Abstract
We characterized a rice dwarf mutant, ebisu dwarf (d2). It showed the pleiotropic abnormal phenotype similar to that of the rice brassinosteroid (BR)-insensitive mutant, d61. The dwarf phenotype of d2 was rescued by exogenous brassinolide treatment. The accumulation profile of BR intermediates in the d2 mutants confirmed that these plants are deficient in late BR biosynthesis. We cloned the D2 gene by map-based cloning. The D2 gene encoded a novel cytochrome P450 classified in CYP90D that is highly similar to the reported BR synthesis enzymes. Introduction of the wild D2 gene into d2-1 rescued the abnormal phenotype of the mutants. In feeding experiments, 3-dehydro-6-deoxoteasterone, 3-dehydroteasterone, and brassinolide effectively caused the lamina joints of the d2 plants to bend, whereas more upstream compounds did not cause bending. Based on these results, we conclude that D2/CYP90D2 catalyzes the steps from 6-deoxoteasterone to 3-dehydro-6-deoxoteasterone and from teasterone to 3-dehydroteasterone in the late BR biosynthesis pathway.
INTRODUCTION
At present, >60 rice dwarf mutants have been identified (Matsuo et al., 1997) but only a few have been used for breeding, because almost all of them are severely dwarfed and/or show unsuitable pleiotropic phenotypes, making their use impractical (Peng et al., 1994). Some of them have good phenotypes that potentially could be useful for breeding if the severity of their dwarfism were controlled or if it were possible to suppress their unsuitable pleiotropic phenotypes. ebisu dwarf (dwarf2 or d2) is a good example of this kind of dwarf mutant, although its dwarfism is slightly stronger than the desirable level. In fact, the erect leaves of d2 allow this cultivar to be planted more densely than the original cultivar, which has bent leaves; consequently, a greater volume of crop products can be harvested in the same cultivation area. Thus, elucidation of the molecular mechanism of the relationship between dwarfism and erect leaves in d2 mutants is important for further molecular breeding for architectural modification.
Various factors cause dwarfism in plants, but recent molecular genetic studies using dwarf mutants of Arabidopsis and other dicot species revealed that gibberellin (GA) and brassinosteroid (BR) are the most important factors in determining plant height (Mandava, 1988; Clouse and Sasse, 1998; Taiz and Zeiger, 2002; Fujioka and Yokota, 2003). It is well known that in rice and other grass plants, GA-deficient or GA-insensitive mutants show the same dwarf phenotype found in dicots (Ashikari et al., 1999; Itoh et al., 2001, 2002). Actually, both of the semidwarf cultivars of rice and wheat used in the “green revolution” are affected by these types of factors, rice being affected by GA metabolism and wheat by insensitivity (Peng et al., 1999; Sasaki et al., 2002; Spielmeyer et al., 2002). By contrast, the relationship between BR and dwarfism in monocot plants has not been well studied. However, we recently characterized a rice dwarf mutant, d61, that shows a pleiotropic abnormal phenotype involving dwarfism and erect leaves. We found that the d61 mutation is caused by the loss of function of OsBRI1, which encodes a putative protein kinase highly similar to Arabidopsis BRI1, the putative BR receptor (Yamamuro et al., 2000). By this pioneer study, it has been revealed that BR is important for stem elongation in monocot plants, and it also served as a cue to investigate the functional role of BR in grass plants.
Besides dwarfism, d61 shows other unique characteristics that are not seen in GA-related dwarf mutants, including inhibition of elongation of the specific internode, erect leaves, and photomorphogenesis in the dark. Using these unique characteristics as screening criteria for BR-related mutants, we found that one of these, BR-deficient dwarf (brd1), was caused by the loss of function of a gene that encodes a protein homologous with the tomato DWARF and Arabidopsis BR6 oxidase proteins, which catalyze the C-6 oxidation step in BR biosynthesis (Shimada et al., 2001, 2003; Hong et al., 2002). The phenotype of brd1 confirmed that the unique characteristics observed in d61 are not specific to the BR-insensitive mutants but are common among BR-insensitive and BR-deficient mutants.
The inhibition of elongation in the second internode and erect leaves found in d2 also were characteristic of the BR-related phenotype; therefore, we inferred that d2 should be related to BR biosynthesis or sensitivity. Here, we report that a novel cytochrome P450, categorized in CYP90D, which has not been described as a protein catalyzing BR biosynthesis, is involved in BR biosynthesis. Rescue experiments with various BR intermediates indicated that the d2 mutation alters the C-3 oxidation step in BR biosynthesis.
RESULTS
Characterization of d2 Mutants
Figure 1A shows the gross morphology of d2 mutants in which the severity of the mutation differs. We confirmed that these two plants with different mutant severity were allelic by the allelism test (Table 1). After heading, the mutant plants reached ∼70% (strong allele, d2-1; right) or 80% (mild allele, d2-2; center) of the height of the wild-type plant (left). When we observed the elongation pattern of internodes of the mutant plants, the second internode from the top was shortened completely in d2-1 and partially in d2-2 (Figure 1B), whereas elongation of the other internodes was affected very little, even by the strong allele, d2-1. Another characteristic phenotype of d2 was its erect leaves (Figure 1C). In wild-type plants, the leaf blade bent away from the vertical axis of the leaf sheath toward the abaxial side (Figure 1C, left). However, in the d2 mutants, almost all of the leaves were appressed to the shoot (Figure 1C, right). In contrast to the second internode's growth inhibition, the neck internode of the mutants was longer than that of wild-type plants (Figure 1D). The mutants showed a further abnormal phenotype in that the grains were shortened slightly (Figure 1E).
Figure 1.
d2 Mutants Displaying Pleiotropic Abnormalities.
(A) Gross morphology at the heading stage of wild-type (left), mild allele (d2-2; center), and strong allele (d2-1; right) plants grown in the field. Bar = 20 cm.
(B) Elongation pattern of the second internode. The second internode from the top was shortened completely in d2-1 and partially in d2-2. From left to right, wild type, d2-2, and d2-1. Arrows indicate the positions of the nodes. Bar = 5 cm.
(C) Leaf morphology. The leaf blade of the wild type (left) bends away from the vertical axis of the leaf sheath toward the abaxial side. The leaf of d2-2 (right) is erect. Arrows show the lamina joint. Bar = 5 cm.
