Abstract
A single dose of tenofovir/emtricitabine (TDF/FTC) during labor significantly reduces peripartum nevirapine-associated viral drug resistance when measured by consensus HIV sequencing. It is unknown whether this effect extends to HIV subpopulations of <25–50%. We conducted a randomized trial of single-dose TDF/FTC added to peripartum nevirapine to reduce drug resistance associated with nonnucleoside reverse transcriptase inhibitors (NNRTIs). To detect mutations for NNRTIs comprising ≥2% of the viral population, we used an oligonucleotide ligation assay (OLA) at codons 103, 106, 181, and 190 of HIV reverse transcriptase. To assess development of drug resistance mutations to our study intervention, OLA was also performed at codons 65 and 184. Among the 328 women included in the 2-week analysis, those receiving TDF/FTC were less likely to have NNRTI resistance by OLA (RR = 0.40, 95% CI = 0.21–0.77). A similar trend was observed among the 315 women included in the 6-week analysis (RR = 0.45, 95% CI = 0.31–0.66). Only two (1%) specimens had detectable K65R by OLA. Both were at 6 weeks postpartum; one was detected in the intervention arm and one in the control arm (p = 0.96). M184V was not detected. The ability of single-dose TDF/FTC to protect against peripartum NVP-induced NNRTI resistance extends to minority populations. This efficacy is achieved without significant selection of TDF- or FTC-resistant viruses.
Introduction
Although effective in reducing mother-to-child transmission of HIV, peripartum single-dose nevirapine (NVP) has been associated with nonnucleoside reverse transcriptase inhibitor (NNRTI) viral drug resistance in the weeks following ingestion.1–3 This complication is attributed to the drug's low genetic barrier for resistance and its slow metabolism and excretion from maternal plasma.4
Administration of additional antiretroviral drugs over the “tail” of NVP exposure has been shown to significantly reduce selection of NNRTI resistance. In South Africa, 4-day and 7-day courses of combination zidovudine/lamivudine (ZDV/3TC) following NVP administration led to considerable reductions in NNRTI resistance when compared to courses in which no additional drugs were given (13% vs. 9% vs. 57%, respectively).5 In Côte d'Ivoire, women who received a 3-day course of ZDV/3TC postpartum, following antenatal ZDV/3TC and intrapartum NVP, had detectable NNRTI resistance in only 1% when measured 4 weeks after delivery.6 In a randomized, controlled trial in Zambia, we demonstrated that a single dose of adjuvant tenofovir/emtricitabine (TDF/FTC) reduced NNRTI viral drug resistance by 73% at 2 weeks postpartum and by 53% at 6 weeks postpartum.7
Studies of NVP-associated viral drug resistance––including those cited above––have typically used standard consensus sequencing, a process in which prevalent HIV strains are evaluated for genetic mutations known to confer resistance to specific agents. Because this technique detects mutations in HIV variants comprising >25–50% of the circulating viral population,8 the true incidence of viral drug resistance may be underestimated.9–12 To determine more fully the impact of TDF/FTC on the selection of NNRTI resistance mutations, we evaluated minority populations of drug-resistant viruses using specimens from a previously reported clinical trial.
Materials and Methods
To better characterize the impact of TDF/FTC on the selection of drug-resistant viruses, we measured drug resistance for NNRTIs, TDF, and FTC using an oligonucleotide ligation assay (OLA) capable of detecting minority HIV populations down to a concentration of 2%. We analyzed specimens from our previously reported trial of peripartum TDF/FTC to reduce NNRTI resistance associated with single-dose NVP use in Lusaka, Zambia. The study methodology for this trial has been described elsewhere.7,13 Briefly, HIV-infected pregnant women were screened for eligibility between 28 and 38 weeks of completed gestation. We targeted women who did not immediately require HIV treatment according to local guidelines14 and were offered short-course zidovudine and peripartum NVP for perinatal HIV prevention.15 Those reporting use of antiretroviral drugs prior to the index pregnancy were excluded. We obtained informed consent from all participants during antenatal care, but participants were randomized to receive either the study intervention (TDF 300 mg/FTC 200 mg) or no intervention when they presented to the delivery ward in labor. Maternal specimens were collected at 2 weeks and 6 weeks postpartum. The trial's primary outcome was NNRTI resistance by consensus sequencing at 6 weeks.
