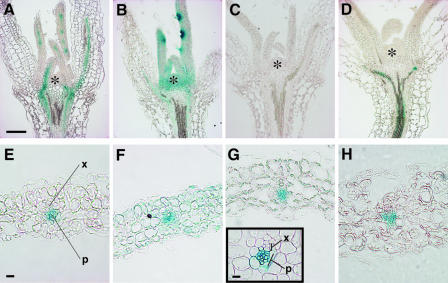
Figure 2.
Histological Analyses of GUS Expression Patterns.
(A) to (D) Longitudinal sections through 8-day-old seedlings of gCO:GUS (A), gTFL2:GUS (B), and pFT:GUS (C) in Col and pFT:GUS in tfl2-2 (D).
(E) to (H) Transverse sections through leaves of gCO:GUS (E), gTFL2:GUS (F), and pFT:GUS (G) in Col and pFT:GUS in tfl2-2 (H).
Asterisks indicates the SAM. P, phloem; X, xylem. Bars = 0.1 mm in (A) for (A) to (D), 10 μm in (E) for (E) to (H), and 10 μm for the inset in (G).
gTFL2:GUS was expressed in proliferating cells of leaves, SAMs, roots, and hypocotyls (Figures 1C, 1G, 1K, 1O, and 2B), as described previously (Kotake et al., 2003). gTFL2:GUS also was expressed in the vascular tissues of leaves and cotyledons (Figures 1C, 1K, and 1O).
gCO:GUS expression was detected in the vascular tissues of cotyledons and leaves (Figures 1B, 1F, 1J, and 1N). In leaves, GUS expression was detected first in the provascular tissues of leaf primordia and was detected later in the vascular tissues of whole leaves (Figures 1J, 1N, and 2A). gCO:GUS expression in roots depended on the line: 5 of 10 lines showed GUS expression in root tips (data not shown). Like endogenous CO, gCO:GUS expression was not changed under short-day conditions (Figure 1R) (Suarez-Lopez et al., 2001; Yanovsky and Kay, 2002).
The expression patterns of both gCO:GUS and gTFL2:GUS in leaves were consistent with the results obtained by in situ hybridization (Simon et al., 1996; Kotake et al., 2003). gCO:GUS and pFT:GUS were expressed strongly in phloem within the vascular tissues (Figures 2E, 2G, and 2H). gTFL2:GUS was expressed in the epidermis, mesophyll, and phloem tissues of leaves (Figure 2F). These results suggest that, to regulate FT expression, TFL2 and CO must function in the phloem cells of leaves, because this is the only region in which all three of these genes are expressed.
tfl2 Causes an Increase in pFT:GUS Expression
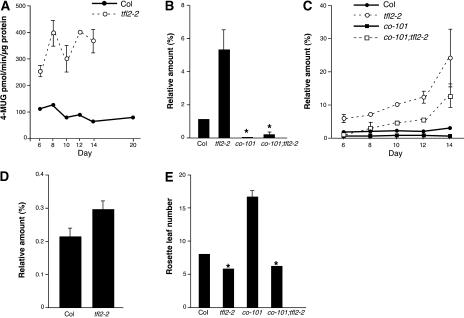
To examine whether the expression of pFT:GUS was responsive to TFL2, lines expressing pFT:GUS in a tfl2 background were crossed with the wild type, and total GUS expression was analyzed in F2 progeny. In tfl2, GUS activities were at least twice as high as those in the wild-type background on any day examined, showing that expression levels of pFT:GUS were repressed by TFL2 (Figure 3A). This result also suggests that the expression of pFT:GUS mimics endogenous FT expression.
Figure 3.
tfl2 Affects Both the CO-Dependent and CO-Independent Expression of FT.
(A) Quantitative GUS expression analysis of pFT:GUS in Col (solid line) and tfl2-2 (dashed line). GUS activity is shown as the mean ± se of 4-methylumbelliferyl glucuronide·min−1·μg−1 protein of three independent experiments.
(B) to (D) Real-time quantitative RT-PCR analysis of FT ([B] and [C]) and CO (D) expression. The same RNA extract was used for the experiments shown in (B) and (D). These data are normalized to the amount of ACT2 (set as 100%) and are means ± se of three independent experiments.
(B) FT expression levels in 6-day-old Col, tfl2-2, co-101, and co-101 tfl2-2 seedlings.
(C) FT expression over time in Col, tfl2-2, co-101, and co-101 tfl2-2 seedlings.