(D) Panicle morphology of the wild type (left) and d2-2 (right). Arrows indicate the positions of the nodes. Bar = 5 cm.
(E) Grain morphology. The mutant (right) has slightly shortened grains. Bar = 1 cm.
Table 1.
Allelism Test between d2-1, d2-2, and d61
| Phenotype of the Offspring after Cross
|
|||
|---|---|---|---|
| Mutant Lines | d2-1 | d2-2 | d61-1a |
| d2-1 | — | Dwarf | Wild |
| d2-2 | Dwarf | — | Wild |
| d61-1 | Wild | Wild | — |
A BR-insensitive mutant impaired in the OsBRI1 gene (Yamamuro et al., 2000).
The abnormal phenotype of d2 described above was similar to that of the BR-deficient brd1 or BR-insensitive d61 mutants; consequently, we suspected that d2 is deficient in BR biosynthesis or sensitivity. To confirm this possibility, we examined the morphogenesis of d2 seedlings grown in complete darkness. It has been reported that Arabidopsis BR-insensitive or BR-deficient mutants show a deetiolated phenotype characterized by less hypocotyl elongation, the opening of cotyledons, and the emergence of primary leaves (Chory et al., 1991; Szekeres et al., 1996; Li and Chory, 1997). The rice BR-insensitive and BR-deficient mutants that we observed showed a photomorphogenic phenotype when grown in the dark: the mesocotyl and internodes were not elongated (Yamamuro et al., 2000; Hong et al., 2002). Thus, this unique phenotype is a good criterion with which to determine whether or not a novel dwarf mutant is related to BR. We grew d2-2, wild-type, and GA-deficient (d18) plants in the dark. The mesocotyl and internodes of the wild-type and d18 plants elongated under dark conditions (Figure 2, left and center, respectively), but elongation did not occur in d2 (Figure 2, right). The failure of mesocotyl and internode elongation in the dark strongly supports the notion that the d2 plant is a BR-related mutant.
Figure 2.

Photomorphogenic Phenotype of d2 Grown in the Dark.
Wild-type (left), d18 (GA-deficient mutant; center), and d2-2 (right) plants were grown in complete darkness. Arrows indicate the nodes, and arrowheads indicate the mesocotyls. Bars = 1 cm.
Rescue of the Dwarf Phenotype of d2 by Brassinolide Treatment
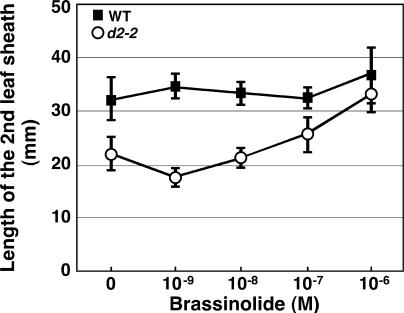
The phenotypic analyses of d2 strongly suggested that d2 is a BR-deficient or BR-insensitive mutant. To determine whether d2 is BR deficient or BR insensitive, we treated the mutants with the most bioactive BR compound, brassinolide (BL). There was no difference between the lengths of the second leaf sheath in untreated d2-2 plants and those given 10−8 M or lower concentrations of BL (Figure 3). However, when the d2 plants were treated with 10−6 M BL, the length of the second leaf sheath was almost the same as that of the wild-type plants. This result indicates that d2 can respond to exogenous BL to rescue the dwarf phenotype and therefore that d2 may be deficient in active BRs.
Figure 3.
Elongation of the Second Leaf Sheath in Wild-Type and d2-2 Plants after Treatment with BL.
The length of the second leaf sheath was measured 2 weeks after wild-type (WT; closed squares) and d2 (open circles) seeds were germinated on MS medium (Murashige and Skoog, 1962) containing various concentrations of BL. n = 25.
D2 Encodes a Novel Cytochrome P450
Rescue of the dwarf phenotype of d2 by BL treatment indicated that this mutant is deficient in the synthesis of bioactive BRs. We have isolated rice genes that are homologous with Arabidopsis BR biosynthesis genes—such as STE1 (DWF7), DWF5, DWF1 (DIM), DET2 (DWF6), DWF4, and CPD (DWF3)—and mapped these homologous genes in the rice genome (T. Sakamoto and M. Matsuoka, unpublished results). We also isolated the rice DWARF gene and mapped it in the rice genome (Hong et al., 2002). We surmised that D2 would correspond to one of these homologous genes because these rice genes included almost all of the Arabidopsis BR biosynthesis genes characterized to date; otherwise, D2 would encode a novel BR biosynthesis enzyme.
To identify the map position of d2, linkage analysis was performed using the F2 population derived from a cross between the d2 japonica mutant and an indica strain, Kasalath. We selected 30 individuals showing dwarfism similar to that of the original d2 plant and used them for identification of the map position of d2. The d2 mutation was located on the short arm of chromosome 1 near the mutation of the GA-deficient dwarf mutant d18 (data not shown). We found no genes homologous with the Arabidopsis BR biosynthesis genes near this position. Thus, D2 may encode a novel BR biosynthesis enzyme.
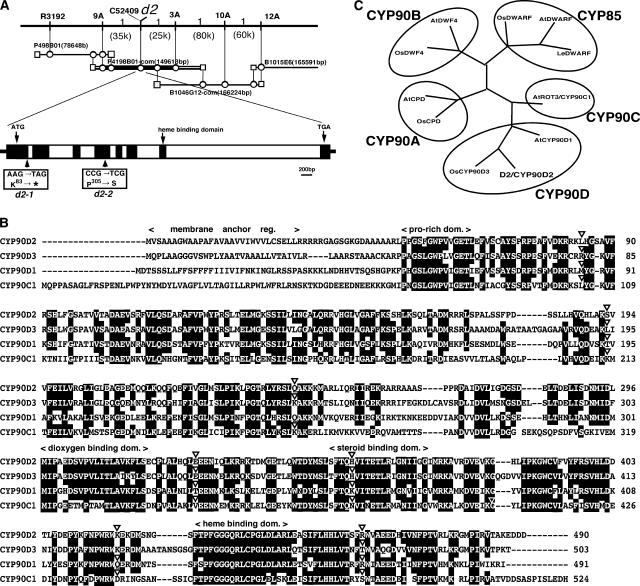
We attempted to isolate D2 by positional cloning using F2 plants from a cross between d2 and SL5. The SL5 plant basically contained the japonica genome except on the short arm of chromosome 1, where the indica genome from Kasalath was present. This plant, with the different short arm of chromosome 1, the putative site of d2, was provided to us by M. Yano of the National Institute of Agrobiological Resources (Tsukuba, Japan). The F2 plants were segregated into two groups showing the normal culm length similar to that of the wild-type japonica cultivar and the dwarf phenotype of d2 in a 3:1 ratio. Approximately 3000 F2 seeds were used for positional mapping of the d2 locus; we found that the d2 mutation was linked completely with a cleaved amplified polymorphic sequence marker, C52409 (Figure 4A). Around this marker in the japonica and indica cultivars, we found two single nucleotide polymorphisms, 9A and 3A, within a distance of ∼60 kb, both of which were located in the same BAC clone, P4198B01. One genetically recombinant plant was found with d2 between 9A and 3A, and the sequence of C52409 was located almost centrally between 9A and 3A (Figure 4A). Thus, we conclude that D2 is located near the central part of this 60-kb region.
