Abstract
In the Arabidopsis accession Columbia, 5S rDNA is located in the pericentromeric heterochromatin of chromosomes 3, 4, and 5. Both a major and some minor 5S rRNA species are expressed from chromosomes 4 and 5, whereas the genes on chromosome 3 are not transcribed. Here, we show that 5S rDNA methylation is reduced in 2-day-old seedlings versus 4-day-old or older aerial plant tissues, and the minor 5S rRNA species are expressed most abundantly at this stage. Similarly, when 5S rDNA is demethylated by 5-azacytidine treatment or via the decrease in DNA methylation1 (ddm1) mutation, the expression of minor 5S rRNA species is increased. We also show that in leaf nuclei of mature wild-type plants, the transcribed fraction of 5S rDNA forms loops that emanate from chromocenters. These loops, which are enlarged in nuclei of mature ddm1 plants, are enriched for histone H3 acetylated at Lys-9 and methylated at Lys-4 compared with the heterochromatic chromocenters. Up to 4 days after germination, heterochromatin is not fully developed: the 5S rDNA resides in prechromocenters, does not form conspicuous loops, and shows the lowest transcription level. Our results indicate that the expression and chromatin organization of 5S rRNA genes change during heterochromatin establishment.
INTRODUCTION
Heterochromatin is cytologically defined as highly condensed chromatin (Heitz, 1928). Arabidopsis chromosomes (n = 5) display small, conspicuous heterochromatin segments (chromocenters) that mark the position of each (peri-)centromere and of the nucleolus-organizing regions of chromosomes 2 and 4 (Fransz et al., 1998, 2002). Chromocenters contain most of the repetitive DNA sequences, constituting ∼15% of the Arabidopsis genome. The heterochromatin of many eukaryotes, including Arabidopsis, is marked by the methylation of DNA and of histone H3 at Lys-9 (H3K9) (Zhang and Reinberg, 2001; Gendrel et al., 2002; Johnson et al., 2002; Soppe et al., 2002; Jasencakova et al., 2003), whereas euchromatin is marked by H3 at Lys-4 (H3K4) methylation and H3K9 acetylation. At the level of the genes, the acetylation of histone tails and the methylation of histone H3 at Lys-4 (H3mK4) correspond to “open” chromatin that is potentially active in transcription, whereas the deacetylation of histones, cytosine methylation, and the methylation of Lys-9 of histone H3 (H3mK9) correspond to “closed” chromatin that is repressed for transcription (Bird and Wolffe, 1999; Turner, 2000; Gendrel et al., 2002; Kouzarides, 2002; Lachner and Jenuwein, 2002).
Soppe et al. (2002) have shown that DECREASE IN DNA METHYLATION1 (DDM1) and METHYLTRANSFERASE1 (MET1) control heterochromatin assembly at chromocenters by their influence on DNA maintenance methylation at CG sites and subsequently on the methylation of H3K9. The DDM1 gene encodes a chromatin-remodeling protein (Jeddeloh et al., 1999; Brzeski and Jerzmanowski, 2003), and MET1 encodes a CG-specific maintenance methyltransferase (Finnegan and Kovac, 2000). The reduction of H3mK9 in the ddm1 background has been correlated to the transcriptional derepression of genes, transgenes, and transposons using chromatin immunoprecipitation (ChIP) assays and cytological analysis (Gendrel et al., 2002; Johnson et al., 2002; Probst et al., 2003). Nuclei from ddm1 and met1 mutants show a reduction of heterochromatin content as a result of the dispersion of pericentromeric sequences away from heterochromatic chromocenters (Soppe et al., 2002). The ddm1 mutation also is known to cause a striking decondensation of chromocenters (Probst et al., 2003).
Arabidopsis contains ∼1000 copies of 5S rDNA per 1C genome arranged in tandem arrays (Campell et al., 1992) within the pericentromeric heterochromatin of chromosomes 3, 4, and 5 (with a large locus on the left arm and a small locus on the right arm of chromosome 5) in the Columbia accession (Murata et al., 1997; Fransz et al., 1998). We have shown that 5S rDNA is, on average, methylated at 79% of cytosines in CG, CNG, and asymmetrical contexts (Mathieu et al., 2002b) and that the density of DNA methylation increases from euchromatin toward pericentromeric heterochromatin (Mathieu et al., 2002a). Arabidopsis expresses a major 5S rRNA transcript and some minor species that differ from the major transcript by one or two base substitutions. The 5S rDNA units homologous with major and minor 5S rRNA transcripts have been identified and belong to chromosome 4 as well as to the large block of chromosome 5 (Cloix et al., 2002).
Here, we report that 5S rRNA genes undergo changes in chromatin organization and transcription during the early development of the leaf. In adult plant leaf nuclei, the 5S rDNA is found both in condensed chromocenters and in loops that emanate from the chromocenters. The loops are undermethylated and enriched for histones modified with H3K9 acetylation and H3K4 methylation; therefore, they presumably correspond to the transcribed fraction of major 5S rRNA genes. This chromatin organization of the 5S rRNA genes occurs between 2 and 4 days after germination. In 2-day-old seedling nuclei, only small prechromocenters are visible and the overall expression of 5S rRNA genes is low. 5S rDNA is undermethylated compared with later stages of development, and there is enriched expression of the minor 5S rRNA genes. Similarly, demethylation of 5S rDNA by 5-azacytidine or by the ddm1 mutation enriches minor 5S rRNA gene expression. These findings suggest that the major and minor 5S rRNA genes have different patterns of chromatin organization and correspondingly different patterns of expression.
RESULTS
5S rDNA Methylation
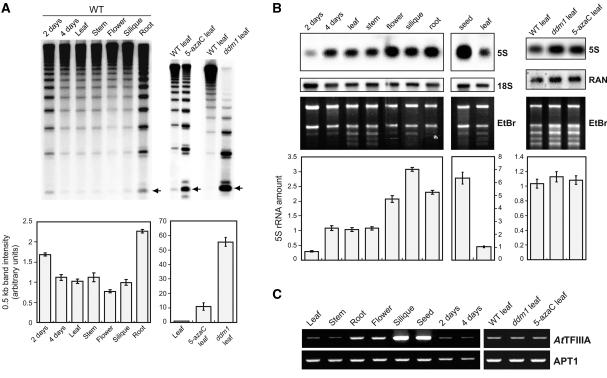
In plants and mammals, DNA of heterochromatin contains a significant amount of 5-methylcytosine (5mC), and 5S rDNA is highly methylated in Arabidopsis (Mathieu et al., 2002b). The methylation frequency of 5S rDNA cytosine residues was estimated by DNA gel blot analysis of genomic DNA after digestion with HpaII and quantifying the 0.5-kb band that corresponds to monomeric units of 5S rDNA and whose intensity is inversely proportional to the methylation frequency (Figure 1A; five repetitions per experimental point). HpaII is inhibited by the methylation of either C in the sequence 5′-CCGG-3′. As expected, in mature leaves of the ddm1 mutant (Vongs et al., 1993) and of wild-type plants treated with the cytosine methylation inhibitor compound 5-azacytidine (5-azaC) (Jones, 1985; Haaf, 1995), 5S rDNA was methylated much less significantly than in leaves of untreated wild-type plants (Mann-Whitney U test [U], 1% level of significance). In adult wild-type plants, 5S rDNA was methylated equally in leaves, stems, and green siliques (U, P > 0.01). 5S rDNA was less methylated in roots and more methylated in flowers compared with 3-week-old wild-type leaves (U, P < 0.01). Four-day-old leaves (roots removed from 4-day-old seedlings) showed a 5S rDNA methylation level comparable to that of 3-week-old leaves (U, P > 0.01). However, soon after germination (2 days), 5S rDNA methylation was reduced significantly relative to that in 4-day-old and 3-week-old leaves (U, P < 0.01).