(D) CO expression levels in 6-day-old Col and tfl2-2 seedlings.
(E) Flowering time measured as the mean number (±se) of rosette leaves at flowering in Col, co-101, tfl2-2, and co-101 tfl2-2 (n = 27, 7, 27, and 9, respectively). All plants were grown under long-day conditions. Asterisks indicate data that do not show statistically significant differences.
pFT:GUS expression was detected earlier in tfl2 than in the wild type (90% of first two true leaves in tfl2 [n = 30] versus 27% in wild-type [n = 18] 6-day-old seedlings). Moreover, pFT:GUS was expressed ectopically in the primary veins of tfl2 leaves (Figure 1M, arrowhead) (86.7% of first two true leaves in 6-day-old seedlings [n = 30]). This expression was never observed in the wild type (Figures 1L and 1P) (0% of first two true leaves in 6-day-old seedlings [n = 18]). In the tfl2 background, the ectopic expression of pFT:GUS also was detected in hypocotyls (Figure 2D) but not in SAMs, young leaves, or roots (Figure 2D and data not shown). FT upregulation in tfl2 hypocotyls suggested that pFT:GUS also is expressed in wild-type hypocotyls but at undetectable levels, consistent with previous studies showing that FT can be detected in wild-type hypocotyls using highly sensitive reverse transcriptase–mediated (RT) PCR techniques (Kardailsky et al., 1999; Kobayashi et al., 1999). In our in situ hybridization experiments, FT expression was not detected in the leaf vascular tissues in the wild type or tfl2, but it was detected in the vascular tissues of tfl2 cotyledons, in which the strongest pFT:GUS expression was observed (Figures 1I and 1U). These results indicate that FT expression is upregulated with the same tissue specificity in tfl2 and wild-type plants, suggesting that the upregulation of FT in the vascular tissues of leaves may be sufficient to cause early flowering.
co Affects the Upregulation of FT in tfl2
To assess the effect of CO on the upregulation of FT in a tfl2 plant, we quantified FT expression in co tfl2 double mutants (see Methods). Real-time quantitative RT-PCR analyses showed that the upregulation of FT in tfl2 was largely suppressed by co in 6-day-old seedlings (Figure 3B), suggesting that CO and TFL2 antagonistically affect FT expression. In co tfl2, expression levels of FT were greater than in the wild type after 8 days (Figure 3C). co tfl2 flowered as early as tfl2 (Table 2, Figure 3E), suggesting that this upregulation of FT expression in co tfl2 after 8 days is sufficient for early flowering. This result also supports the notion that the early flowering of tfl2 is not attributable to the upregulation of CO (Kotake et al., 2003).
Table 2.
Flowering Time of Mutant and Transgenic Lines
| Plant | No. of Rosette Leaves | se | n |
|---|---|---|---|
| Wild type (Col) | 8.1 | 0.15 | 27 |
| tfl2-2 | 5.9 | 0.15 | 27 |
| co-101 | 16.7 | 0.87 | 7 |
| co-101 tfl2-2 | 6.3 | 0.17 | 9 |
| 35S:CO | 4.6 | 0.14 | 20 |
| 35S:CO tfl2-2a | 3.5 | 0.33 | 8 |
| 35S:TFL2 #16 | 6.9 | 0.4 | 8 |
| 35S:TFL2 #26 | 7.5 | 0.37 | 10 |
| 35S:TFL2 #16/+ 35S:CO/+ | 5 | 0 | 9 |
| 35S:TFL2 #26/+ 35S:CO/+ | 5 | 0.27 | 8 |
| 35S:CO/+ | 5 | 0 | 9 |
Plants were grown on Murashige and Skoog (1962) plates containing 2% sucrose at a density of 9 plants/9-cm-diameter plate. F1 plants were grown on plates containing kanamycin (50 μg/mL).
Includes both 35S:CO/+ tfl2-2 and 35S:CO tfl2-2.
In the co mutant background, pFT:GUS expression was not detected in 69.6% of leaves and was weakly detected in 30.4% of leaves in 12-day-old seedlings (n = 23) (Figures 4B and 4E). In co tfl2, however, pFT:GUS expression was detected in the basal parts of leaves (78.2% of leaves in 12-day-old seedlings [n = 23]) (Figures 4C and 4F). These results suggest that the expression of FT in the apical parts of leaves requires the activity of CO and that CO-independent FT upregulation in tfl2 occurs mainly in the basal parts of leaves.