Figure 4.
Physical Map of the D2 Gene and Structure of the D2 Protein.
(A) High-resolution linkage and physical map of the d2 locus. The vertical bars represent the molecular markers, and the numbers of recombinant plants are indicated above the linkage map. The d2 mutation was tightly linked with a marker, C52409. The physical distances between adjacent markers are shown in parentheses. The D2 gene consists of eight exons and seven introns. Closed and open rectangles indicate exons and introns, respectively. Mutations identified in d2-1 and d2-2 are indicated.
(B) Comparison of amino acid sequences of D2/CYP90D2 and other CYP90C and CYP90D proteins. According to the nomenclature for the P450 superfamily (Nelson et al., 1996), rice D2 and its D2 homolog were named CYP90D2 and CYP90D3, respectively. Dashes indicate gaps introduced to maximize alignment. Identical amino acids are represented by white-on-black letters. Triangles indicate the positions of intron insertions. Multiple sequence alignment was performed using the CLUSTAL W analysis tool in DDBJ.
(C) Phylogenetic relationship between D2 and the BR biosynthetic P450 protein. D2/CYP90D2 is highly similar to the Arabidopsis BR biosynthetic P450 proteins CYP90D1, ROT3/CYP90C1 (Kim et al., 1998), CYP90A1(Szekeres et al., 1996), CYP90B (Choe et al., 1998), CYP85 (Bishop et al., 1996), and CYP85A1 (Shimada et al., 2001). The structural relationship was calculated using CLUSTAL W and illustrated using Treeview (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Around this area, there were several putative genes, but we paid attention to one gene that encoded a putative cytochrome P450 (P450, also called CYP), because some members of P450, such as CPD, DWF4, BR6ox, and DWARF, catalyze BR biosynthesis (Szekeres et al., 1996; Choe et al., 1998; Bishop et al., 1999; Shimada et al., 2001). In fact, the deduced amino acid structure of this rice P450 was similar to that of these BR biosynthesis P450s, although there are numerous members of this family in higher plants (see below). Thus, we performed sequence analysis of this P450 gene in two d2 mutants and found one base substitution for each mutant, which generated either a premature stop codon in exon 1 (d2-1) or one amino acid substitution (Pro to Ser) in exon 4 (d2-2) (Figure 4A). These results indicate that D2 encodes a member of P450.
We isolated the full-length cDNA clone encompassing the putative entire coding sequence of D2 by reverse-transcription PCR (RT-PCR) using the total RNA isolated from young seedlings. The cDNA clone contained a large open reading frame that encodes 490 amino acid residues. BLAST (Basic Local Alignment Search Tool) searches revealed that the deduced amino acid sequence was highly similar to that of the previously reported CYP90D protein from Arabidopsis (54% identity and 75% similarity) and ROT3/CYP90C from Arabidopsis (46% identity and 69% similarity) (Kim et al., 1998) over almost the entire region except the N-terminal portion (Figure 4B). The D2 sequence also showed similarity to known BR biosynthesis enzymes such as CPD/CYP90A (39% identity), DWF4/CYP90B (33% identity), and DWARF/CYP85 (31% identity) (Figure 4C), confirming that D2 is involved in BR biosynthesis. Furthermore, the BLAST search indicated that rice also has a P450 protein that is homologous with D2 (Figure 4B). This rice P450 protein showed the greatest similarity to D2 (66% identity and 79% similarity) among all of these P450 proteins (Figure 4C). We confirmed the expression of this D2 homologous gene (see below) and designated D2 and the D2 homolog CYP90D2 and CYP90D3, respectively, according to the nomenclature of the P450 superfamily (Nelson et al., 1996; see http://drnelson.utmem.edu/CytochromeP450.html).
To confirm that CYP90D2 corresponds to the d2 locus, we performed a complementation experiment using d2-1. A DNA fragment of ∼10 kb, including the entire sequence of the putative D2 gene, was introduced into d2 via Agrobacterium tumefaciens–mediated transformation. The dwarf phenotype of d2-1 was rescued in all plants that were resistant to hygromycin, a selection marker for transformation (Figure 5, right). Transformation with a control vector that contained no insert had no apparent effect on the dwarf phenotype (Figure 5, left). In subsequent DNA gel blot analysis, we detected the cosegregation of T-DNA with the rice genome using the fragment as a probe (data not shown). These results confirmed that the d2 mutation was caused by the loss of function of a new member of the P450 gene family.
Figure 5.
Phenotypic Complementation by the Introduction of D2.
d2-1 mutant plants containing the empty vector (left) and the DNA fragment encompassing the entire D2 gene (right) are shown. Bar = 20 cm.