Figure 1.
5S rDNA Methylation, Quantitative 5S rDNA Transcription, and AtTFIIIA Expression Pattern.
(A) DNA methylation of 5S rDNA. Genomic DNA was prepared from different tissues and developmental stages and from leaves of 5-azaC and ddm1 plants, digested with HpaII, and probed by 5S rDNA on gel blots. 5S rDNA methylation was estimated by quantifying the intensity of the 0.5-kb band (normalized with the whole-line radioactivity), which corresponds to monomeric units of 5S rDNA (arrows). Quantifications of five independent experiments are presented in the histograms below the gels, and the standard deviation of the mean is indicated on each bar. WT, wild type.
(B) 5S rRNA amounts in various organs, developmental stages, and hypomethylated plants. Total RNA was hybridized using a 5S rDNA probe. RNA quantities in each line were normalized using the 18S rRNA or a RAS-related nuclear protein (Haizel et al., 1997) probe together with the ethidium bromide (EtBr) profile, and 5S rRNA amounts were quantified accordingly. The results of five independent experiments are represented in the corresponding histograms, and the standard deviation of the mean is indicated on each bar. Results obtained from seeds are presented separately, because another RNA purification method was used (see Methods). For comparison, total RNA from wild-type mature leaves also was purified using this method.
(C) Expression pattern of AtTFIIIA mRNA. Transcriptional analysis of AtTFIIIA was performed by semiquantitative RT-PCR. Expression of the constitutive APT1 gene was used to normalize the amounts of cDNA.
5S. rRNA Amounts and 5S rRNA Heterogeneity
To investigate 5S rDNA transcription, we performed RNA gel blot experiments together with reverse transcriptase–mediated (RT) PCR analysis. RNA gel blots revealed a constant 5S rRNA amount in mature 3-week-old leaves and stems and 4-day-old leaves (U, P > 0.01), whereas only 30% of this amount appeared in 2-day-old seedlings (U, P < 0.01). Flowers, siliques, and roots had two to three times more and seeds had six times more 5S rRNA than was detected in leaves (U, P < 0.01). In mature leaves of ddm1 and of 5-azaC–treated plants, the same quantity of 5S rRNA was present as in wild-type leaves (Figure 1B) (U, P > 0.01).
To analyze the relative expression levels of the heterogeneous 5S rRNA species, we performed RT-PCR experiments on total RNA samples (Table 1). For each sample, two to four independent RNA purifications followed by RT-PCR were pooled before sequencing. Contaminating 5S genomic rDNA was totally removed by DNase I treatment (see Methods). Mature wild-type leaves, green siliques, flowers, and 4-day-old leaves contained a low proportion (∼3.25%) of minor 5S rRNAs. In mature wild-type 3-week-old leaf tissue, minor 5S rRNA genes were repressed and heterochromatin was fully established (see below); therefore, this stage was used as a reference. Significantly (Fisher's exact test; see Methods), higher minor 5S rRNAs proportions were observed in 2-day-old seedlings, in roots, and in 5-azaC–treated and ddm1 plants (13.2 to 22.7%). In mature seeds, 13.6% of 5S rRNA were minor RNAs accumulated during embryogenesis.
Table 1.
Proportion of Minor 5S rRNAs**,
| Sample | Number of Minor 5S rRNA Clones |
Total Clones a |
Percent of Minor 5S rRNAs |
Significance Level b |
|---|---|---|---|---|
| Wild type | ||||
| 3-week-old leaf | 2 | 103 | 1.9 | Reference |
| 2-day-old seedling | 9 | 53 | 17 | 0.001* |
| 4-day-old leaf | 1 | 56 | 1.8 | 0.717 |
| Seed | 6 | 44 | 13.6 | 0.009* |
| Root | 7 | 53 | 13.2 | 0.008* |
| Silique | 3 | 51 | 5.9 | 0.203 |
| Flower | 2 | 58 | 3.4 | 0.456 |
| ddm1 | ||||
| 3-week-old leaf | 12 | 54 | 22.2 | 0.00005** |
| 2-day-old seedling | 11 | 55 | 20 | 0.0002** |
| 5-azaC (1mM) 3-week-old leaf |
10 | 44 | 22.7 | 0.0001** |
Total clones = number of minor 5S rRNA clones recovered + number of major 5S rRNA clones recovered.
Significance levels are as follows:
, 0.01;
0.001 (Fisher's exact test [see Methods]).
These results reveal that in hypomethylated plants (5-azaC–treated and ddm1) as well as in some tissues (roots) and soon after germination (2 days) when 5S rDNA methylation was lower, additional 5S rRNA genes were transcribed to produce minor 5S rRNA species. By contrast, 4-day-old and 3-week-old wild-type leaves, which shared equivalent 5S rRNA amounts and 5S rDNA methylation levels, contained the same low proportion of minor 5S rRNAs.
Correlation between 5S rRNA Heterogeneity, 5S rDNA Methylation, and Plant Development
The 5S rRNA heterogeneity, 5S rDNA methylation, and 5S rRNA amount results summarized in Table 2 led us to the following conclusions. (1) There was no correlation between 5S rRNA amount and 5S rDNA methylation. (2) There was no correlation between the 5S rRNA amount and the proportion of minor 5S rRNAs. (3) There was an inverse correlation between the proportion of minor 5S rRNAs and the 5S rDNA methylation level. In other words, the less the 5S rDNA was methylated, the higher was the minor 5S rRNAs proportion, although this relationship was not strictly linear. (4) Two-day-old seedlings contained a lower 5S rDNA methylation level and a higher proportion of minor 5S rRNAs compared with 4-day-old or 3-week-old leaves.
Table 2.
Relative 5S rRNA Amounts, Proportions of Minor 5S rRNAs, and Levels of 5S rDNA Methylation
| Wild Type
|
ddm1
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Feature | Root | 2d a | 4d b | Leaf c | Stem | Flower | Silique | Seed | 2d a | 3w d | 5-azaC e |
| Relative 5S rRNA amount | 2.3 f | 0.3 f | 1.1 | 1 | 1.1 | 2 f | 3.1f | 6.3 f | nd g | 1.1 | 1.1 |
| Minor 5S rRNAs (%) | 13.2 h | 17 h | 1.8 | 1.9 | nd | 3.4 | 5.9 | 13.6 h | 20 h | 22.2 h | 22.7 h |
| Relative 5S rDNA methylation (%) i | 44.4 f | 59.5 f | 89.3 | 100 | 88.5 | 125.5 f | 103.9 | nd | 1.1 f | 1.8 f | 9 f |
Two days after germination.
Four days after germination.
Three-week-old leaf, used as a reference in the table.
Three-week-old leaf.
5-Azacytidine–treated (1 mM) 3-week-old leaf.