Specimens with ≥1000 copies/ml of plasma HIV RNA had HIV-1 pol amplicons generated. RNA extracted using the Qiagen viral RNA extraction kit (Qiagen, Inc., Chatsworth, CA) and sequenced using the ViroSeq HIV-1 Genotyping System (Abbott Molecular, Abbott Park, IL) was then tested for viral drug resistance by OLA.16–19 OLA was accomplished by adding the amplicon to a ligation reaction containing probes specific for wild-type and mutant codons labeled at the 5′ end and a probe to the region adjacent to the site of interest that was biotinylated at the 3′ end. Mutant and wild-type oligonucleotides were labeled with digoxygenin; separate ligation reactions and OLA plates were used to test for the mutant and wild-type reaction for each specimen. Following the ligation reaction, the products were bound to a streptavidin-coated microtiter plate and an ELISA was performed using horseradish peroxidase-labeled antibodies to develop color for the mutant and wild-type codons. All participants' specimens and assay controls were analyzed in duplicate, including standards with 0%, 2%, 5%, and 100% mutant. Control plasmids are described at http://depts.washington.edu/idimmweb/faculty/frenkel/OLAmanual1305april04.pdf. Reactions were considered indeterminate when the optical density (OD) readings for the mutant was less than the 2% mutant control and the OD for the wild type was less than 0.5. Specimens with plasma HIV RNA concentrations <1000 copies/ml were not analyzed by OLA.
Nucleotide sequence-specific genetic mutations for NNRTI resistance were detected via OLA at K103N (AAY sequence), V106M (ATG sequence), Y181C (TGY sequence), and G190A (GCA sequence). Similar assays were used to detect mutations conferring TDF (K65R; AGR sequence) and FTC (K65R and M184V; GTG sequence) resistance. We did not test for the M184I mutation, which confers resistance to FTC. Specimens with a concentration of mutant ≥2% were considered OLA positive. In our analysis comparing NNRTI resistance by randomization arm, specimens with an indeterminate OLA reaction at one codon, accompanied by wild-type or indeterminate reactions at other codons, were excluded. Those with a single indeterminate OLA reaction, but detectable mutations at other designated codons, were classified as NNRTI resistant.
Following the convention of our previous report,7 we performed two analyses to determine the effectiveness of TDF/FTC on NNRTI resistance. We first categorized all specimens with <1000 copies/ml as nonresistant, thus accounting for the suppressive effect the study intervention may have on circulating HIV-1. We then considered only those specimens with circulating HIV RNA of ≥1000 copies/ml and a valid OLA result. Relative risks (RR) with 95% confidence intervals (95% CI) were calculated to measure the impact of TDF/FTC on NNRTI resistance. To demonstrate the relative concentrations of mutant virus at each codon, we graphed our OLA results along a normalized scale. Concentration standards of for 0%, 2%, 5%, and 100% were directly measured and included in the analysis. The threshold for the 20% concentration was calculated using logistic regression.
We compared these OLA results with consensus sequencing. In our previous report,7 we performed consensus sequencing on only those specimens with HIV-1 viral concentrations ≥2000 copies/ml, in accordance with the manufacturer's recommendation. To provide a comprehensive comparison to OLA, however, we additionally tested for viral drug resistance among specimens with HIV-1 concentrations between 1000 and 1999 copies/ml here. Our methodology for consensus sequencing has been reported elsewhere.7 Briefly, the pol gene was amplified and bidirectionally sequenced.20 Sequences were assembled and edited using Sequencher (Gene Codes, Ann Arbor, MI), and then analyzed with the Stanford Resistance Database (http://hivdb.stanford.edu). Mutations were considered to be present when detected alone or in combination with wild-type sequences (mixtures). Samples that did not amplify or were of poor quality were reamplified and sequenced using ViroSeq HIV-1 Genotyping System. All analyses were performed using SAS version 9.1 (SAS Institute, Cary, NC). The study was approved by the University of Zambia Research Ethics Committee (Lusaka, Zambia), University of Alabama at Birmingham Institutional Review Board (Birmingham, Alabama), Childrens Hospital Los Angeles (Los Angeles, California), and Seattle Children's Hospital (Seattle, Washington).