Figure 4.
pFT:GUS Expression Requires CO Activity.
pFT:GUS expression is shown in 12-day-old seedlings of wild-type Col (A), co-101 (B), and co-101 tfl2-2 (C) (roots were cut off for genotyping) and in leaves of wild-type Col (D), co-101 (E), and co-101 tfl2-2 (F). Bars = 1 mm.
In 6-day-old seedlings, FT expression depends largely on CO, and the expression levels of CO were similar in tfl2 and wild-type plants (Figure 3D); however, the CO-dependent expression of FT was approximately five times greater in tfl2 than in the wild type (Figure 3B). These findings, together with the fact that FT was upregulated in the CO-expressing tissues of tfl2, indicate that TFL2 may alleviate the effect of CO on FT activation.
tfl2 Enhances the Early-Flowering Phenotype and Upregulation of FT in 35S:CO Plants
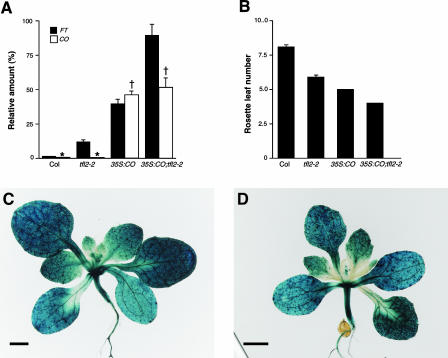
We next examined whether TFL2 could counteract the activity of CO in a CO overexpressor line, 35S:CO, which shows a daylength-independent early-flowering phenotype (Putterill et al., 1995; Simon et al., 1996). Quantitative RT-PCR analysis showed that expression levels of FT were approximately twice as high in 35S:CO tfl2 plants as in 35S:CO plants (Figure 5A). Moreover, 35S:CO tfl2 flowered earlier than 35S:CO (Figure 5B), reflecting the higher expression levels of FT. These findings demonstrate that TFL2 can reduce the expression of FT even in the CO overexpressor line 35S:CO.
Figure 5.
Roles of TFL2 during FT Repression in the CO Overexpressor.
(A) Real-time quantitative RT-PCR analysis of FT (closed bars) and CO (open bars) expression in 12-day-old seedlings of Col, tfl2-2, 35S:CO, and 35S:CO tfl2-2. The data are normalized to the amount of ACT2 (set as 100%) and are means ± se of three independent experiments. There was no statistically significant difference among genotypes marked with the same symbols.
(B) Flowering time measured as the mean number (±se) of rosette leaves at flowering in Col, tfl2-2, 35S:CO, and 35S:CO tfl2-2 (n = 27, 27, 43, and 22, respectively).
(C) and (D) Whole-mount analysis of pFT:GUS expression in 35S:CO (C) and 35S:CO tfl2-2 (D). Bars = 1 mm.
To examine the differences in the expression patterns of FT between 35S:CO and 35S:CO tfl2, the pFT:GUS transgene was introgressed into the 35S:CO and 35S:CO tfl2 backgrounds (see Methods). pFT:GUS was expressed ectopically in hypocotyls, roots, and inflorescences of both 35S:CO and 35S:CO tfl2 (Figures 5C and 5D). GUS expression also was observed in the primary veins of leaves in both the 35S:CO (91.7% of the first two true leaves in 8-day-old seedlings [n = 36]) and 35S:CO tfl2 (95.7% of the first two true leaves in 8-day-old seedlings [n = 47]) backgrounds. We found no significant differences in the expression patterns of FT within tissues between 35S:CO and 35S:CO tfl2 (Figures 5C and 5D), indicating that in 35S:CO, the expression patterns of FT are determined mainly by CO and are independent of the presence of TFL2.
In contrast to the loss of TFL2 expression, overexpression of TFL2 (35S:TFL2) showed little effect on flowering time and the expression levels of FT in both the wild-type and 35S:CO backgrounds (Table 2 and data not shown), suggesting that an increase in TFL2 expression alone does not affect flowering times.
DISCUSSION
Here, we have documented the spatial and temporal expression patterns of FT, which were poorly known previously. Our results suggest that FT expression in these tissues is sufficient for daylength-dependent flowering and is regulated by the activator CO and the repressor TFL2. Although TFL2 is homologous with HP1, TFL2 is not involved in an on/off switch of FT, because TFL2 does not determine the timing or the spatial pattern of FT expression. Rather, TFL2 counteracts the activity of activators such as CO on FT expression to ensure daylength-dependent flowering, because only a small change in the normal expression of FT is sufficient to induce flowering that is independent of daylength.