Significantly different from reference by Mann-Whitney U test (1% level of significance).
nd, not determined.
Significantly different from reference by Fisher's exact test (see Table 1).
Compared with 3-week-old leaf; see Methods for calculation.
AtTFIIIA mRNA Levels Correlate with 5S rRNA Levels
Because the 5S rRNA amount was correlated neither with the proportion of minor 5S rRNAs nor with 5S rDNA methylation, we explored whether the 5S rDNA–specific transcription factor AtTFIIIA (Mathieu et al., 2003) could act as a limiting factor for 5S rDNA expression. This transcript cannot be detected by RNA gel blot analysis. Instead, AtTFIIIA mRNA quantities were estimated by semiquantitative RT-PCR for the same RNA samples that were analyzed for 5S rDNA transcription, with five independent replicates of each experiment.
The results revealed a good correlation between AtTFIIIA mRNA (Figure 1C) and overall 5S rRNA amounts (Figure 1B). For example, siliques and seeds, which contained the highest 5S rRNA amounts, also accumulated the highest amounts of AtTFIIIA mRNA. Flowers and roots contained slightly higher AtTFIIIA mRNA levels and two times more 5S rRNA than leaves. Although in ddm1 and 5-azaC–treated leaves additional minor 5S rRNA genes were transcribed, no significant increase in AtTFIIIA mRNA amounts was observed between these hypomethylated backgrounds and wild-type leaves. This result is in agreement with the presence of equivalent 5S rRNA quantities in ddm1, 5-azaC–treated, and wild-type leaves (Figure 1B). Together, these results suggest that AtTFIIIA is a limiting factor for the transcription of 5S rRNA genes. The one exception to this general pattern was found in 2-day-old seedlings, which contained similar levels of AtTFIIIA mRNA to more mature tissues but which expressed only 30% as much 5S rRNA.
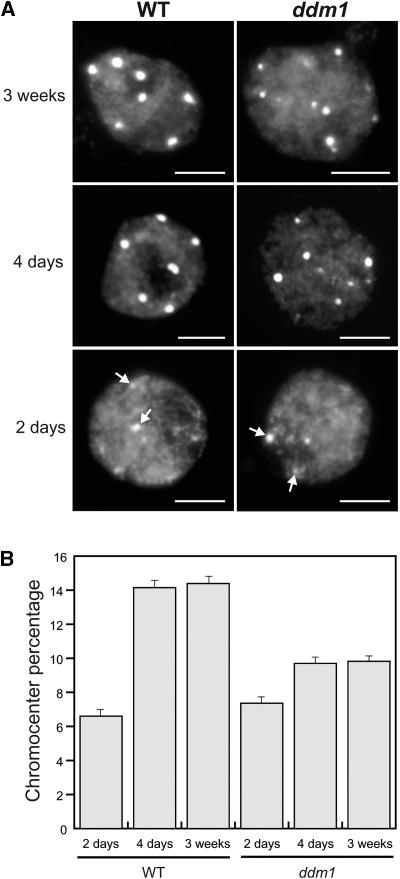
Nuclei of 2-Day-Old Seedlings Contain Prechromocenters and Reduced Amounts of Heterochromatin
A possible explanation for the reduced expression of total 5S rRNA transcripts in 2-day-old seedlings is that the appropriate chromatin structure for optimal 5S rDNA transcription is not fully formed at this stage. Therefore, we used cytological methods to investigate the chromatin structure of 5S rDNA at 2 days after germination versus later stages. Four-day-old and 3-week-old leaf nuclei of ddm1 showed on average smaller chromocenters (CCs), based on their 4′,6-diamidino-2-phenylindole (DAPI) staining intensity, than did corresponding wild-type nuclei. In 2-day-old nuclei from roots as well as from cotyledons of wild-type and ddm1 plantlets, the CCs were smaller than those in corresponding 4-day-old and 3-week-old leaf nuclei and were considered to represent prechromocenters (pre-CCs) (Figure 2A). The heterochromatin content was quantified by measuring the area and staining intensity of CCs/pre-CCs in relation to that of the entire nucleus (Figure 2B). In agreement with previous data (Soppe et al., 2002), a chromocenter fraction reduced by nearly one-third compared with that of wild-type nuclei was found in ddm1 leaf nuclei of 3-week-old plants. The observation that the chromocenter fraction of wild-type and ddm1 2-day-old nuclei amounts to approximately half of that measured in leaf nuclei of 4-day-old and 3-week-old wild-type plants (Figure 2B) reveals a developmental dynamic for constitutive heterochromatin and suggests that CCs become fully developed at 4 days after germination in Arabidopsis leaves. Our results also suggest that there is a comparable development of heterochromatin in the wild type and ddm1 between 2 and 4 days, although this process is less efficient in ddm1, in which mature CCs remained smaller.
Figure 2.
Nuclei from 2-Day-Old Postgermination Seedlings Contain Less Heterochromatin Than Nuclei from Leaves of 4-Day-Old and 3-Week-Old Plants.
(A) Representative DAPI-stained leaf nuclei from 2-day-old seedlings and 4-day-old seedlings and leaves of 3-week-old wild-type (WT) and ddm1 plants. ddm1 leaf nuclei of 3-week-old plants contain smaller chromocenters than corresponding wild-type nuclei. In nuclei from 2-day-old seedlings, small, weakly stained prechromocenters are visible (arrows). Bars = 5 μm.
(B) Chromocenter fractions of 2-day-old, 4-day-old, and 3-week-old nuclei of Arabidopsis wild type and ddm1. Percentages are derived from measurements of 60 to 68 nuclei each, and the standard error of the mean is indicated on each bar. In the wild type and ddm1, 4-day-old and 3-week-old nuclei have equivalent chromocenter fractions (P > 0.05), whereas the chromocenter fraction of 2-day-old nuclei is significantly different (P < 0.05). All ddm1 values are significantly different from wild-type values (P < 0.05) except for those from wild-type 2-day-old versus ddm1 2-day-old plants (P > 0.05).
Because 2-day-old plantlets contained a lower 5S rDNA methylation level, a lower total 5S rRNA amount, a higher minor 5S rRNA proportion, and a smaller heterochromatin fraction compared with 4-day-old and 3-week-old leaf nuclei, we conclude that there is a switch in 5S rDNA transcriptional activity between days 2 and 4 after germination, correlating with the establishment of mature heterochromatin.