Results
From March 2005 to February 2007, 400 HIV-infected pregnant women were enrolled and randomized. Of these, three were incorrectly randomized and thus excluded from the analysis. Maternal and infant characteristics did not differ significantly according to study arm (data previously reported).7 Overall, 360 of 397 (91%) specimens were available at 2 weeks; 356 of 397 (90%) specimens were available at 6 weeks. Of these 716 cumulative specimens, 282 (39%) had viral load <1000 copies/ml of circulating HIV and were not tested for viral drug resistance. Of the remaining 434 specimens with viral load ≥1000 copies/ml, 44 (10%) were excluded due to failed PCR amplification. The remaining 390 specimens were tested by OLA, of which 29 (7%) were excluded because of indeterminate OLA results.
When we evaluated specimens with viral load <1000 copies/ml and those with viral load ≥1000 copies/ml with successful OLA reactions, 328 of 397 participants (83%) were included in the 2-week analysis. Those excluded were younger (median 23 years vs. 25 years; p = 0.02), less likely to initiate short-course zidovudine antenatally (68% vs. 84%; p < 0.01), and experienced lengthier labor (median 13.3 h vs. 10.4 h; p = 0.05). When we used similar criteria at 6 weeks, 315 participants were included in the analysis, a figure that represented 79% of the original 397 randomized. Women in the excluded group were again younger (median 23 years vs. 26 years; p < 0.01) but also less likely to have detectable NVP levels in their cord blood (70% vs. 84%; p < 0.01). Otherwise these groups did not differ by other demographic, medical, or obstetric characteristics.
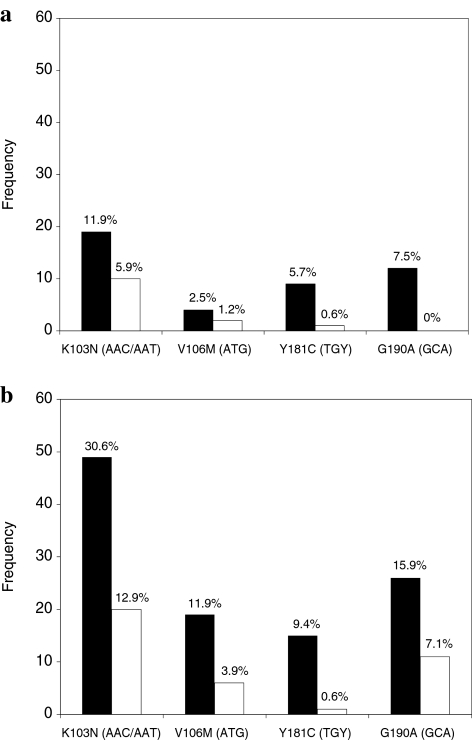
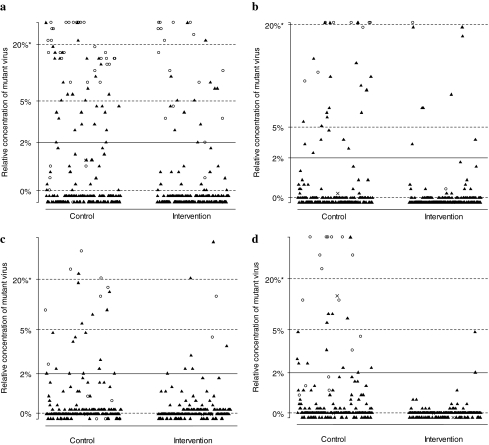
When specimens with viral load <1000 copies/ml were assumed to be nonresistant, women receiving TDF/FTC were less likely to develop NNRTI resistance at 2 weeks [12/169 (7%) vs. 28/159 (18%); RR = 0.40, 95% CI = 0.21–0.77; p = 0.004] and at 6 weeks postpartum [29/155 (19%) vs. 68/160 (41%); RR = 0.45, 95% CI = 0.31–0.66; p < 0.0001]. Similar trends were observed when only specimens with ≥1000 HIV RNA copies/ml and a valid OLA result were analyzed (Table 1). Subjects in the intervention arm had lower rates of NNRTI resistance at 2 weeks and 6 weeks across all tested codons (Fig. 1). The relative concentration of NNRTI mutant variants is shown according to study arm in Fig. 2. We compared OLA and consensus sequencing results (Table 2) by analyzing results from the 390 specimens with viral load ≥1000 copies/ml.
Table 1.