Expression Analysis of D2
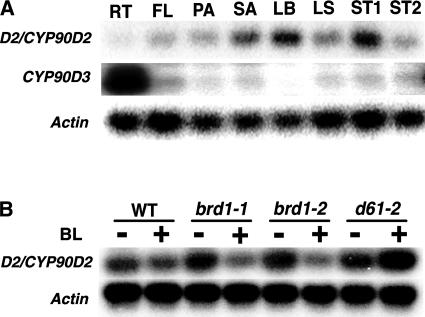
We examined the expression pattern of D2 in various organs. RNA gel blot analysis using the entire length of the cDNA fragment as a probe did not result in any bands, indicating that the level of D2 expression was low in the organs we tested (data not shown). Thus, we performed semiquantitative RT-PCR analysis to estimate the level of the D2 transcript. Because D2/CYP90D2 and CYP90D3 have very similar sequences, we had to carefully distinguish the PCR products from each mRNA. To confirm the identity of the PCR products, we digested them with HindIII or SmaI, because the products of the D2 mRNA contained a HindIII site, whereas the products of CYP90D3 contained a SmaI site. RNAs extracted from the leaf blade and elongating stem produced the strongest bands derived from the D2 mRNA (Figure 6A). Bands of intermediate intensity were amplified with RNAs from the shoot apical region and leaf sheath, whereas RNAs from the root, flower, rachis, and elongated stem produced only faint bands. The preferential expression of D2 in the leaf and elongating stem corresponded to the abnormal phenotype of the leaf structure and shortened stem. We also examined the expression pattern of the D2 homologous gene (CYP90D3). The expression level of CYP90D3 was much less than that of D2/CYP90D2, and the PCR product of CYP90D3 was barely detected in any organs under conditions identical to those used for D2/CYP90D2 (25 cycles). However, when the number of cycles was increased to 37, strong bands were observed in the root and faint bands were seen in the stem, leaf sheath, and flower.
Figure 6.
Expression Pattern of D2/CYP90D2 and CYP90D3 in Various Organs, and the Negative Feedback Effect of BL on D2/CYP90D2 Expression.
(A) Organ-specific expression of D2/CYP90D2 and CYP90D3 in a wild-type plant. Total RNA was isolated from root (RT), flower (FL), panicle (PA), shoot apex (SA), leaf blade (LB), leaf sheath (LS), elongating stem (ST1), and elongated stem (ST2), and RT-PCR was performed. Signals were detected with the 32P-labeled cDNA clone indicated at left. Expression of the Actin gene was used as a control.
(B) Negative feedback regulation of D2/CYP90D2 by BL. Total RNA was prepared from 10-day-old seedlings of the wild type (WT), brd1-1, brd1-2, and d61-2 with or without exogenous application of 10−6 M BL. Expression of the Actin gene was used as a control.
We also studied the effect of BL on D2 expression, because the transcription of other BR biosynthesis P450s, such as CPD/CYP90A and BR6ox/CYP85, is regulated in a feedback manner by the end product of the BR biosynthesis pathway, BL. A high level of D2 expression was detected in the BR-deficient mutant brd1, whereas D2 was expressed at a low level in wild-type plants (Figure 6B). The expression of D2 in the mutants was reduced dramatically by treatment with BL. A higher level of D2 expression also was seen in d61-2, which is partially defective in the BR signaling pathway (Yamamuro et al., 2000). In this mutant, the exogenous BL treatment did not decrease D2 expression (Figure 6B).
The d2 Mutation Lies Downstream in the BR Biosynthesis Pathway
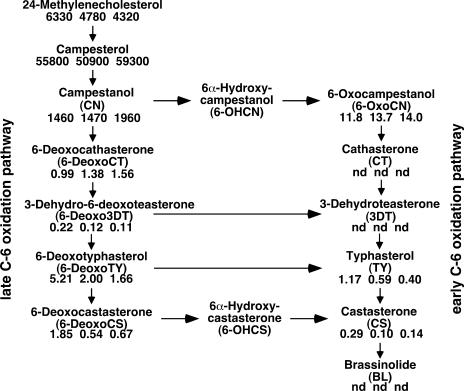
As described previously, the addition of exogenous BL promoted the elongation of the leaf sheath of d2 plants, and the D2 protein sequence is similar to those of BR biosynthesis enzymes such as CPD, DWF4, and DWARF. However, a comparative study indicated that D2 should be placed in the CYP90D group, whose function has not yet been determined. Thus, we expect that D2 catalyzes a step(s) in BR biosynthesis that is different from the steps catalyzed by CPD, DWF4, and DWARF. To investigate this possibility, we measured the BRs in d2-1, d2-2, and wild-type plants by gas chromatography–mass spectrometry analysis. BL was not detected in the young seedling shoots of either the mutant or the wild-type plants (Figure 7). Another bioactive BR, castasterone, was detected in both wild-type and mutant plants, but its level in the mutants was reduced to one-third or one-half of that in the wild type, confirming that the mutants are deficient in the biosynthesis of active BR. This trend of reduced levels of BRs also was observed in 6-deoxocastasterone, typhasterol, 6-deoxotyphasterol, and 3-dehydro-6-deoxoteasterone (6-Deoxo3DT). By contrast, the levels of 6-deoxocathasterone (6-DeoxoCT) and 6-oxocampestanol were slightly greater in the mutants than in the wild type. This finding suggests that D2 may catalyze one or a few steps in the pathway from 6-DeoxoCT to 6-Deoxo3DT in the late C-6 oxidation pathway and also the corresponding steps in the early C-6 oxidation pathway. We also measured the level of 6-deoxoteasterone (6-DeoxoTE) and teasterone (TE) but could not obtain reliable results, because the levels of these compounds were quite low even in the wild-type plants (data not shown).
Figure 7.
Quantitative Analysis of the Endogenous BR Intermediates in Wild-Type, d2-1, and d2-2 Plants.
Sterol and BR levels (ng/g fresh weight) in wild-type (left), d2-2 (middle), and d2-1 (right) plants are shown below each product. nd, not detected.