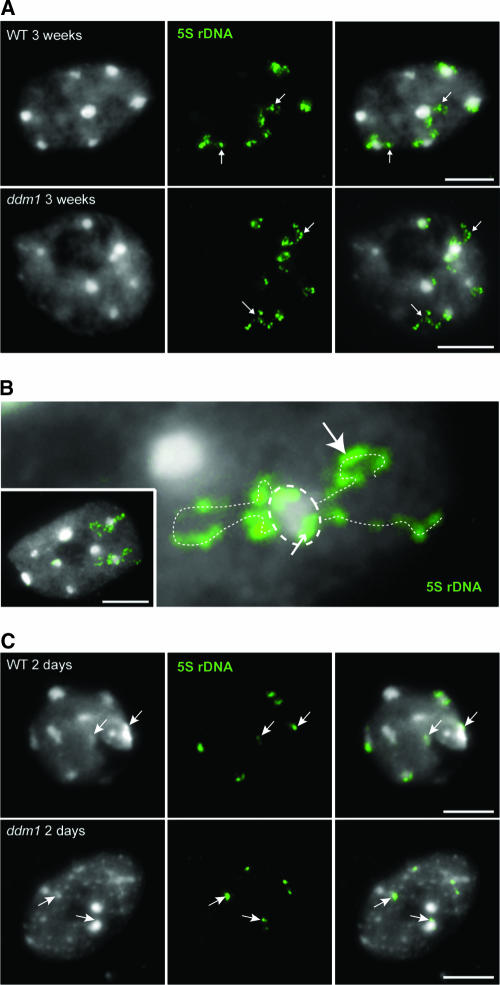
5S. rDNA “Loops” Emanate from the CCs in 3-Week-Old but Not in 2-Day-Old Nuclei
The smaller CCs of ddm1 3-week-old leaf nuclei were shown to contain less DNA as a result of the dispersion of low-copy pericentromeric sequences away from the CCs (Soppe et al., 2002). The higher proportion of minor 5S rRNAs and the lower level of 5S rDNA methylation together with the smaller size of CCs in ddm1 versus wild-type 3-week-old nuclei led us to ask whether 5S rDNA also was dispersed away from CCs in ddm1 3-week-old plants. Fluorescent in situ hybridization (FISH) with 5S rDNA yielded signals both within and outside the CCs in ddm1 3-week-old nuclei but also, surprisingly, in wild-type 3-week-old nuclei (Figures 3A and 3B). Counting of 5S rDNA signals at CCs versus signals outside CCs indicated that loop-forming 5S rDNA signals were significantly (χ2 test, P < 1%) more frequent in ddm1 (n = 54 nuclei) than in the wild type (n = 58 nuclei) (ratio of nonloop to loop 5S rDNA signals = 1:3.1 and 1:1.8, respectively). Such loops could be observed for CC4 (chromocenter 4), identified by the presence of both 5S rDNA and 45S rDNA signals, as well as for other 5S rDNA–bearing CCs, such as CC3 and CC5, that could not be discriminated cytologically.
Figure 3.
5S rDNA Loops Emanate from Chromocenters in Nuclei of 3-Week-Old Plants.
FISH with a 5S rDNA probe (green) on 3-week-old (A) and 2-day-old (C) nuclei from wild-type (WT) and ddm1 plants (loops indicated by arrows in [A]). (B) shows an enlargement of a wild-type nucleus (inset) from a 3-week-old plant with 5S rDNA loops (interrupted white lines) emanating from a chromocenter (circled). Parts of the 5S rDNA signal are located in the heterochromatic chromocenter (small arrow), and other parts form loops within euchromatin (large arrow). In 2-day-old postgermination seedling nuclei (C), 5S rDNA is located at prechromocenters (arrows). Counterstaining with DAPI (left in [A] and [C]), FISH with the 5S rDNA probe (middle in [A] and [C]), and the merge of both ([B] and right in [A] and [C]) are shown. Bars = 5 μm.
In nuclei of wild-type and ddm1 2-day-old plantlets, the 5S rDNA probe hybridized only to the heterochromatic pre-CCs (Figure 3C). The absence of loop-forming signals suggests that no significant part of 5S rDNA disperses away from heterochromatin in these nuclei. This observation is consistent with the view that 5S rDNA expression was reduced in 2-day-old plantlets because the appropriate chromatin structure had not yet formed.
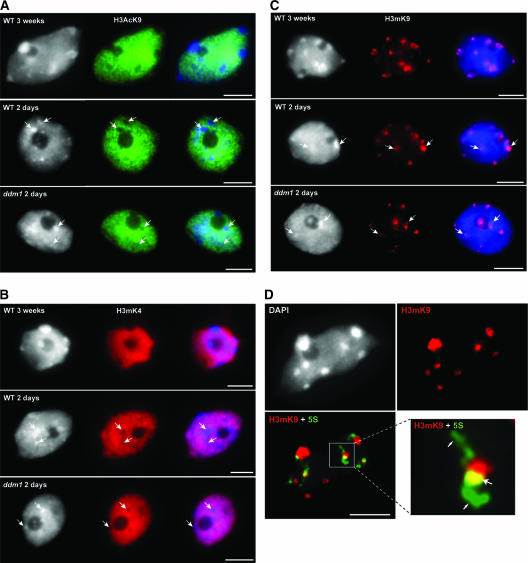
H3K9 Acetylation and H3K4 Methylation Are Not Detectable at 2-Day Pre-CCs
Histone H3 acetylated at Lys-9 (H3AcK9) and H3mK4 are two histone modifications often associated with transcriptionally active chromatin. Immunolabeling of 2-day-old and 3-week-old nuclei from wild-type and ddm1 plants using antibodies recognizing specifically H3AcK9 or H3mK4 revealed no signals at CCs/pre-CCs (Figures 4A and 4B). The size of the area free of signals correlated with the size of CCs and pre-CCs. The remaining chromatin, which likely includes the 5S rDNA loops in 3-week-old leaf nuclei, revealed consistently stronger H3AcK9 and H3mK4 signals. In nuclei of 2-day-old wild-type and ddm1 plantlets, although the heterochromatin was not fully developed (as shown above), the labeling pattern indicated that in both cases pre-CCs were less acetylated at H3K9 and less methylated at H3K4 than was euchromatin, as were the larger CCs of mature leaves.
Figure 4.
Chromatin Modifications in Nuclei of 2-Day-Old and 3-Week-Old Seedlings and at 5S rDNA Loops.
(A) Histone H3K9 acetylation (green). As in leaf nuclei of 3-week-old plants, heterochromatin of 2-day-old nuclei from both the wild type (WT) and ddm1 were unlabeled by the H3AcK9 antibody.
(B) Histone H3K4 dimethylation (red). As for H3AcK9, heterochromatin of nuclei from 2-day-old seedlings of the wild type and ddm1 remained unlabeled, whereas euchromatin showed strong labeling intensity.
(C) Histone H3K9 dimethylation (red). In 2-day-old nuclei from both the wild type and ddm1 as well as in 3-week-old nuclei, H3mK9 immunosignals localized preferentially to heterochromatin.
In (A) to (C), DAPI staining (left), immunosignals (middle), and the merge of both (right) are shown. Arrows indicate prechromocenters. Bars = 5 μm.
(D) 5S rDNA loops are devoid of H3K9 dimethylation. H3mK9 labeling of a 3-week-old wild-type nucleus shows strong immunosignals on chromocenters. A merge of the H3mK9 immunosignal (red) and the 5S rDNA FISH signal (green) is shown. The enlargement of a 5S rDNA loop reveals that euchromatic 5S rDNA loops are weakly associated or not associated with dimethylated H3K9 (small arrows), whereas heterochromatic 5S rDNA that associated with dimethylated H3K9 (large arrow) appears yellow (resulting from the superposition of green and red signals). Bar = 5 μm.