Resistance to Nonnucleoside Reverse Transcriptase Inhibitor Drugsa
| Intervention | Control | Relative risk (95% CI) | p | |
|---|---|---|---|---|
| All specimensb | ||||
| 2 weeks | 12/169 (7%) | 28/159 (18%) | 0.40 (0.21–0.77) | 0.004 |
| 6 weeks | 29/155 (19%) | 66/160 (41%) | 0.45 (0.31–0.66) | <.0001 |
| Specimens with viral load ≥1000 copies/ml | ||||
| 2 weeks | 12/35 (34%) | 28/52 (54%) | 0.64 (0.38–1.07) | 0.083 |
| 6 weeks | 29/137 (21%) | 66/137 (48%) | 0.44 (0.30–0.63) | <.0001 |
By study randomization arm as assessed by an oligonucleotide ligation assay of HIV-1 pol encoding reverse transcriptase codons 103, 106, 181, and 190.
Specimens with viral load ≤1000 copies/ml were classified as nonresistant. CI = confidence interval.
FIG. 1.
Proportion of participants with HIV-1 drug resistance to nonnucleotide reverse transcriptase inhibitors as assessed by oligonucleotide ligation assay at 2 (a) and 6 (b) weeks postpartum. Results from codons 103, 106, 181, and 190 are shown; the specific nucleotide sequences detected are included in parentheses. Black columns represent the control arm. White columns represent the intervention arm.
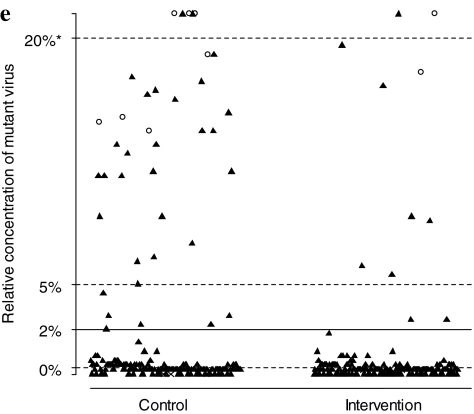
FIG. 2.
Relative concentrations of HIV-1 variants resistant to nonnucleotide reverse transcriptase inhibitors by the oligonucleotide ligation assay (OLA). The following mutations were tested: (a) K103N, sequence AAC, (b) K103N, AAT, (c) V106M, ATG, (d) Y181C, TGY, and (e) G190A, GCA. Thresholds for 0%, 2%, 5%, and 20% of circulating mutant concentrations are based on optical density readings from OLA and logistic regression (see Materials and Methods section). Results are categorized by study arm and are cumulative across 2- and 6-week visits. Black triangles indicate specimens with no resistance mutations detected by consensus sequencing. Open circles represent specimens with resistance mutations detected by consensus sequencing. “X” indicates that the specimens were missing a consensus sequence or the OLA was indeterminate.
Table 2.
Comparison of Consensus Sequencing (CS) and Oligonucleotide Ligation Assay (OLA) Resultsa
| |
K103N |
V106M |
Y181C |
|||
|---|---|---|---|---|---|---|
| CS | OLA | CS | OLA | CS | OLA | |
| Wild type | 329 (84%) | 284 (73%) | 370 (95%) | 356 (91%) | 368 (94%) | 340 (87%) |
| Mutation detected | 59 (15%) | 98 (25%) | 17 (4%) | 31 (8%) | 15 (4%) | 26 (7%) |
| Indeterminate | 0 | 8 (2%) | 0 | 3 (1%) | 0 | 24 (6%) |
| Missing | 2 (1%) | 0 | 3 (1%) | 0 | 7 (2%) | 0 |
| |
G190A |
M184V |
K65R |
|||
|---|---|---|---|---|---|---|
| CS | OLA | CS | OLA | CS | OLA | |
| Wild type | 373 (96%) | 333 (85%) | 385 (99%) | 381 (98%) | 388 (99%) | 387 (99%) |
| Mutation detected | 10 (2%) | 49 (13%) | 0 | 0 | 1 (<1%) | 2 (<1%) |
| Indeterminate | 0 | 8 (2%) | 0 | 9 (2%) | 0 | 1 (<1%) |
| Missing | 7 (2%) | 0 | 5 (1%) | 0 | 1 (<1%) | 0 |
Of specified resistance mutations on the reverse transcriptase gene among specimens ≥1000 copies/ml of HIV-1 from both 2 and 6 weeks postpartum.