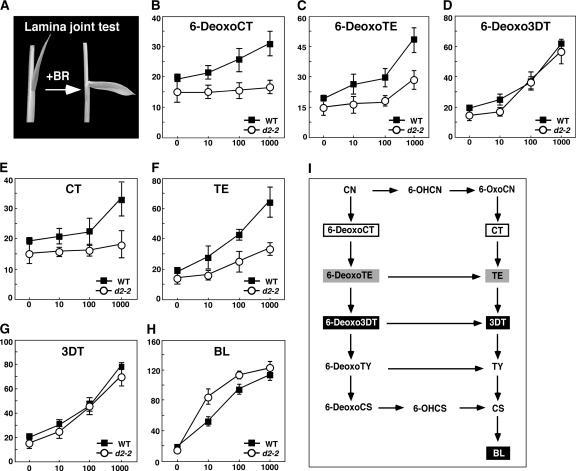
We also performed a feeding experiment with BR biosynthetic intermediates to identify the D2 catalyzing step(s) in the BR biosynthetic pathway. For this feeding experiment, we used the lamina joint test. It is well known that the degree of bending between the rice leaf sheath and blade is highly sensitive to the bioactive BRs exogenously applied (Figure 8A). Using this unusual characteristic, Wada et al. (1981) developed the lamina joint test to measure the content of bioactive BRs. We used this BR bioassay and applied the BR intermediates to wild-type and d2-2 plants. Application of all of the intermediates we tested increased the lamina joint bending of the wild-type plants (Figures 8B to 8H). By contrast, low responsiveness to CT, 6-DeoxoCT, TE, and 6-DeoxoTE was observed in the d2 plants (Figures 8B, 8C, 8E, and 8F), whereas other BR intermediates, such as 3DT, 6-Deoxo3DT, and BL, effectively increased the lamina joint bending of the mutants (Figures 8D, 8G, and 8H). These findings indicate that the conversion of TE to 3DT or 6-DeoxoTE to 6-Deoxo3DT does not occur effectively in the mutants; consequently, D2 may catalyze these steps (see Discussion).
Figure 8.
Effect of BR Intermediates on the Degree of Inclination of the Leaf Lamina in Wild-Type and d2 Plants.
(A) Typical response of the second leaf lamina joint in wild-type plants to treatment with BR intermediates.
(B) to (H) The dose response to BR intermediates (ng/plant) of the bending angle in wild-type (WT; closed squares) and d2-2 (open circles) plants. Data presented are means from 10 plants. Error bars indicate standard deviations.
(I) Pathway of late BR biosynthesis. BR intermediates that effectively, partially, or slightly increased the bending angle in d2-2 plants are indicated by closed, gray, or open boxes, respectively. Abbreviations are as in Figure 7.
DISCUSSION
This study describes the molecular characterization of a classic rice dwarf mutant, d2, which first was described as ebisu dwarf in an article published in 1925 (Matsuo et al., 1997). This dwarf mutant has drawn attention because of its unusual phenotypic characteristics, such as its erect leaves and the specific inhibition of second internode elongation (Figure 1). On the basis of the following observations, we conclude that these abnormal phenotypes of the d2 mutants are caused by a defect in BR biosynthesis. First, the level of active BR (castasterone) in the mutants was reduced relative to that in the wild-type plants (Figure 7). Second, the exogenous application of BL or some BR intermediates restored the dwarfism of the second leaf sheath and/or the bending angle of the lamina joint (Figures 3 and 8). Third, the D2 gene encodes a P450 protein that is classified in the CYP90D group that is highly similar to other BR biosynthesis P450 proteins, such as CPD/CYP90A, DWF4/CYP90B, and DWARF/CYP85 (Figure 4C).
D2 Encodes a Novel Cytochrome P450
The positional cloning of the d2 mutation locus revealed that D2 encodes a putative P450 protein. P450 proteins are heme binding enzymes that have mono-oxygenase activity such as oxidation, hydroxylation, isomerization, and dehydration (Werck-Reichhart and Feyereisen, 2000). Comparative studies revealed that D2 is categorized in the CYP90 group (Figure 4C). There are four reported members of the CYP90 group: CYP90A (CPD), CYP90B (DWF4), CYP90C (ROT3), and CYP90D. Two of them, CPD/CYP90A and DWF4/CYP90B, have been identified as BR biosynthesis enzymes that catalyze BR C-23 and BR C-22 hydroxylation, respectively. Although the remaining two proteins have not had their enzymatic functions identified to date, Goda et al. (2002) predicted that these two P450s also are involved in BR biosynthesis, because the expression of these genes in Arabidopsis was repressed by BL treatment in a manner similar to the BL-dependent suppression of CPD and DWF4 expression (Mathur et al., 1998; Asami et al., 2001; Choe et al., 2001).
By contrast, Kim et al. (1998), in their discussion, expressed doubt about the possibility of the involvement of Arabidopsis ROT3 in BR biosynthesis based on the phenotype of the rot3 mutants. In fact, the hypocotyls of dark-grown rot3 seedlings elongated and the cotyledons did not open, indicating that the rot3 mutants show the normal skotomorphogenesis under dark conditions, unlike other BR-deficient or BR-insensitive mutants. Moreover, none of the rot3 mutants exhibited dwarfism (Tsuge et al., 1996), and the application of exogenous BR did not rescue the mutant phenotype (Kim et al., 1998). Consequently, it is difficult to determine whether or not CYP90D proteins are involved in BR biosynthesis, but our results clearly demonstrate that at least the rice D2/CYP90D2 protein is involved in BR biosynthesis.
Our BLAST search revealed that there is another CYP90D protein (CYP90D3) in rice that is highly similar to D2 (Figure 4C). This finding corresponds to the fact that the putative null allele of d2, d2-1, still does not show severe dwarfism with abnormal morphology, which was seen in the loss-of-function mutants of BR C-6 oxidase, brd1 (Hong et al., 2002). In this BLAST search, we found no gene classified in the CYP90C group in the rice genome. On the other hand, there is one gene classified in CYP90D in the Arabidopsis genome. Thus, when we made the phylogenetic tree using all of the rice and Arabidopsis CYP90 proteins, the CYP90C group contained only one protein, ROT3, whereas the CYP90D group contained three proteins, including D2 (Figure 4C).
This intertwining relationship between rice and Arabidopsis proteins in the CYP90C and CYP90D groups is unusual. Actually, other CYP90 proteins show a clear orthologous relationship between rice and Arabidopsis in each group (Figure 4C), although there are two Arabidopsis proteins in the CYP85 group. However, both CYP85 proteins in Arabidopsis function as BR C-6 oxidases; therefore, the loss of function of one of the genes does not induce any abnormal phenotypic traits (Shimada et al., 2003). The reason why there is no protein orthologous with Arabidopsis ROT3 in rice may be explained by one or both of the following explanations. First, ROT3/CYP90C functions specifically for the polar elongation of leaf cells in dicot plants (Kim et al., 1998); therefore, monocot plants, including rice, which have a much different leaf structure from that of dicot plants, do not contain a comparable protein. The other possibility is that the functions of ROT3/CYP90C and Arabidopsis CYP90D overlap in the BR biosynthesis pathway and the restricted phenotype observed in the rot3 mutants is caused by the redundant function of CYP90D. With this possibility, the functions of CYP90C and CYP90D overlap completely or partially in terms of BR biosynthesis, and both rice and Arabidopsis have two redundant enzymes in this group. At present, we cannot say which possibility is plausible. Analyses of the double mutants of ROT3/CYP90C and CYP90D in Arabidopsis or D2/CYP90D2 and CYP90D3 in rice probably are necessary to elucidate the functional relationship between CYP90C and CYP90D.