H3mK9 Is Present at Pre-CCs on 2-Day-Old Nuclei but Reduced at 5S rDNA Loops in 3-Week-Old Leaf Nuclei
Strong methylation of H3K9 has been found to be a typical feature of wild-type heterochromatin (see Introduction). At 3 weeks old, immunosignals obtained with antibodies directed against dimethylated K9 of H3 were clustered at the CCs (Figure 4C), whereas the area and intensity of signals were reduced strongly in ddm1 CCs (Soppe et al., 2002) compared with the wild type. 5S rDNA loops were not labeled or were less labeled with H3mK9, suggesting a euchromatic structure (Figure 4D). At 2 days old, both wild-type and ddm1 nuclei pre-CCs revealed heterochromatic features such as H3mK9 and the absence of both H3mK4 and H3AcK9 (Figures 4A to 4C). At this stage, 5S rDNA did not form loops and largely colocalized with the heterochromatic pre-CCs. This chromatin organization may underlie the less efficient 5S rRNA transcription observed in 2-day-old plantlets (Figure 1B).
Small 2-Day-Old Pre-CCs Are Already DNA Methylated
We compared the distribution patterns of methylated DNA in 2-day-old and 3-week-old nuclei of both wild-type and ddm1 plants using antibodies against 5mC. Wild-type 3-week-old nuclei showed strong signals, especially at CCs, whereas in ddm1 3-week-old nuclei, the immunosignals were dispersed and not clustered at CCs, as observed previously (Soppe et al., 2002). In 2-day-old nuclei of wild-type and ddm1 plantlets, the immunosignals were distributed nearly uniformly and not restricted to pre-CCs (Figure 5). Although DNA methylation was present at 2 days after germination, pre-CCs were not fully developed and were smaller than those in adult leaves.
Figure 5.
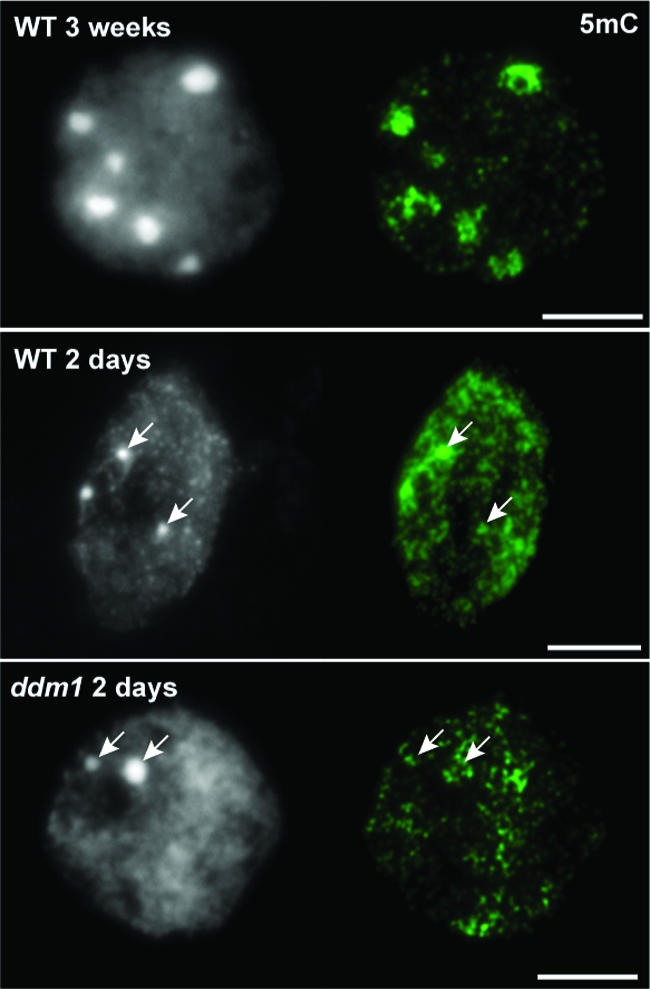
DNA Methylation Pattern in 2-Day-Old Nuclei.
Immunosignals for 5-methylcytosine (5mC; green) are strongly clustered at chromocenters in wild-type (WT) 3-week-old nuclei (top), whereas in ddm1 3-week-old nuclei, the immunosignal is dispersed and no longer clustered at chromocenters (not shown; see Soppe et al., 2002). In 2-day-old nuclei from the wild type and ddm1, the immunosignals were not clustered at prechromocenters. DAPI staining (left) and immunosignals (right) are shown. Arrows indicate prechromocenters. Bars = 5 μm.
Transcribed 5S rDNA Loci Are Less Associated with H3mK9 in ddm1 Nuclei
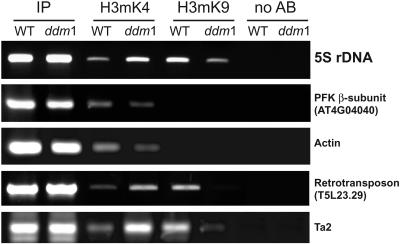
The hypomethylation of 5S rDNA in the ddm1 background (Figure 1A) was correlated with an increased proportion of minor 5S rRNAs (Table 1), indicating the transcription of additional 5S rRNA genes. It was shown previously that in ddm1 heterochromatin, DNA methylation is reduced strongly and H3mK9 is replaced by H3mK4. This pattern has been observed at reactivated genes and transposons (Gendrel et al., 2002; Johnson et al., 2002) and for regions that are dispersed from CCs (Soppe et al., 2002). We analyzed histone H3 methylation patterns of 5S rDNA from wild-type and ddm1 mature plants by ChIP using antibodies raised against H3 dimethylated at K9 and H3 dimethylated at K4 peptides. To validate the immunoprecipitation step, four different controls were used: the phosphofructokinase β-subunit gene (At4g04040), the Cinful-like retrotransposon (T5L23.29), the ACTIN gene, and the Ta2 transposon. Because our ChIP-PCRs yielded the previously described patterns for all four sequences (Gendrel et al., 2002; Johnson et al., 2002), we used the immunoprecipitations for subsequent 5S rDNA–specific PCR procedures.
In wild-type plants, 5S rDNA was associated mostly with H3mK9, whereas a smaller fraction was associated with H3mK4 (Figure 6). In ddm1, a shift in the pattern of histone methylation was observed, with a decreased association with H3mK9 and an increased association with H3mK4 compared with the wild type. We then investigated whether changes in histone H3 methylation were distributed equally through the 5S rDNA regions, with the potential to be transcribed on chromosome 4 and the large locus of chromosome 5 versus nontranscribed regions on chromosome 3. For this purpose, ChIP PCR products from three independent ChIP experiments were sequenced and assigned to the corresponding 5S rDNA loci through the T-stretch signature present downstream of the transcribed region (Table 3) (Cloix et al., 2002). This analysis revealed that 5S rRNA genes from transcribed loci were significantly (Z test, P = 0.05) more associated with H3mK9 in the wild type (89.7%) than in ddm1 (70.8%). By contrast, H3mK4 ChIP-PCR results showed that 5S rDNA from these loci was significantly (Z test, P = 0.05) more associated with H3mK4 in ddm1 (82.5%) than in the wild type (60%). These results indicate that in ddm1, more 5S rRNA genes from transcribed loci acquire euchromatin features, in agreement with the presence of larger/more euchromatic 5S rDNA loops in ddm1. Thus, in ddm1, additional minor 5S rRNA genes, usually present in heterochromatin in the wild type, likely become integrated in the euchromatic 5S rDNA loops that provide a permissive context for transcription.
Figure 6.
ChIP Analysis of 5S rRNA Genes.