TFL2 Counteracts the Activation of the Transcription of FT
Three lines of evidence suggest that TFL2 counteracts the activation of FT by CO: (1) CO-dependent FT expression is enhanced in tfl2; (2) FT expression is upregulated in the CO-expressing tissues of tfl2; and (3) overexpression of CO induces higher levels of FT in the absence of TFL2 than in its presence. However, the fact that FT was still upregulated in co tfl2 (in 8-day-old seedlings and in the later stages) implies that activators other than CO also are involved in the activation of FT. These factors have been proposed in previous studies, because FT expression increases gradually in later seedling development even in co mutants (Kardailsky et al., 1999; Kobayashi et al., 1999; Samach et al., 2000). In the tfl2 background, FT is upregulated in the presence or absence of CO. A simple model to explain this CO-independent FT upregulation is that TFL2 directly represses FT expression.
How does TFL2 reduce the effect of transcriptional activators in the cells? TFL2 is homologous with HP1, which represses gene expression in heterochromatin by binding directly to histone H3 tails (methylated K9) to form higher order chromatin structures (Lachner et al., 2001); therefore, TFL2 may localize to the chromatin of FT to control transcription. In contrast to the proposed function of HP1 in heterochromatin formation in fly and mammals, however, TFL2 cannot completely inhibit but can only counteract the activities of transcription factors. This “offsetting” of the activity of the activator, as opposed to the stable repression observed (e.g., in the repression of FLC by the Polycomb-group protein VRN2), suggests a novel role of HP1-type repressors. In other words, TFL2 may constitute a component of the dynamic structure of silent chromatin that does not completely inhibit but only reduces the accessibility of transcription factors, as proposed recently in the “site-exposure model” (Ahmad and Henikoff, 2002).
TFL2 Is a Regulator of the Expression Levels of FT
Our results suggest that TFL2 does not determine the timing of floral transition in wild-type Arabidopsis, because the expression of TFL2 was not downregulated before flowering and 35S:TFL2 did not show late flowering (Table 1). Moreover, the expression level of TFL2 was not altered between long-day and short-day conditions, and TFL2 expression did not show circadian oscillations, suggesting that TFL2 activity itself does not change in response to daylength (our unpublished data). Rather, TFL2 functions to maintain a low level of FT both before and after the floral transition, because the expression levels of FT in the wild type never reached those in tfl2 even after the floral transition (Figure 3C). This mechanism of regulation differs from that used by FLC, whose expression must be diminished to induce flowering (Sheldon et al., 2000). Moreover, overexpression of FLC causes a strong repression of both FT and SOC1 even in the 35S:CO background (Hepworth et al., 2002), whereas TFL2 can only reduce the expression level of FT and has little role in determining the tissues that express FT in the 35S:CO background. Therefore, unlike other HP1 proteins, TFL2 seems to generally reduce gene expression levels and does not distinguish between active and silent states.
What is the biological relevance of the TFL2-mediated repression of FT? One interpretation is that only a small amount of FT is needed to induce flowering; therefore any enhanced response to CO will cause an accumulation of FT before floral induction and will disrupt the long-day-regulated flowering of plants. Thus, TFL2 may be expressed continuously to offset any excess activity of CO. In addition, even 35S:CO tfl2 could not induce pFT:GUS expression in young leaves, root tips, or 8-day-old SAMs (data not shown), suggesting that the existence of other repressors or the absence of coactivators may limit the activity of CO on FT expression in these tissues. EARLY BOLTING IN SHORT DAYS (EBS), a putative chromatin-remodeling factor that specifically represses FT expression (Gomez-Mena et al., 2001; Pineiro et al., 2003), is a candidate for another repressor that modulates the CO responsiveness of FT. In our yeast two-hybrid screening, however, TFL2 did not interact with EBS (our unpublished data), suggesting that TFL2 and EBS may form a different protein complex and may function independently. The existence of at least two repressors suggests that the strict regulation of FT expression levels is critical for the correct flowering time during plant development.