The D2 Protein and BR Metabolism in Rice
Feeding experiments, providing BR intermediates to the d2 seedlings, suggested that D2 catalyzes the steps of BR C-3 oxidation (i.e., reactions changing 6-DeoxoTE to 6-Deoxo3DT and TE to 3DT). By contrast, C-3 oxidation in mammalian steroid biosynthesis is catalyzed by 3β-hydroxysteroid dehydrogenase (Payne et al., 1997). At present, the reason why plant and animal cells use different kinds of enzymes for C-3 oxidation is unknown. We searched for a gene that is homologous with the mammalian 3β-hydroxysteroid dehydrogenase in the rice genome but found no such gene. Thus, it is possible that rice does not have a comparable enzyme.
The bending angle of the lamina joint of d2 was increased sensitively by treatments with 3DT and 6-Deoxo3DT, whereas TE and 6-DeoxoTE did not effectively increase this angle, although these latter components effectively increased the bending angle of the wild-type plants (Figure 8). Treatments with CT and 6-DeoxoCT were less effective even in wild-type plants, with only 1000 ng of CT per plant efficaciously increasing it, whereas the d2 plants responded very slightly to the same amount of these compounds (Figures 8B and 8E). The application of TE and 6-DeoxoTE weakly but effectively increased the bending angle of the lamina joint of d2. This result probably was attributable to the redundant function of another CYP90D protein, CYP90D3, which may catalyze the same steps of D2 protein synthesis. This alternative CYP90D protein should function in leaves and stems, because d2-1, which is predicted to be a null allele (see above), still can cause the elongation of these parts; however, the severe allele brd1, which produces a loss of function of BR C-6 oxidase, produces very severe dwarfism of leaves and stems, with abnormal morphology relative to that in d2-1 (Hong et al., 2002).
Actually, a small amount of the transcript of CYP90D3 was detected in the leaves of wild-type plants, supporting the prediction that CYP90D3 is expressed in leaves and partially rescues the protein deficiency in the loss of function of D2. We found that BL treatment caused a d2 mutant to be hypersensitive in the lamina joint test (Figure 8H). In rice BR-deficient mutants such as d2 and brd1, the expression of the BR receptor kinase, OsBRI1, is increased relative to that in wild-type plants (M. Ueguchi-Tanaka and M. Matsuoka, unpublished results). Thus, it is very possible that d2 plants become more sensitive to BL than do wild-type plants. This may be why d2 showed greater sensitivity to the BL treatment than did wild-type plants.
The most conclusive means to determine the biochemical function of D2 is to directly examine the enzymatic activity of the D2 protein produced in yeast cells, which carry a clone for NADPH-P450 reductase (Urban et al., 1997). The enzymatic activities of some P450 enzymes, including BR C-6 oxidase, have been determined by this assay (Bishop et al., 1999; Shimada et al., 2001, 2003). We attempted to examine the enzymatic activity of D2 using this system, but we failed to detect its activity (data not shown). This is not unusual, because the detection of enzymatic activity of any CYP90 protein has never been reported. Thus, it may be necessary to produce double mutants from D2/CYP90D2 and CYP90D3 to perform precise phenotypic analyses, such as accumulation and feeding experiments with BR intermediates, to confirm BR C-3 oxidation by D2.
Feedback Regulation of D2 Expression by BL
In previous studies on the rice DWARF gene, we demonstrated that the expression of this gene, which encodes BR C-6 oxidase, was regulated in a feedback manner by the level of the bioactive BR, BL (Hong et al., 2002). Similarly, the expression of D2 was negatively regulated by the level of the bioactive BR. D2 was highly expressed in the BR-deficient mutant brd1, and its expression was downregulated by treatment with BL (Figure 6B). Recently, Goda et al. (2002) reported that the suppression of P450 genes such as DWF4/CYP90B, CPD/CYP90A, BR6ox/CYP85, ROT3/CYP90C, and CYP90D occurred when Arabidopsis was treated with BL. Such feedback regulation of BR-related gene expression also was seen in rice genes homologous with Arabidopsis CPD and DWF4 (Sakamoto et al., unpublished results), indicating that negative feedback regulation of the genes related to BR biosynthesis is a common phenomenon in both dicot and monocot plants.
The failure of the downregulation of D2 expression in the BR-insensitive mutant d61-2 indicates that such feedback regulation of D2 expression by exogenously applied BL is mediated by the BR signaling pathway (Figure 6B). Higher levels of D2 expression in the nontreated d2 mutants than in the wild-type plants support the idea that BR signaling is important for the negative feedback regulation of D2 expression. An interesting follow-up question is whether the regulation of the negative feedback expression of BR-related genes in rice and Arabidopsis functions by a similar mechanism. Further studies are needed to investigate the molecular mechanism of the regulatory expression of BR biosynthesis genes.
METHODS
Plant Materials and Growth Conditions
Two d2 mutant lines, d2-1 and d2-2, were used in this study. A backcross of the original d2 (ebisu dwarf) with T65 was used to produce d2-2, whereas d2-1 was screened from a mutant library produced by N-methyl-N-nitrosourea. Rice plants (Oryza sativa) were grown in a greenhouse at 30°C during the day and 24°C at night. For deetiolated analysis, the wild type, d2, and the gibberellin-deficient mutant d18 (Itoh et al., 2001) were germinated and grown on MS medium (Murashige and Skoog, 1962) in complete darkness at 25°C for 3 weeks.
Brassinolide Induction in Shoot Elongation
Rice seeds were sterilized with 1.7% NaClO and grown on 0.8% agar medium containing various concentrations of brassinolide. The seeds were incubated at 30°C under continuous light. After 2 weeks, the length of the second leaf sheath was measured. A total of 25 plants were used for each treatment.