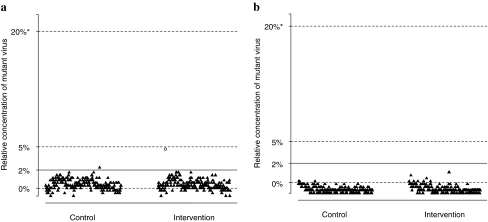
We examined by OLA the frequency of mutations associated with TDF and FTC resistance. Of the 390 specimens that were amplified, 387 (99%) were found to be wild type, 2 (1%) were found to be resistant, and 1 (<1%) was found to be indeterminate at codon 65. The K65R mutation was not detected by OLA in 2-week specimens. At 6 weeks, there was no difference noted in the frequency between the intervention arm [1 of 154 (1%)] and the control arm [1 of 143 (1%); p = 0.96]. When we examined the concentration of mutant detected by OLA for K65R (Fig. 3), the concentration of the one positive result in the intervention arm was estimated to be near 5%. The mutant concentration of the positive result in the control arm was lower, at approximately 2%. This could represent a random spontaneous mutation, TAQ error, carryover contamination, or an artifact from an abnormally high background reaction peculiar to this specimen. Of the same 390 specimens, 381 (98%) were found to be wild type and 7 (2%) were found to be indeterminate at codon 184. None had the M184V resistance mutation by OLA.
FIG. 3.
Relative concentrations of variants resistant to tenofovir and emtricitabine by the oligonucleotide ligation assay. We tested for mutations (a) K65R, AGR sequence and (b) M184V, GTG. Thresholds for 0%, 2%, 5%, and 20% of circulating mutant concentrations are based on optical density readings from OLA and logistic regression (see Materials and Methods section). Results are categorized by study arm and are cumulative across 2- and 6-week visits. Black triangles indicate specimens with no resistance mutations detected by consensus sequencing. Open circles represent specimens with resistance mutations detected by consensus sequencing. “X” indicates that the specimens were missing a consensus sequence or the OLA was indeterminate.
Discussion
Our analysis of four codon mutations known to impart high-level NNRTI resistance demonstrates that the TDF/FTC intervention reduced the frequency of mutations by 60% at 2 weeks and a 55% at 6 weeks postpartum. Across the four codons tested for NNRTI resistance, between 3% and 11% had detectable mutant virus by OLA but not by consensus sequencing, suggesting substantial selection of mutant variants within HIV populations of <25–50%. Notably, no M184V mutations were detected among study participants and mutations at K65R were detected only rarely.
While NNRTI resistance was more frequently detected by OLA, the impact of our TDF/FTC intervention remained comparable to that of our previous report. When consensus HIV sequencing was used to measure NNRTI-related viral drug resistance, TDF/FTC was associated with a reduced risk at 2 weeks [6/178 (3%) vs. 21/169 (12%); RR = 0.27, 95% CI = 0.11–0.66) and at 6 weeks [20/173 (12%) vs. 41/166 (25%); RR = 0.47, 95% CI = 0.29–0.76].7 When OLA was used, the protective effect at 2 weeks appeared attenuated (RR = 0.40, 95% CI = 0.21–0.77), while the risk reduction at 6 weeks––the primary outcome of the trial––was virtually identical (RR = 0.45, 95% CI = 0.31–0.66).
Two factors contribute to the effectiveness of the TDF/FTC intervention. Addition of TDF/FTC raises the genetic barrier to NNRTI resistance following NVP ingestion. When all three agents reach inhibitory levels within infected cells, the risk for selection of preexisting mutant strains decreases dramatically. Although single-dose NVP reduces intrapartum/early postpartum plasma HIV RNA concentrations––a function of the drug's long half-life––TDF/FTC further limits viral replication at a rate greater than seen with NVP alone.7 Since viral replication is critical to the selection for resistant mutant strains, its inhibition confers some degree of indirect protection. In fact, it was due in part to the higher viral suppression associated with TDF/FTC (63% vs. 53% with <400 copies/ml; p = 0.04) that such a small proportion of specimens at 2 weeks (27%) met the HIV-1 threshold for resistance testing. The borderline significance of the 2-week comparison among specimens ≥1000 copies/ml (34% vs. 54%; p = 0.083) was likely due to the low statistical power associated with the reduced sample size.