TFL2 and CO Regulate FT Expression in the Vascular Tissues of Leaves
Classic experiments have suggested that inductive photoperiods are recognized by leaves and that signaling molecules produced in the leaves are transmitted into SAMs to induce flowering (reviewed by Colasanti and Sundaresan, 2000). In addition, the expression of the maize INDETERMINATE gene only in leaves has been shown to be sufficient for floral promotion (Colasanti et al., 1998), indicating the importance of the expression of flowering genes in leaves. The expression of FT in leaves suggests that FT may be involved in a floral promotion signaling pathway in leaves; FT activation by CO may cause the generation of unidentified signal molecules in the vascular tissues of leaves, and these signals (or FT itself) may be transmitted via vascular tissues to SAMs to induce flowering. In potatoes, the Arabidopsis CO gene was found to act non-cell-autonomously to delay tuber formation, which is induced in short days (Martinez-Garcia et al., 2002). Our results show that in Arabidopsis, CO itself seems not to act non-cell-autonomously, because CO activates FT expression only in CO-expressing tissues, raising the possibility that FT and/or its downstream gene(s) may act non-cell-autonomously to induce flowering.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia (Col) was used as the wild type. We searched T-DNA insertion lines provided by the Torrey Mesa Research Institute and found co and ft mutant alleles in the Col background, named co-101 and ft-101, respectively. ft-1 was introgressed into Col and tfl2-2 is a null allele, as described previously (Kotake et al., 2003). 35S:CO transgenic plants in the Col background were a gift from G. Coupland (Max Planck Institute for Plant Breeding, Cologne, Germany).
To obtain co-101 tfl2-2 double mutants (CO and TFL2 loci are only ∼2 centimorgan apart), co-101 was crossed with tfl2-2. In F2 progeny, co-101/+ tfl2-2 seedlings were identified on the basis of BASTA resistance and the tfl2 phenotype, and co-101 tfl2-2 was obtained in the next generation. Genotypes were confirmed by PCR, and F4 plants were used for analysis.
To obtain 35S:CO tfl2-2 plants, 35S:CO was crossed with tfl2-2. 35S:CO tfl2-2 was identified by screening kanamycin-resistant F2 seedlings for a curled-leaf phenotype, and homozygosity for tfl2-2 was confirmed by PCR. Because of the low fertility of 35S:CO tfl2-2, F2 plants were used for flowering-time and expression analyses.
For flowering-time and expression analyses, plants were grown on Murashige and Skoog (1962) agar plates with 2% sucrose in long-day conditions (16 h of light/8 h of dark) under white fluorescent lights (∼50 μmol·m−2·s−1) or short-day conditions (10 h of light/14 h of dark; ∼50 μmol·m−2·s−1). Sown seeds were kept for 2 days at 4°C and then moved to 22°C, which was defined as day 0 after sowing.
Plasmid Construction and Transgenic Plants
gCO:co-101
An 8420-bp CO genomic fragment containing a region encompassing 3576 bp upstream of the start codon to 3489 bp downstream of the stop codon was excised from BAC clone F14F8, cloned into the pCGN1547 plant transformation vector (Calgene, Davis, CA), and transformed into co-101. Among T1 plants, 13 of 20 lines flowered earlier than co-101 and produced no more than 10 leaves. Two T2 lines were used to count the number of rosette leaves at flowering.
gCO:GUS
To make gCO:GUS, the start codon of the 8420-bp CO genomic fragment was replaced with uidA cDNA. Whole-mount GUS expression analysis revealed that 10 of 11 T2 lines showed basically the same expression pattern. Four strong lines, carrying a single locus insertion of the transgene, were sectioned. T2 seedlings were used for analysis.
gFT:ft-101
An 11,781-bp FT genomic fragment extending from 8,902 bp upstream of the start codon to 699 bp downstream of the stop codon was excised from BAC clone F5I14, cloned into the pCGN1547 vector, and used to transform ft-101. Two independent T2 lines were obtained and used for flowering-time analysis.
pFT:FT/ft-1
Introns were removed from the 11.8-kb FT genomic fragment by reference to the FT cDNA, and the 3′ downstream sequence was replaced with the nopaline synthase terminator. Thus, this construct contained 8.9 kb of 5′ upstream sequence and the coding region of the FT gene. In T1 plants, 12 of 20 independent lines flowered earlier than ft-1 and produced no more than 10 leaves. Two T2 lines were used to count the number of rosette leaves at flowering.