Mapping, Isolation, and Sequencing of the D2 Gene
For rough mapping of d2, we used an indica strain, Kasalath, whereas for fine mapping, we used a substitution line, SL5. In both strains, chromosomes of Kasalath were substituted on the outside of the short arm of chromosome 1. Approximately 3000 F2 plants of a cross between d2-2 and SL5 were used for positional cloning of D2. To identify the mutation sites of the d2 alleles, we amplified D2 using genomic DNA extracted from the two alleles. The amplified DNA fragments were sequenced directly with appropriate primers without cloning. To isolate a genomic D2 clone, we performed PCR screening of the BAC library produced at the Clemson University Genomics Institute (CUGI) using two pairs of primers (5′-CACTTGCGTAGGAGCCTAG-3′ and 5′-CATGCATGCACACCCATGC; and 5′-CTGCTGATCCATCCATTGC-3′ and 5′-CTTGGTGTGTGCAACTTGGC-3′). A CUGI BAC clone, XBH13-4F02, contains the entire coding region and the 5′ and 3′ flanking regions of D2. D2 full-length cDNA was amplified using primers containing the BamHI and SmaI sites (5′-GCGGATCCATGGTGTCGGCGGCCG-3′ and 5′-CCCCCGGGCTAGTCGTCGTCCTCC-3′) by reverse-transcription PCR (RT-PCR). For RT-PCR, we used the total RNA isolated from young seedlings as a template. The amplified fragment was fully sequenced to confirm that no nucleotide substitution occurred during PCR and cloned into pBluescript SK+ vector (Stratagene, La Jolla, CA).
Complementation Test
The DNA fragment containing a full-length genomic D2 gene was obtained by digesting the CUGI clone, XBH13-4F02, with EcoRI and SmaI. The digested fragment was inserted into pBluescript vector at the EcoRI and SmaI sites, and then the fragment (∼10.2 kb) was redigested with HindIII and SmaI, blunted, and inserted at the SmaI site in a hygromycin-resistant binary vector, pBI-Hm12, which was kindly provided by Hiroyuki Hirano (Tokyo University). This fragment was introduced into the d2-1 plant by Agrobacterium tumefaciens–mediated transformation. The empty pBI-Hm12 vector also was transformed into the d2-1 plant as a control.
RNA Isolation and RT-PCR Analysis
Total RNA was extracted with the RNeasy plant mini kit (Qiagen, Hilden, Germany) from various rice organs or whole seedlings of wild-type, brd1-1, brd1-2, and d61-2 plants, which were grown on half-strength MS medium (Murashige and Skoog, 1962) under continuous light with or without 10−6 M brassinolide. Semiquantitative RT-PCR analysis was performed to estimate the level of the D2 transcript. The first strand of cDNA was synthesized from 1 mg of total RNA using an Omniscript reverse transcription kit (Qiagen). The primers 5′-TTCAACCCATGGAGGTGGAA-3′ and 5′-GCACGGTGGGGAAGTTGACGA-3′ were used to amplify the cDNA fragments of both D2 and the D2 homolog. According to the sequences of D2 and the D2 homolog, the amplified fragment of D2 should be 187 bp, whereas that of the D2 homolog should be 205 bp. The conditions used during PCR were 99°C for 10 min and then 94°C for 30 s, 55°C for 50 s, and 72°C for 40 s for 25 or 37 cycles. The PCR product of 25 cycles was digested with HindIII to distinguish the D2 product from the D2 homolog, and then it was hybridized with the D2 probe. The product of 37 cycles was digested with SmaI to distinguish the D2 homolog from D2, and then it was hybridized with the D2 homolog probe. The hybridization was performed at 65°C in 0.25 M Na2HPO4, 1 mM EDTA, and 7% SDS. Filters were washed twice with 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS at 65°C for 30 min and once with 0.2× SSC and 0.1% SDS at 65°C for 10 min. The PCR primers 5′-TCCATCTTGGCATCTCTCAG-3′ and 5′-GTACCCGCATCGGCATCTG-3′ were used to amplify the Actin fragment as a control. Hybridization conditions were as described above.
Quantification of Endogenous Brassinosteroids
Shoots from wild-type and mutant plants were harvested during the 8th week after germination and lyophilized immediately at −80°C. To analyze the endogenous brassinosteroids (BRs), lyophilized shoots (equivalent to 20 g fresh weight) were extracted twice with 250 mL of methanol:CHCl3 (4:1, v/v). BR purification and quantification were performed according to the method described by Fujioka et al. (2002) and He et al. (2003).
Lamina Joint Inclination Assay
Germinated seeds were selected for uniformity of coleoptile length, transplanted onto 1% agar medium, and grown at 30°C for 3 days. The seedlings were injected at the top of the lamina with 1 μL of ethanol solution containing 0, 10, 100, or 1000 ng of BR intermediates. After incubation for 3 days, the angle between the lamina and its leaf sheath was measured (Fujioka et al., 1998). Ten plants were used for each treatment.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact M. Matsuoka, makoto@nuagr1.agr.nagoya-u.ac.jp.
Accession Numbers
The GenBank accession numbers for the sequences mentioned in this article are AP003244 (D2/CYP90D2), AC130732 (OsCYP90D3), AB066286 (AtCYP90D1), AB008097 (AtROT3/CYP90C1), X87367 (AtCPD), AB035868 (AtDWARF), AF044216 (AtDWF4), U54770 (LeDWARF), AB084385 (OsDWARF), AC104473 (OsDWF4), and AC123526 (OsCPD) (T. Sakamoto and M. Matsuoka, unpublished results).
Acknowledgments
We thank Masayo Sekimoto, and Makoto Kobayashi (all of RIKEN) for their technical assistance. Masako Hattori and Ikuko Aichi (both of Nagoya University) helped with preliminary preparation. We thank David Nelson of the P450 nomenclature committee, who kindly classified the putative D2 and D2 homologous proteins as CYP90D2 and CYP90D3, Yukihisa Shimada (RIKEN, Japan), and Yue-ie Hsing (Institute of Botany, Taiwan) for helpful discussion. This work was supported in part by a Grant-in-Aid from the Program for the Promotion of Basic Research Activities for Innovative Biosciences (M.M., M.U.-T., and H.K.), a Grant-in-Aid from the Center of Excellence (M.M. and M.A.), and a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Rice Genome Project IP-1003; to M.A.).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.014712.