To account for both mechanisms associated with TDF/FTC effectiveness, we considered specimens with HIV-1 concentrations below 1000 copies/ml in two separate ways. First, we made the assumption that all specimens under this threshold have no viral drug resistance to NNRTIs. This approach allowed us to consider the collateral benefit of viral suppression associated with the study intervention, but risked misclassification of those specimens with NNRTI resistance. We then excluded those specimens under this viral concentration threshold entirely. We observed comparable risk reductions at 6 weeks postpartum (the study's primary outcome measure) for both analytical approaches.
The impact of prior NVP use on future treatment outcomes remains controversial, but growing evidence suggests that the interval between NVP use and subsequent ART initiation plays an important role. Several studies indicate that recent exposure to NVP is associated with compromised treatment outcomes, with either 6 months or 12 months commonly cited as relevant clinical thresholds.21–23 Risk appears to decline as the interval between exposure and ART initiation increases, presumably due to the decay of mutant virus concentrations over time.
Detectable antiretroviral drug resistance is also an important risk factor for later virologic failure on ART. In a follow-up analysis of the PHPT-2 cohort, for example, women with NVP resistance detected by consensus sequencing were more likely to fail therapy when started on NNRTI-based ART (52% vs. 38%); however, this difference did not meet statistical significance (p = 0.08).3 While sensitive assays (e.g., OLA, LigAmp, real-time PCR) have been used to measure NNRTI resistance patterns within HIV minority populations, the clinical impact of low-concentration mutations is not yet certain.24 Recent work by Coovadia et al. showed that minority subpopulations with detectable K103N mutation were a risk factor for poor virologic outcomes on NNRTI-based ART, regardless of prior NVP exposure.25 However, further studies are needed to understand clinically significant concentration thresholds of mutant subpopulations and how their detection may be used to optimize HIV treatment.
OLA is more sensitive than consensus HIV sequencing and can reliably detect mutations in subpopulations comprising as little as 2% of circulating virus; however, each reaction detects only the specified nucleotide triplet(s). When an amino acid is encoded by several closely related sequences, two or more oligonucleotide reagents can be combined as a mixture (e.g., the type-common oligonucleotides for K65R, Y181C). In addition, polymorphisms within two nucleotides of the ligation site can preclude ligation and give indeterminate results. Multiple polymorphisms within the regions where the probes bind can also lead to a high degree of indeterminate OLA reactions, as observed in our codon 181 results.
We note several limitations to our analysis. Our assumptions about specimens with low-level viremia (<1000 copies/ml) could introduce bias into our analyses, a problem that we attempted to address through the parallel analytical approaches noted above. Although we used a similar approach as our primary analysis of the trial,7 the threshold under which specimens were not tested for viral drug resistance was lowered from 2000 copies/ml to 1000 copies/ml, to facilitate comparisons between the two assays. Finally, we recognize that a significant proportion of test results was unavailable because of follow-up losses, lost specimens, inadequate amplifications, and indeterminate assays. Most differences noted between the included and excluded populations were not associated with selection for NNRTI resistance and were therefore unlikely to impact the external validity of our results. The one exception was the higher rate of NVP adherence observed among individuals included in the 6-week analysis.
In summary, when combined with intrapartum NVP, single-dose intrapartum TDF/FTC reduces selection for NNRTI resistance at both high and low concentrations. Although the overall prevalence of NNRTI resistance was higher when the more sensitive OLA was used, risk reduction associated with TDF/FTC use was comparable with resistance assessed by consensus sequencing.7 A notable finding of this study is that mutations associated with TDF and FTC resistance were rare (K65R) or not detected (M184V). Simple and effective, single-dose TDF/FTC should be considered alongside other proven interventions to reduce NNRTI-associated resistance after intrapartum NVP.5
Acknowledgments
We thank our study participants and study staff for their hard work and cooperation. We would also like to acknowledge Mark Giganti for his assistance with the statistical analysis and Ram Parvataneni, Dwight Rouse, John Hauth, Alice Goepfert, and Sue Cliver for their roles in project oversight. The clinical trial (Clinical trials.gov registration number: NCT00204308) described in this report was supported by the Elizabeth Glaser Pediatric AIDS Foundation (EGSA 19-02). Oligonucleotide ligation assays were supported by NIH awards R01 AI058723 (LMF) and U01 AI068632 (LMF for a Virology Specialty Laboratory of IMPAACT). Additional investigator salary or trainee support was provided by the National Institutes of Health (K01-TW06670; K01-TW05708; K23-AI01411) and the Doris Duke Clinical Scientist Award (2007061). This work was presented in part at the 15th Conference for Retroviruses and Opportunistic Infections held on February 3–6, 2008 in Boston, Massachusetts (Poster Abstract 631).