pFT:GUS
An 8.9-kb region upstream of the start codon of FT (the same region as in pFT:FT) was fused to the GUS coding sequence followed by the nopaline synthase terminator in the pCGN1547 vector. This construct was used to transform Col and tfl2-2. In the Col background, 9 of 15 T2 lines showed a similar expression pattern. In the tfl2-2 mutant background, 8 of 8 T2 lines showed a similar expression pattern. In general, GUS staining was stronger in tfl2-2 than in Col, and GUS expression in the primary veins of leaves was rarely observed in Col background, whereas all eight lines in the tfl2-2 background showed GUS expression in the primary veins. Two strong lines in the tfl2-2 background were sectioned, and four strong lines in the Col background were sectioned. For histological GUS staining, T2 seedlings were used. Two lines in the tfl2-2 background, carrying a single locus insertion of the transgene, were used for crossing with Col. GUS activity was compared between pFT:GUS/Col and pFT:GUS/tfl2-2 segregated in F2 progeny. To examine the expression pattern of pFT:GUS in the 35S:CO and 35S:CO tfl2-2 backgrounds, a pFT:GUS line in the tfl2-2 background was crossed with 35S:CO, and the expression patterns of GUS were analyzed in F2 progeny. To obtain pFT:GUS co-101 and pFT:GUS co-101 tfl2-2, a pFT:GUS line in the Col background was crossed with co-101 and co-101 tfl2-2, respectively, and kanamycin-resistant F2 seedlings were genotyped by PCR.
35S: TFL2
To obtain 35S:TFL2, TFL2 cDNA was inserted between the 35S promoter of Cauliflower mosaic virus and the nopaline synthase terminator in the pCGN1547 vector and used to transform Col. Two independent T3 homozygous lines, showing overexpression of TFL2 (data not shown), were used for analysis. To obtain 35S:TFL2/+ 35S:CO/+, 35S:TFL2 lines were crossed with 35S:CO, and F1 plants were used for flowering-time analysis.
Expression Analyses
Samples were collected at dusk, when the expression of FT is the highest (Suarez-Lopez et al., 2001; Kotake et al., 2003).
Real-Time Quantitative PCR
Real-time quantitative PCR using TaqMan probes was performed as described previously (Kotake et al., 2003) with the following modifications. Total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen, Valencia, CA) and was treated with RNase-free DNase (Qiagen) according to the manufacturer's instructions. Two micrograms of total RNA was reverse-transcribed using Omniscript reverse transcriptase (Qiagen). cDNA was resuspended in 200 μL of water, and 2.5-μL aliquots were analyzed.
Histological Analysis of GUS Staining
GUS staining was performed as described previously (Honma and Goto, 2000). Samples were embedded in paraffin and sectioned at a thickness of 8 μm with a microtome. For the section shown in the inset of Figure 2G, samples were embedded in Technovit 7100 (Heraeus Kulzer, Wehrheim, Germany) and sectioned at a thickness of 5 μm.
GUS Activity Measurement
For quantitative GUS measurements, we used an assay based on the substrate 4-methylumbelliferyl glucuronide, as described (Jefferson et al., 1987). The protein concentrations of samples were determined using a bicinchoninic acid assay kit (Pierce).
In Situ Hybridization
In situ hybridization was performed as described by Kotake et al. (2003). The full-length FT cDNA clone kindly provided by T. Araki (Kyoto University) was used as template for the FT probe.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact K. Goto, kgoto@v004.vaio.ne.jp.
Acknowledgments
We thank K. Nagae and Y. Nakamichi for technical assistance; T. Kotake, K. Nakahigashi, and T. Demura for discussion and technical advice; and the ABRC for BAC clones. This work was supported, in part, by grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.016345.
