Abstract
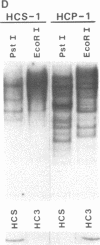
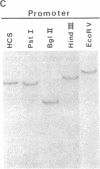
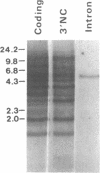
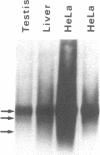
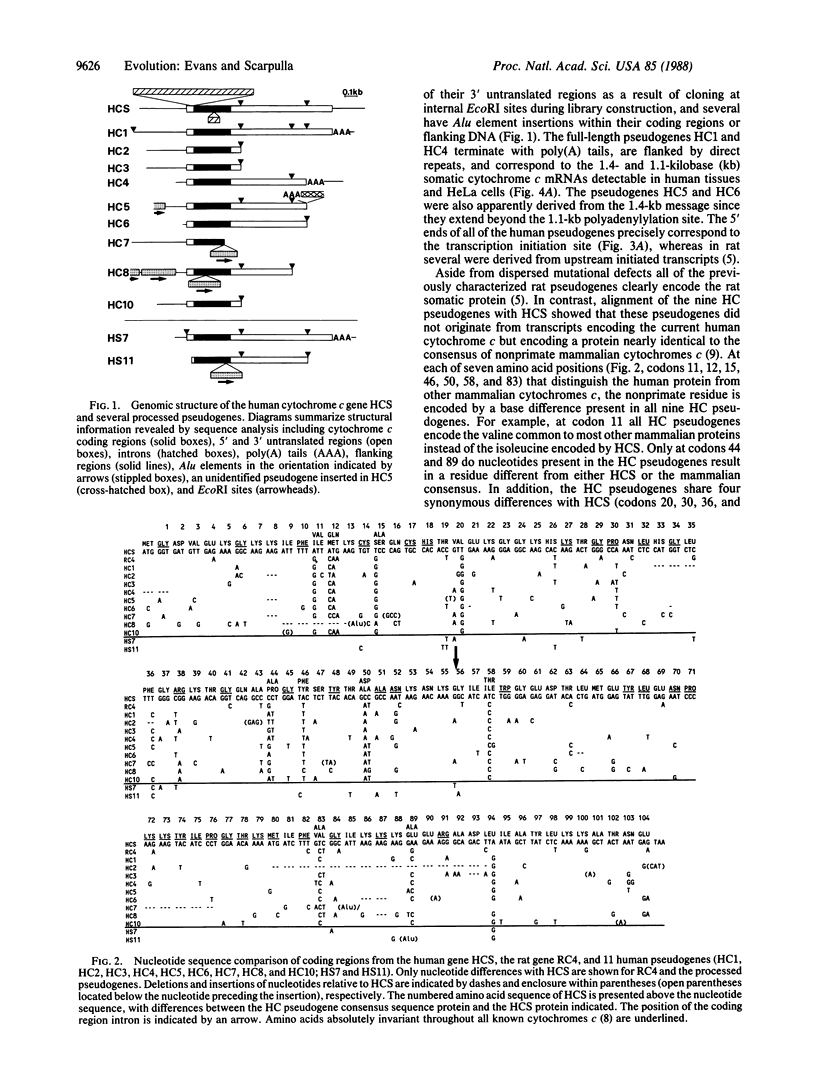
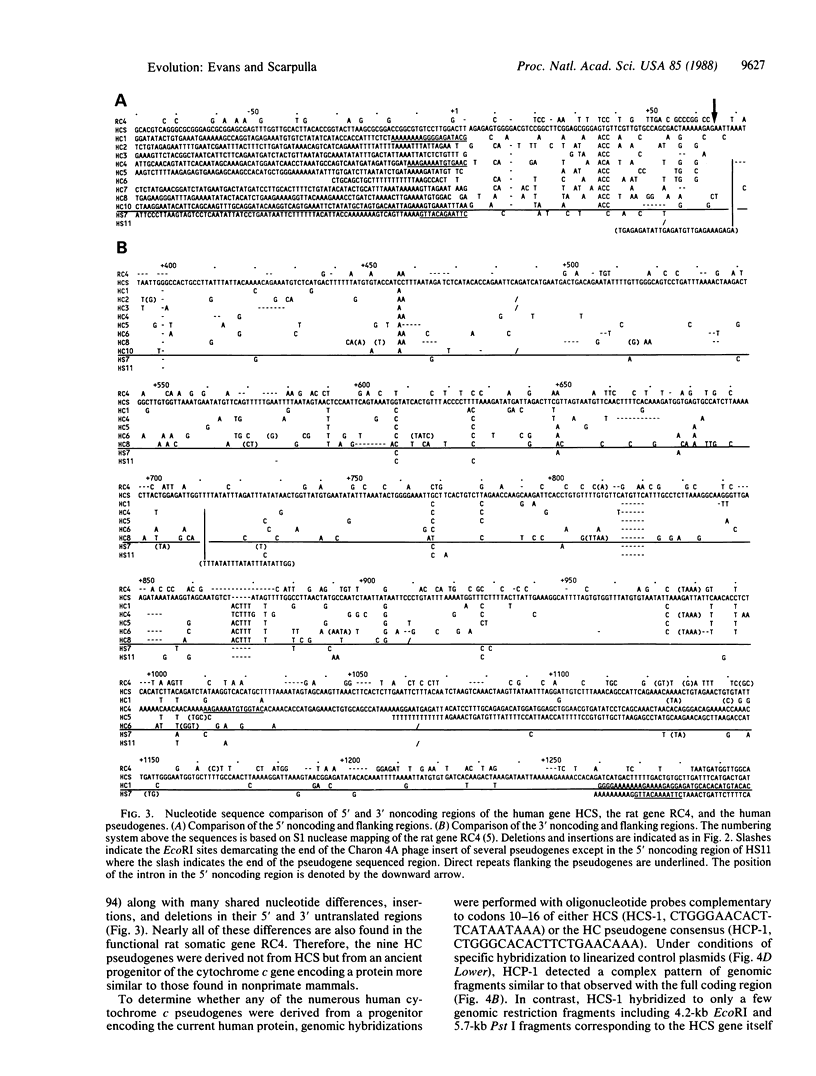
We have isolated and determined the DNA sequences of the human somatic cytochrome c gene (HCS) and 11 processed pseudogenes. HCS is the functional homologue to the previously characterized rat somatic gene because it correctly encodes the human heart protein, is present in single copy in the human genome, is nearly identical in both size and intron/exon structure to rodent somatic genes, and shares a high degree of sequence homology with its rat counterpart including a well-conserved promoter region (77% over 250 nucleotides). In contrast to the rodent system, however, where the known pseudogenes all originated from a locus encoding the present day cytochrome c, the human pseudogenes are of two types. A predominant class of older pseudogenes came from a progenitor of HCS that encoded an ancestral form of the protein, while a second group of only a few young pseudogenes originated from a recent parent of HCS that encoded the current cytochrome c polypeptide. These two distinct classes of human pseudogenes provide a molecular record of the history of cytochrome c evolution in primates and demarcate a short period of rapid evolution of the functional gene.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baba M. L., Darga L. L., Goodman M., Czelusniak J. Evolution of cytochrome C investigated by the maximum parsimony method. J Mol Evol. 1981;17(4):197–213. doi: 10.1007/BF01732758. [DOI] [PubMed] [Google Scholar]
- Bisson R., Azzi A., Gutweniger H., Colonna R., Montecucco C., Zanotti A. Interaction of cytochrome c with cytochrome c oxidase. Photoaffinity labeling of beef heart cytochrome c oxidase with arylazido-cytochrome c. J Biol Chem. 1978 Mar 25;253(6):1874–1880. [PubMed] [Google Scholar]