ChIP assays performed on mature plantlets from both wild-type (WT) and ddm1 plants to analyze the H3K4 or H3K9 dimethylation state of 5S rRNA genes (top gel). A representative image from three independent ChIP experiments is shown. In the wild type, the 5S rDNA is much more associated with H3mK9 than with H3mK4. The association of 5S rDNA with H3mK9 decreases dramatically in ddm1 compared with the wild type, whereas the association with H3mK4 increases. The phosphofructokinase (PFK) β-subunit gene (At4g04040), the Cinful-like retrotransposon (T5L23.29), the ACTIN gene, and the Ta2 transposon were used as controls with known H3mK4 and H3mK9 patterns. In each case, total chromatin extract (input; IP) and no antibody-precipitated extracts (no AB) are presented.
Table 3.
Histone H3 Dimethylation Pattern on 5S rDNA Loci in Wild-Type and ddm1 Backgrounds
| H3mK4
|
H3mK9
|
|||
|---|---|---|---|---|
| 5S rDNA loci | Wild type | ddm1 | Wild type | ddm1 |
| Chromosome 3 (not transcribed) |
40% | 17.5% | 10.3% | 29.2% |
| Chromosomes 4 and 5 (transcribed) |
60% | 82.5% | 89.7% | 70.8% |
| Total | 100% | 100% | 100% | 100% |
| Number of clones sequenced | 40 | 57 | 58 | 48 |
A higher proportion of 5S rDNA sequences from the units on chromosome 3 were recovered by H3mK9 ChIP in the ddm1 versus the wild-type background. Reciprocally, a higher proportion of 5S rDNA sequences from chromosome 3 were recovered by H3mK4 ChIP in the wild-type versus the ddm1 background. Because 5S rDNA loci from chromosome 3 are not transcribed in vivo because of the presence of numerous mutations (Cloix et al., 2002), the ChIP results for this locus cannot be correlated with 5S rDNA expression.
DISCUSSION
In this study, we report that 5S rRNA genes undergo changes in chromatin organization and transcription during leaf development. In leaves, heterochromatin formation at chromocenters becomes fully established between days 2 and 4 after germination, accompanied by changes in DNA methylation, chromatin organization, and 5S rDNA transcription. At 2 days after germination, the heterochromatin fraction is composed of less-condensed pre-CCs that are much smaller than the CCs observed in 4-day-old and 3-week-old leaf nuclei (Figure 2). This result provides evidence that the appearance of constitutive heterochromatin is dynamic during leaf development in Arabidopsis. Interestingly, there also is a correlation between the formation of pericentric heterochromatin and cell differentiation during mammalian early development (Cammas et al., 2002; Rangasamy et al., 2003).
The chromatin organization of 5S rDNA in 2-day-old nuclei (in roots as well as in cotyledons) is different from that in 3-week-old leaves. 5S rDNA loops are not present and 5S rDNA is undermethylated compared with later stages of development. At 2 days old, major and minor 5S rRNA genes, both located in pre-CCs, are transcribed but produce only 30% as much total rRNA as is detected in mature wild-type leaves. From 4 days on, mostly major 5S rRNA is recovered from leaf nuclei. In adult plant leaf nuclei, the 5S rDNA is found both in condensed CCs and in loops that extend outward from the CCs. The loops show the same H3mK4 and H3AcK9 immunosignals compared with the surrounding euchromatin and no enrichment for 5mC or H3mK9 immunosignals, suggesting that they represent the transcribed fraction of 5S rRNA genes. The silent 5S rDNA presumably resides in condensed CCs. In support of this view, we reported previously that a small proportion of 5S rDNA units have reduced DNA methylation levels (Mathieu et al., 2002b). These hypomethylated units could correspond to 5S rRNA gene located in the loops. We also reported previously that the in vitro transcription of a naked 5S rRNA gene is not impaired by DNA methylation (Mathieu et al., 2002b). Similarly, Santoro and Grummt (2001) reported that the DNA methylation–mediated transcriptional repression of mouse rDNA is efficient on chromatin but not on naked DNA templates. These findings suggest the requirement for a specific chromatin state associated with DNA methylation for the repression of a subset of 5S rRNA genes. Therefore, we propose that between 2 and 4 days after germination, changes in chromatin organization cause major 5S rRNA genes to be incorporated preferentially in 5S rDNA loops and transcribed and cause minor genes to become condensed and repressed.
Demethylation of 5S rDNA by 5-azaC or by the ddm1 mutation enriches minor 5S rRNA genes expression. The ddm1 mutation is known to cause a striking decondensation of CCs (Probst et al., 2003). Like other sequences usually located inside CCs (Soppe et al., 2002), a fraction of 5S rDNA is dispersed from CCs, yielding larger 5S rDNA loops than in wild-type nuclei. Because the expression of minor 5S rRNAs is increased in ddm1 leaves, the minor 5S rRNA genes presumably are included in the larger loops. Consistent with this view, the ChIP data for the transcribed 5S rDNA blocks on chromosomes 4 and 5 show an increased association with H3mK4 and a decreased association with H3mK9 in ddm1 versus the wild type. The ChIP results for the nontranscribed 5S rDNA units on chromosome 3 present the inverse pattern. A possible explanation for this pattern is that the histone methylation marks on the nontranscribed loci do not actually vary between ddm1 and the wild type, but the relative proportions of these loci recovered by ChIP is altered by changes in the recovery of the chromosome 4 and 5 loci. This view is supported by the data of Johnson et al. (2002), who found that in various DNA methylation mutants, nontranscribed sequences retained H3K9 methylation.
5S rDNA transcription revealed quantitative and qualitative differences between organs and developmental stages. The proportion of minor 5S rRNAs showed a negative correlation with DNA methylation but no correlation with the overall 5S rRNA transcript level (Figure 1, Table 1). Instead, the overall level of 5S rRNA transcripts correlated with AtTFIIIA amounts. Because of the unavailability of an AtTFIIIA-specific antibody, we monitored AtTFIIIA mRNA accumulation by semiquantitative RT-PCR and found a good correlation between AtTFIIIA mRNA and 5S rRNA amounts. Although hypomethylated ddm1 and 5-azaC–treated adult plants transcribe additional minor 5S rRNA genes, they contain the same total quantity of 5S rRNA as untreated wild-type plants. This finding supports the idea that AtTFIIIA is a limiting factor for 5S rDNA transcription.
In conclusion, we have shown that during heterochromatin establishment in Arabidopsis, variations in epigenetic marks at the level of DNA methylation and histone modifications accompany 5S rDNA chromatin organization, resulting in the transcriptional repression of minor 5S rRNA genes. In ddm1, heterochromatin does not undergo complete development, yielding smaller mature CCs and resulting in the derepression of minor 5S rRNA gene transcription.
METHODS
Plant Materials
Arabidopsis thaliana wild-type and ddm1 plants were from the Columbia ecotype. Wild-type seeds (Col-4) were obtained from the ABRC (stock number CS933; Ohio State University, Columbus), and Eric J. Richards (Washington University, St. Louis, MO) provided the seed stock for the ddm1 mutant. Seeds were grown on a germination medium (MS Salt [Sigma] supplemented with 3% sucrose and 0.8% bacto-agar) for 3 weeks in a growth chamber using a 16-h-light (120 μmol·m−2·s−1)/8-h-dark regime at 23°C. 5-Azacytidine (5-azaC) treatments were performed as described previously (Mathieu et al., 2002b). To obtain postgermination plantlets, seeds were germinated in water in a growth chamber (16 h of light/8 h of dark) and collected directly at 2 days after germination or transferred on germination medium and collected at 4 days after germination. Roots were removed from 4-day-old plantlets.