Disclosure Statement
No competing financial interests exist.
References
- 1.Eshleman SH. Mracna M. Guay LA, et al. Selection and fading of resistance mutations in women and infants receiving nevirapine to prevent HIV-1 vertical transmission (HIVNET 012) AIDS. 2001;15:1951–1957. doi: 10.1097/00002030-200110190-00006. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro RL. Thior I. Gilbert PB, et al. Maternal single-dose nevirapine versus placebo as part of an antiretroviral strategy to prevent mother-to-child HIV transmission in Botswana. AIDS. 2006;20:1281–1288. doi: 10.1097/01.aids.0000232236.26630.35. [DOI] [PubMed] [Google Scholar]
- 3.Jourdain G. Ngo-Giang-Huong N. Le Coeur S, et al. Intrapartum exposure to nevirapine and subsequent maternal responses to nevirapine-based antiretroviral therapy. N Engl J Med. 2004;351:229–240. doi: 10.1056/NEJMoa041305. [DOI] [PubMed] [Google Scholar]
- 4.Cressey TR. Jourdain G. Lallemant MJ, et al. Persistence of nevirapine exposure during the postpartum period after intrapartum single-dose nevirapine in addition to zidovudine prophylaxis for the prevention of mother-to-child transmission of HIV-1. J Acquir Immune Defic Syndr. 2005;38:283–288. [PubMed] [Google Scholar]
- 5.McIntyre J. Martinson N. Gray G, et al. Addition of short-course combivir (CBV) to single dose viramune (sdNVP) for the prevention of mother-to-child transmission (pMTCT) of HIV-1 can significantly decrease the development of maternal and paediatric NNRTI-resistant virus; In the 3rd IAS Conference on HIV Pathogenesis and Treatment; Rio de Janiero, Brazil. 24–27 July; 2005. [Abstract TuFo0204]. [Google Scholar]
- 6.Chaix ML. Ekouevi DK. Rouet F, et al. Low risk of nevirapine resistance mutations in the prevention of mother-to-child transmission of HIV-1: Agence Nationale de Recherches sur le SIDA Ditrame Plus, Abidjan, Cote d'Ivoire. J Infect Dis. 2006;193:482–487. doi: 10.1086/499966. [DOI] [PubMed] [Google Scholar]
- 7.Chi BH. Sinkala M. Mbewe F, et al. Single-dose tenofovir and emtricitabine for reduction of viral resistance to non-nucleoside reverse transcriptase inhibitor drugs in women given intrapartum nevirapine for perinatal HIV prevention: An open-label randomised trial. Lancet. 2007;370:1698–1705. doi: 10.1016/S0140-6736(07)61605-5. [DOI] [PubMed] [Google Scholar]
- 8.Schuurman R. Demeter L. Reichelderfer P. Tijnagel J. de Groot T. Boucher C. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J Clin Microbiol. 1999;37:2291–2296. doi: 10.1128/jcm.37.7.2291-2296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flys T. Nissley DV. Claasen CW, et al. Sensitive drug-resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevirapine (NVP) resistance mutation in some women and infants after the administration of single-dose NVP: HIVNET 012. J Infect Dis. 2005;192:24–29. doi: 10.1086/430742. [DOI] [PubMed] [Google Scholar]
- 10.Johnson JA. Li JF. Morris L, et al. Emergence of drug-resistant HIV-1 after intrapartum administration of single-dose nevirapine is substantially underestimated. J Infect Dis. 2005;192:16–23. doi: 10.1086/430741. [DOI] [PubMed] [Google Scholar]
- 11.Church JD. Jones D. Flys T, et al. Sensitivity of the ViroSeq HIV-1 genotyping system for detection of the K103N resistance mutation in HIV-1 subtypes A, C, and D. J Mol Diagn. 2006;8:430–432. doi: 10.2353/jmoldx.2006.050148. quiz 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Troyer RM. Lalonde MS. Fraundorf E, et al. A radiolabeled oligonucleotide ligation assay demonstrates the high frequency of nevirapine resistance mutations in HIV type 1 quasispecies of NVP-treated and untreated mother-infant pairs from Uganda. AIDS Res Hum Retroviruses. 2008;24:235–250. doi: 10.1089/aid.2007.0138. [DOI] [PubMed] [Google Scholar]