- Brown G. G., Simpson M. V. Novel features of animal mtDNA evolution as shown by sequences of two rat cytochrome oxidase subunit II genes. Proc Natl Acad Sci U S A. 1982 May;79(10):3246–3250. doi: 10.1073/pnas.79.10.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann R. L., Brown W. M., Wilson A. C. Polymorphic sites and the mechanism of evolution in human mitochondrial DNA. Genetics. 1984 Mar;106(3):479–499. doi: 10.1093/genetics/106.3.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. J., Scarpulla R. C. Both upstream and intron sequence elements are required for elevated expression of the rat somatic cytochrome c gene in COS-1 cells. Mol Cell Biol. 1988 Jan;8(1):35–41. doi: 10.1128/mcb.8.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey D. M., Das G., Sherman F. Amino acid replacements in yeast iso-1-cytochrome c. Comparison with the phylogenetic series and the tertiary structure of related cytochromes c. J Biol Chem. 1986 Mar 5;261(7):3259–3271. [PubMed] [Google Scholar]
- Hennig B. Change of cytochrome c structure during development of the mouse. Eur J Biochem. 1975 Jun 16;55(1):167–183. doi: 10.1111/j.1432-1033.1975.tb02149.x. [DOI] [PubMed] [Google Scholar]
- Koop B. F., Goodman M., Xu P., Chan K., Slightom J. L. Primate eta-globin DNA sequences and man's place among the great apes. Nature. 1986 Jan 16;319(6050):234–238. doi: 10.1038/319234a0. [DOI] [PubMed] [Google Scholar]
- Lawn R. M., Fritsch E. F., Parker R. C., Blake G., Maniatis T. The isolation and characterization of linked delta- and beta-globin genes from a cloned library of human DNA. Cell. 1978 Dec;15(4):1157–1174. doi: 10.1016/0092-8674(78)90043-0. [DOI] [PubMed] [Google Scholar]
- MARGOLIASH E. PRIMARY STRUCTURE AND EVOLUTION OF CYTOCHROME C. Proc Natl Acad Sci U S A. 1963 Oct;50:672–679. doi: 10.1073/pnas.50.4.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margoliash E., Fitch W. M. Evolutionary variability of cytochrome c primary structures. Ann N Y Acad Sci. 1968 Jun 14;151(1):359–381. doi: 10.1111/j.1749-6632.1968.tb11901.x. [DOI] [PubMed] [Google Scholar]
- Margoliash E., Schejter A. Cytochrome c. Adv Protein Chem. 1966;21:113–286. doi: 10.1016/s0065-3233(08)60128-x. [DOI] [PubMed] [Google Scholar]
- Osheroff N., Speck S. H., Margoliash E., Veerman E. C., Wilms J., König B. W., Muijsers A. O. The reaction of primate cytochromes c with cytochrome c oxidase. Analysis of the polarographic assay. J Biol Chem. 1983 May 10;258(9):5731–5738. [PubMed] [Google Scholar]