References
- Ahmad, K., and Henikoff, S. (2002). Epigenetic consequences of nucleosome dynamics. Cell 111, 281–284. [DOI] [PubMed] [Google Scholar]
- Araki, T. (2001). Transition from vegetative to reproductive phase. Curr. Opin. Plant Biol. 4, 63–68. [DOI] [PubMed] [Google Scholar]
- Blazquez, M.A., Ahn, J.H., and Weigel, D. (2003). A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat. Genet 33, 168–171. [DOI] [PubMed] [Google Scholar]
- Blazquez, M.A., and Weigel, D. (2000). Integration of floral inductive signals in Arabidopsis. Nature 404, 889–892. [DOI] [PubMed] [Google Scholar]
- Colasanti, J., and Sundaresan, V. (2000). ‘Florigen’ enters the molecular age: Long-distance signals that cause plants to flower. Trends Biochem. Sci. 25, 236–240. [DOI] [PubMed] [Google Scholar]
- Colasanti, J., Yuan, Z., and Sundaresan, V. (1998). The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93, 593–603. [DOI] [PubMed] [Google Scholar]
- Eissenberg, J.C., and Elgin, S.C. (2000). The HP1 protein family: Getting a grip on chromatin. Curr. Opin. Genet. Dev. 10, 204–210. [DOI] [PubMed] [Google Scholar]
- Gaudin, V., Libault, M., Pouteau, S., Juul, T., Zhao, G., Lefebvre, D., and Grandjean, O. (2001). Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development 128, 4847–4858. [DOI] [PubMed] [Google Scholar]
- Gendall, A.R., Levy, Y.Y., Wilson, A., and Dean, C. (2001). The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107, 525–535. [DOI] [PubMed] [Google Scholar]
- Gomez-Mena, C., Pineiro, M., Franco-Zorrilla, J.M., Salinas, J., Coupland, G., and Martinez-Zapater, J.M. (2001). early bolting in short days: An Arabidopsis mutation that causes early flowering and partially suppresses the floral phenotype of leafy. Plant Cell 13, 1011–1024. [PMC free article] [PubMed] [Google Scholar]
- Hepworth, S.R., Valverde, F., Ravenscroft, D., Mouradov, A., and Coupland, G. (2002). Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 21, 4327–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2000). The Arabidopsis floral homeotic gene PISTILLATA is regulated by discrete cis-elements responsive to induction and maintenance signals. Development 127, 2021–2030. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (2001). Polycomb repression of flowering during early plant development. Proc. Natl. Acad. Sci. USA 98, 14156–14161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Kotake, T., Takada, S., Nakahigashi, K., Ohto, M., and Goto, K. (2003). Arabidopsis TERMINAL FLOWER 2 gene encodes a Heterochromatin Protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol. 44, 555–564. [DOI] [PubMed] [Google Scholar]
- Lachner, M., O'Carroll, D., Rea, S., Mechtler, K., and Jenuwein, T. (2001). Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120. [DOI] [PubMed] [Google Scholar]
- Larsson, A.S., Landberg, K., and Meeks-Wagner, D.R. (1998). The TERMINAL FLOWER2 (TFL2) gene controls the reproductive transition and meristem identity in Arabidopsis thaliana. Genetics 149, 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H., Suh, S.S., Park, E., Cho, E., Ahn, J.H., Kim, S.G., Lee, J.S., Kwon, Y.M., and Lee, I. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Kirschmann, D.A., and Wallrath, L.L. (2002). Does heterochromatin protein 1 always follow code? Proc. Natl. Acad. Sci. USA 99 (suppl. 4), 16462.–16469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Virgos-Soler, A., and Prat, S. (2002). Control of photoperiod-regulated tuberization in potato by the Arabidopsis flowering-time gene CONSTANS. Proc. Natl. Acad. Sci. USA 99, 15211–15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, Y.H., Chen, L., Pan, R.L., Chang, H.S., Zhu, T., Maffeo, D.M., and Sung, Z.R. (2003). EMF genes maintain vegetative development by repressing the flower program in Arabidopsis. Plant Cell 15, 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov, A., Cremer, F., and Coupland, G. (2002). Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell 14 (suppl.), S111.–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Pineiro, M., Gomez-Mena, C., Schaffer, R., Martinez-Zapater, J.M., and Coupland, G. (2003). EARLY BOLTING IN SHORT DAYS is related to chromatin remodeling factors and regulates flowering in Arabidopsis by repressing FT. Plant Cell 15, 1552–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. [DOI] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sheldon, C.C., Rouse, D.T., Finnegan, E.J., Peacock, W.J., and Dennis, E.S. (2000). The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. USA 97, 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, R., Igeno, M.I., and Coupland, G. (1996). Activation of floral meristem identity genes in Arabidopsis. Nature 384, 59–62. [DOI] [PubMed] [Google Scholar]
- Simpson, G.G., and Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289. [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. [DOI] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419, 308–312. [DOI] [PubMed] [Google Scholar]
- Yoshida, N., Yanai, Y., Chen, L., Kato, Y., Hiratsuka, J., Miwa, T., Sung, Z.R., and Takahashi, S. (2001). EMBRYONIC FLOWER2, a novel Polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13, 2471–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]