Nucleic Acids Isolation and Gel Blot Analysis
Total genomic DNA was isolated from different tissues of 2-day-old and 4-day-old plantlets using the DNeasy kit (Qiagen, Valencia, CA). Genomic DNA (500 ng) was digested with 20 units of restriction enzyme in the recommended buffer (New England Biolabs, Beverly, MA). Digested DNA was electrophoresed on 0.8% agarose gels overnight, depurinated in 0.25 N HCl, and capillary blotted onto Hybond-N+ membranes (Amersham).
Total RNA was extracted according to Logemann et al. (1987) with minor modifications (Mathieu et al., 2002b). For seeds, total RNA was purified according to Vicient and Delseny (1999). For RNA gel blot analysis, 500 ng of total RNA per lane was fractionated on 1% agarose/1.9% formaldehyde gels and capillary blotted onto Hybond-N membranes (Amersham).
DNA probes were labeled with α-32P-dCTP using a random hexamer priming method (Megaprime DNA labeling system; Amersham). For quantification of the 0.5-kb band intensity, the 0.5-kb band radioactivity (which corresponds to monomeric units of 5S rDNA) was normalized with the whole-line radioactivity (Figure 1A). For the calculation of relative 5S rDNA methylation indicated in Table 2, the ratio obtained in 3-week-old wild-type leaves (used as reference) was divided by the ratio obtained for each sample tested and expressed as a percentage. Quantifications were performed on a phosphorimager (Molecular Imager FX; Bio-Rad).
Reverse Transcription PCR and Sequencing
Total RNA was isolated as described above, and contaminating genomic DNA was removed by DNase I treatment (Roche, Meylan, France) followed by phenol:chloroform:isoamyl alcohol (25:24:1) extraction. Reverse transcription reactions were performed in 20 μL with 1 μg of total RNA using 1 μL of Expand reverse transcriptase (Roche) and 2 μL of hexanucleotide mix (for 5S rRNA reverse transcription; Roche) or 25 to 50 pmol of the AtTFIIIA-specific primer AtTFIIIA2 (5′-CTAGCAAGTTTCGTGTTCTTCTG-3′) plus 25 to 50 pmol of the apt2 primer (5′-CCTTTCCCTTAAGCTCTG-3′), according to the manufacturer's instructions (Roche). Complete removal of the 5S genomic rDNA in the reverse-transcribed samples was controlled by verifying that a PCR step (40 cycles) using primers located in the nontranscribed spacer region of the 5S rRNA genes (5Suniv1, 5′-CTTTTCGGGCNTTTTNGTG-3′; 5Suniv2, 5′-CGAAAAGGTATCACATGCC-3′) yielded no amplification.
5S cDNA amplification was performed on 50 ng of the cDNA samples using the primers RTPCR5S1 (5′-GGATGCGATCATACCAG-3′) and RTPCR5S2 (5′-GAGGGATGCAMCACSAG-3′) amplifying the entire transcribed region of the 5S rRNA genes. For AtTFIIIA cDNA amplification, PCR was performed on 350 ng of the cDNA samples with the primers AtTFIIIA4 (5′-CTTACACATAAAGGGAAGCTC-3′) and AtTFIIIA5 (5′-CCGTAGATGTCTCTTGATG-3′) spanning the third intron. As an internal control, the APT1 cDNA encoding the adenine phosphoribosyl transferase (Moffatt et al., 1994) was amplified from 150 ng of the cDNA samples with the primers apt1 (5′-TCCCAGAATCGCTAAGATTGC-3′) and apt2 spanning the fourth intron. PCR conditions were as follows: 4 min at 94°C, 20 (APT1) or 30 (AtTFIIIA) cycles of 45 s at 94°C, 1 min at 54°C (AtTFIIIA) or 52°C (APT1), and 90 s at 72°C, and 10 min at 72°C. PCR conditions for 5S cDNAs were 4 min at 94°C, 35 cycles of 45 s at 94°C, 45 s at 50°C, and 45 s at 72°C, and 5 min at 72°C.
Statistics
Minor 5S rRNA frequencies were compared with Fisher's exact test for a 2 × 2 contingency table (van Tassel, 1981). The probabilities were calculated from a one-tailed test. Statistical analyses of 5S rDNA methylation and 5S rRNA amounts were performed using the nonparametric Mann-Whitney U test with mean values comparison.
Cytogenetics
For heterochromatin quantification and fluorescent in situ hybridization (FISH) with 5S rDNA and 45S rDNA probes, ethanol:acetic acid (3:1) fixation was used. For immunodetection of modified histones (coupled with 5S rDNA FISH), the tissue was fixed in 4% paraformaldehyde in Tris buffer (10 mM Tris, 10 mM Na2EDTA, and 100 mM Triton X-100, pH 7.5). Nuclear suspensions were produced as described (Jasencakova et al., 2000, 2001). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI).
FISH
The following probes were used: Arabidopsis BAC T15P10 (AF167571) containing 45S rDNA repeats labeled with biotin-16-dUTP using the nick translation kit (Roche), and 5S rDNA labeled with digoxigenin-11-dUTP by PCR using gene-specific primers. FISH experiments were performed according to Schubert et al. (2001). When the digoxigenin-labeled 5S rDNA probe was used alone, mouse anti-digoxigenin (1:250; Roche) followed by goat anti-mouse antibody conjugated with biotin (1:500; Jackson ImmunoResearch, West Grove, PA) and avidin conjugated with Texas Red (1:1000; Vector Laboratories, Burlingame, CA) were used for the detection. When digoxigenin-labeled 5S rDNA and biolabeled 45S rDNA were used together, avidin conjugated with Texas Red (1:1000; Vector Laboratories) followed by goat anti-avidin conjugated with biotin (1:200; Vector Laboratories) and avidin–Texas Red (1:1000) were used for the detection of the biotin-labeled probe, and sheep anti-digoxigenin fluorescein isothiocyanate (FITC) (1:20; Roche) followed by rabbit anti-sheep FITC (1:100; Jackson ImmunoResearch) were used for the detection of the digoxigenin-labeled probe.
Histone and 5-Methylcytosine Immunodetection
The following primary antibodies were used: anti-acetyl-histone H3 (Lys-9), anti-dimethyl-histone H3 (Lys-4), and anti-dimethyl-histone H3 (Lys-9) (1:100; all from Upstate Biotechnology, Lake Placid, NY). The immunolabeling procedure for histones was as described (Jasencakova et al., 2000, 2001). After postfixation in 4% paraformaldehyde/PBS, washes in PBS, and blocking at 37°C, the slides were exposed to primary antiserum overnight at 4°C. After washes in PBS, incubation with the secondary antibody anti-rabbit FITC (1:80; Sigma) or anti-rabbit rhodamine (1:100; Jackson ImmunoResearch) was performed at 37°C.