References
- Asami, T., Mizutani, M., Fujioka, S., Goda, H., Min, Y.K., Shimada, Y., Nakano, T., Takatsuto, S., Matsuyama, T., Nagata, N., Sakata, K., and Yoshida, S. (2001). Selective interaction of triazole derivatives with DWF4, a cytochrome P450 monooxygenase of the brassinosteroid biosynthetic pathway, correlates with brassinosteroid deficiency in planta. J. Biol. Chem. 276, 25687–25691. [DOI] [PubMed] [Google Scholar]
- Ashikari, M., Wu, J., Yano, M., Sasaki, T., and Yoshimura, A. (1999). Rice gibberellin-insensitive dwarf mutant gene Dwarf1 encodes the α-subunit of GTP-binding protein. Proc. Natl. Acad. Sci. USA 96, 10284–10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, G.J., Harrison, K., and Jones, J.D. (1996). The tomato Dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell 8, 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, G.J., Nomura, T., Yokota, T., Harrison, K., Noguchi, T., Fujioka, S., Takatsuto, S., Jones, J.D.G., and Kamiya, Y. (1999). The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc. Natl. Acad. Sci. USA 96, 1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Dilkes, B.P., Fujioka, S., Takatsuto, S., Sakurai, A., and Feldmann, K.A. (1998). The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10, 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Fujioka, S., Noguchi, T., Takatsuto, S., Yoshida, S., and Feldmann, K.A. (2001). Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 26, 573–582. [DOI] [PubMed] [Google Scholar]
- Chory, J., Nagpal, P., and Peto, C.A. (1991). Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3, 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse, S.D., and Sasse, J.M. (1998). Brassinosteroids: Essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 427–451. [DOI] [PubMed] [Google Scholar]
- Fujioka, S., Noguchi, T., Takatsuto, S., and Yoshida, S. (1998). Activity of brassinosteroids in the dwarf rice lamina inclination bioassay. Phytochemistry 49, 1841–1848. [Google Scholar]
- Fujioka, S., Takatsuto, S., and Yoshida, S. (2002). An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol. 130, 930–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka, S., and Yokota, T. (2003). Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54, 137–164. [DOI] [PubMed] [Google Scholar]
- Goda, H., Shimada, Y., Asami, T., Fujioka, S., and Yoshida, S. (2002). Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 130, 1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, J.X., Fujioka, S., Li, T.S., Kang, S.G., Seto, H., Takatsuto, S., Yoshida, S., and Jang, J.C. (2003). Sterols regulate development and gene expression in Arabidopsis. Plant Physiol. 131, 1258–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Z., et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32, 495–508. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14, 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sentoku, N., Kitano, H., and Matsuoka, M. (2001). Cloning and functional analysis of two gibberellin 3β-hydroxylase genes that are differently expressed during the growth of rice. Proc. Natl. Acad. Sci. USA 98, 8909–8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, G.T., Tsukaya, H., and Uchimiya, H. (1998). The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 12, 2381–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Mandava, N.B. (1988). Plant growth-promoting brassinosteroids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 23–52. [Google Scholar]
- Mathur, J., et al. (1998). Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 14, 593–602. [DOI] [PubMed] [Google Scholar]
- Matsuo, T., Futsuhara, Y., Kikuchi, F., and Yamaguchi, H. (1997). Science of the Rice Plant, Vol. 3. (Tokyo, Japan : Nobunkyo), pp. 302–303.
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nelson, D.R., Koymans, L., Kamataki, T., Stegeman, J.J., Feyereisen, R., Waxman, D.J., Waterman, M.R., Gotoh, O., Coon, M.J., Estabrook, R.W., Gunsalus, I.C., and Nebert, D.W. (1996). P450 superfamily: Update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 6, 1–42. [DOI] [PubMed] [Google Scholar]
- Payne, A.H., Abbaszade, I.G., Clarke, T.R., Bain, P.A., and Park, C.H. (1997). The multiple murine 3β-hydroxysteroid dehydrogenase isoforms: Structure, function, and tissue- and developmentally specific expression. Steroids 62, 169–175. [DOI] [PubMed] [Google Scholar]
- Peng, J., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261. [DOI] [PubMed] [Google Scholar]
- Peng, S., Khush, G.S., and Cassman, K.G. (1994). Evolution of the new plant idiotype for increased yield potential. In Breaking the Yield Barrier: Proceedings of a Workshop on Rice Yield Potential in Favorable Environments, K.G. Cassman, ed (Manila, The Philippines: International Rice Research Institute), pp. 5–16.
- Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Swapan, D., Ishiyama, K., Saito, T., Kobayashi, M., Khush, G.S., Kitano, H., and Matsuoka, M. (2002). A mutant gibberellin-synthesis gene in rice. Nature 416, 701–702. [DOI] [PubMed] [Google Scholar]
- Shimada, Y., Fujioka, S., Miyauchi, N., Kushiro, M., Takatsuto, S., Nomura, T., Yokota, T., Kamiya, Y., Bishop, G.J., and Yoshida, S. (2001). Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 126, 770–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, Y., Goda, H., Nakamura, A., Takatsuto, S., Fujioka, S., and Yoshida, S. (2003). Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol. 131, 287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmeyer, W., Ellis, M.H., and Chandler, P.M. (2002). Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 99, 9043–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres, M., Nemeth, K., Koncz-Kalman, Z., Mathur, J., Kauschmann, A., Altmann, T., Redei, G.P., Nagy, F., Schell, J., and Koncz, C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182. [DOI] [PubMed] [Google Scholar]
- Taiz, L., and Zeiger, E. (2002). Plant Physiology, 3rd Ed. (Sunderland, MA: Sinauer Associates), pp. 461–488.
- Tsuge, T., Tsukaya, H., and Uchimiya, H. (1996). Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122, 1589–1600. [DOI] [PubMed] [Google Scholar]
- Urban, P., Mignotte, C., Kazmaier, M., Delorme, F., and Pompon, D. (1997). Cloning, yeast expression and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J. Biol. Chem. 272, 19176–19186. [DOI] [PubMed] [Google Scholar]
- Wada, K., Marumo, S., Ikekawa, N., Morisaki, M., and Mori, K. (1981). Brassinolide and homobrassinolide promotion of lamina inclination of rice seedlings. Plant Cell Physiol. 22, 323–325. [Google Scholar]
- Werck-Reichhart, D., and Feyereisen, R. (2000). Cytochromes P450: A success story. Genome Biol. 1, 3003.1–3003.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., Ashikari, M., Kitano, H., and Matsuoka, M. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12, 1591–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]