- Ramharack R., Deeley R. G. Structure and evolution of primate cytochrome c oxidase subunit II gene. J Biol Chem. 1987 Oct 15;262(29):14014–14021. [PubMed] [Google Scholar]
- Rothfus J. A., Smith E. L. Amino acid sequence of rhesus monkey heart cytochrome c. J Biol Chem. 1965 Nov;240(11):4277–4283. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R. C., Agne K. M., Wu R. Cytochrome c gene-related sequences in mammalian genomes. Proc Natl Acad Sci U S A. 1982 Feb;79(3):739–743. doi: 10.1073/pnas.79.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R. C., Agne K. M., Wu R. Isolation and structure of a rat cytochrome c gene. J Biol Chem. 1981 Jun 25;256(12):6480–6486. [PubMed] [Google Scholar]
- Scarpulla R. C. Association of a truncated cytochrome c processed pseudogene with a similarly truncated member from a long interspersed repeat family of rat. Nucleic Acids Res. 1985 Feb 11;13(3):763–775. doi: 10.1093/nar/13.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R. C. Processed pseudogenes for rat cytochrome c are preferentially derived from one of three alternate mRNAs. Mol Cell Biol. 1984 Nov;4(11):2279–2288. doi: 10.1128/mcb.4.11.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R. C., Wu R. Nonallelic members of the cytochrome c multigene family of the rat may arise through different messenger RNAs. Cell. 1983 Feb;32(2):473–482. doi: 10.1016/0092-8674(83)90467-1. [DOI] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W. Genetics and biosynthesis of cytochrome c. Annu Rev Genet. 1971;5:257–296. doi: 10.1146/annurev.ge.05.120171.001353. [DOI] [PubMed] [Google Scholar]
- Virbasius J. V., Scarpulla R. C. Structure and expression of rodent genes encoding the testis-specific cytochrome c. Differences in gene structure and evolution between somatic and testicular variants. J Biol Chem. 1988 May 15;263(14):6791–6796. [PubMed] [Google Scholar]
- Wu C. I., Li W. H., Shen J. J., Scarpulla R. C., Limbach K. J., Wu R. Evolution of cytochrome c genes and pseudogenes. J Mol Evol. 1986;23(1):61–75. doi: 10.1007/BF02100999. [DOI] [PubMed] [Google Scholar]