- 13.Chi BH. Chintu N. Cantrell RA, et al. Addition of single-dose tenofovir and emtricitabine to intrapartum nevirapine to reduce perinatal HIV transmission. J Acquir Immune Defic Syndr. 2008;48:220–223. doi: 10.1097/QAI.0b013e3181743969. [DOI] [PubMed] [Google Scholar]
- 14.Stringer JS. Zulu I. Levy J, et al. Rapid scale-up of antiretroviral therapy at primary care sites in Zambia: Feasibility and early outcomes. JAMA. 2006;296:782–793. doi: 10.1001/jama.296.7.782. [DOI] [PubMed] [Google Scholar]
- 15.Chi BH. Chintu N. Lee A. Stringer EM. Sinkala M. Stringer JS. Expanded services for the prevention of mother-to-child HIV transmission: Field acceptability of a pilot program in Lusaka, Zambia. J Acquir Immune Defic Syndr. 2007;45:125–127. doi: 10.1097/QAI.0b013e318050d28f. [DOI] [PubMed] [Google Scholar]
- 16.Beck IA. Mahalanabis M. Pepper G, et al. Rapid and sensitive oligonucleotide ligation assay for detection of mutations in human immunodeficiency virus type 1 associated with high-level resistance to protease inhibitors. J Clin Microbiol. 2002;40:1413–1419. doi: 10.1128/JCM.40.4.1413-1419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellis GM. Mahalanabis M. Beck IA, et al. Comparison of oligonucleotide ligation assay and consensus sequencing for detection of drug-resistant mutants of human immunodeficiency virus type 1 in peripheral blood mononuclear cells and plasma. J Clin Microbiol. 2004;42:3670–3674. doi: 10.1128/JCM.42.8.3670-3674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frenkel LM. Wagner LE., 2nd Atwood SM. Cummins TJ. Dewhurst S. Specific, sensitive, and rapid assay for human immunodeficiency virus type 1 pol mutations associated with resistance to zidovudine and didanosine. J Clin Microbiol. 1995;33:342–347. doi: 10.1128/jcm.33.2.342-347.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edelstein RE. Nickerson DA. Tobe VO. Manns-Arcuino LA. Frenkel LM. Oligonucleotide ligation assay for detecting mutations in the human immunodeficiency virus type 1 pol gene that are associated with resistance to zidovudine, didanosine, and lamivudine. J Clin Microbiol. 1998;36:569–572. doi: 10.1128/jcm.36.2.569-572.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tobin NH. Learn GH. Holte SE, et al. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: Expression of archival virus and replication of virus. J Virol. 2005;79:9625–9634. doi: 10.1128/JVI.79.15.9625-9634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lockman S. Shapiro RL. Smeaton LM, et al. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. N Engl J Med. 2007;356:135–147. doi: 10.1056/NEJMoa062876. [DOI] [PubMed] [Google Scholar]
- 22.Chi BH. Sinkala M. Stringer EM, et al. Early clinical and immune response to NNRTI-based antiretroviral therapy among women with prior exposure to single-dose nevirapine. AIDS. 2007;21:957–964. doi: 10.1097/QAD.0b013e32810996b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weidle PJ. Stringer JS. McConnell MS, et al. Effectiveness of non-nucleoside reverse transcriptase inhibitator-containing antiretroviral therapy in women previously exposed to a single dose of nevirapine: A multi-country cohort study; In the 14th Conference on Retroviruses and Opportunistic Infections; Boston, MA. 2007. [Google Scholar]
- 24.Peuchant O. Thiebaut R. Capdepont S, et al. Transmission of HIV-1 minority-resistant variants and response to first-line antiretroviral therapy. AIDS. 2008;22:1417–1423. doi: 10.1097/QAD.0b013e3283034953. [DOI] [PubMed] [Google Scholar]
- 25.Coovadia A. Hunt G. Abrams EJ, et al. Persistent minority K103N mutations among women exposed to single-dose nevirapine and virologic response to nonnucleoside reverse-transcriptase inhibitor-based therapy. Clin Infect Dis. 2009;48(4):462–472. doi: 10.1086/596486. [DOI] [PMC free article] [PubMed] [Google Scholar]