For the combined detection of H3-dimethylK9 and 5S rDNA, immunodetection of histones was performed first. After evaluation and image capture, the slides were processed for subsequent FISH as described previously (Jasencakova et al., 2000, 2001). The detection of 5-methylcytosine was performed using a monoclonal antibody against 5-methylcytidine (1:100; Eurogentec, Seraing, Belgium) in 100 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.5% blocking reagent (Roche), followed by rabbit anti-mouse FITC (1:1000; Sigma) and goat anti-rabbit Alexa-488 (1:200; Molecular Probes, Eugene, OR). The nuclei were counterstained with DAPI (2 μg/mL in Vectashield; Vector Laboratories).
Microscopy and Image Processing
For microscopic analysis, a Zeiss Axiophot 2 epifluorescence microscope (Jena, Germany) equipped with a cooled charge-coupled device camera (Photometrics, Tucson, AZ) was used. Fluorescence images were captured for each fluorochrome separately through the appropriate excitation filters. The images were pseudocolored, merged, and processed using Adobe Photoshop (Adobe Systems, Mountain View, CA).
Measuring of Chromocenter Fractions
Digital images in gray scale were analyzed with the freeware program NIH Image 1.62. Special macros were written to measure the size and average staining intensity of nuclei and chromocenters. The chromocenter value was divided by the whole-nucleus value and yielded the chromocenter fraction.
Chromatin Immunoprecipitation Assays
Two-week-old plants (wild type and ddm1) were harvested and immersed in 1% formaldehyde under vacuum for 15 min. Chromatin immunoprecipitation (ChIP) was performed as described (Gendrel et al., 2002; Johnson et al., 2002) using anti-dimethyl-histone H3 (Lys-4) or anti-dimethyl-histone H3 (Lys-9) antibodies (07-030 or 07-212, respectively; Upstate Biotechnology). Each of the immunoprecipitations was performed three independent times.
For ChIP-PCR, the primer pairs used were as follows: for 5S rDNA, RTPCR5S1 (5′-GGATGCGATCATACCAG-3′) and 5Suniv2; for the phosphofructokinase β-subunit gene (At4g04040), T24H24.15F (5′-GCCACGAAAACCAAACAGAC-3′) and T24H24.15R (5′-CCGGAATTTCGATCAATCCT-3′); for the Cinful-like retrotransposon (T5L23.29), T5L23.29F (5′-CTCGATGTCGTATTCGCTGA-3′) and T5L23.29R (5′-GCAACCTATCAACGCTTCGT-3′); for the ACTIN gene, Actin2/7-F (5′-CGTTTC-GCTTTCCTTAGTGTTAGCT-3′) and Actin2/7-R (5′-AGCGAACGGATCTAGAGACTCACCTTG-3′); and for the Ta2 transposon, Ta2-F (5′-AAACGATGCGTTGGGATAGGTC-3′) and Ta2-R (5′-ATACTCTCCACTTCCCGTTTTTCTTTTTA-3′). PCR was performed on 5 μL of a 1:10 dilution of ChIP samples (At4g04040, T5L23.29, ACTIN, and Ta2) or 2 μL of a 1:100 dilution (5S rDNA) in a final volume of 25 μL. PCR conditions were as follows: 5 min at 95°C, 25 (5S rDNA) or 30 cycles (At4g04040, T5L23.29, ACTIN, and Ta2) of 45 s at 94°C, 1 min at 54°C (5S rDNA) or 60°C (At4g04040, T5L23.29, ACTIN, and Ta2), and 30 s (5S rDNA) or 1 min (At4g04040, T5L23.29, ACTIN, and Ta2) at 72°C, and 5 min at 72°C.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Sylvette Tourmente, sylvette.tourmente@univ-bpclermont.fr.
Acknowledgments
We thank Achim Bruder, Martina Kühne, Nathalie Maroncle, and Claudine Cuvillier for helpful assistance, Armin Meister and Isabelle Jouan for help with statistics and flow cytometry, and Judith Bender and Charles White for critical reading of the manuscript. This work was supported by the Centre National de la Recherche Scientifique (CNRS), the epigenetic Groupement De Recherche CNRS, and the Université Blaise Pascal. V.C. was supported by Genopole and by a specific fund from the CNRS. Z.J. and I.S. were supported by a grant from the Land Sachsen-Anhalt (3233A/0020T). O.M., I.V., and A.-V.G. were supported by a fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.017467.
References
- Bird, A.P., and Wolffe, A.P. (1999). Methylation-induced repression: Belts, braces, and chromatin. Cell 99, 451–454. [DOI] [PubMed] [Google Scholar]
- Brzeski, J., and Jerzmanowski, A. (2003). Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J. Biol. Chem. 278, 823–828. [DOI] [PubMed] [Google Scholar]
- Cammas, F., Oulad-Abdelghani, M., Vonesch, J.L., Huss-Garcia, Y., Chambon, P., and Losson, R. (2002). Cell differentiation induces TIF1β association with centromeric heterochromatin via an HP1 interaction. J. Cell Sci. 115, 3439–3448. [DOI] [PubMed] [Google Scholar]
- Campell, B.R., Song, Y., Posch, T.E., Cullis, C.A., and Town, C.D. (1992). Sequence and organization of 5S ribosomal RNA-encoding genes of Arabidopsis thaliana. Gene 112, 225–228. [DOI] [PubMed] [Google Scholar]
- Cloix, C., Tutois, S., Yukawa, Y., Mathieu, O., Cuvillier, C., Espagnol, M.C., Picard, G., and Tourmente, S. (2002). Analysis of the 5S RNA pool in Arabidopsis thaliana: RNAs are heterogeneous and only two of the genomic 5S loci produce mature 5S RNA. Genome Res. 12, 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, E.J., and Kovac, K.A. (2000). Plant DNA methyltransferases. Plant Mol. Biol. 43, 189–201. [DOI] [PubMed] [Google Scholar]
- Fransz, P., Armstrong, S., Alonso-Blanco, C., Fischer, T.C., Torres-Ruiz, R.A., and Jones, G. (1998). Cytogenetics for the model system Arabidopsis thaliana. Plant J. 13, 867–876. [DOI] [PubMed] [Google Scholar]
- Fransz, P., De Jong, J.H., Lysak, M., Castiglione, M.R., and Schubert, I. (2002). Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl. Acad. Sci. USA 99, 14584–14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel, A.V., Lippman, Z., Yordan, C., Colot, V., and Martienssen, R. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 20, 1871–1873. [DOI] [PubMed] [Google Scholar]
- Haaf, T. (1995). The effects of 5-azacytidine and 5-azadeoxycytidine on chromosome structure and function: Implications for methylation-associated cellular processes. Pharmacol. Ther. 65, 19–46. [DOI] [PubMed] [Google Scholar]
- Haizel, T., Merkle, T., Pay, A., Fejes, E., and Nagy, F. (1997). Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. Plant J. 11, 93–103. [DOI] [PubMed] [Google Scholar]
- Heitz, E. (1928). Das Heterochromatin der Moose. Jahrb. Wiss. Botanik 69, 762–818. [Google Scholar]
- Jasencakova, Z., Meister, A., and Schubert, I. (2001). Chromatin organization and its relation to replication and histone acetylation during the cell cycle in barley. Chromosoma 110, 83–92. [DOI] [PubMed] [Google Scholar]
- Jasencakova, Z., Meister, A., Walter, J., Turner, B.M., and Schubert, I. (2000). Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell 12, 2087–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasencakova, Z., Soppe, W.J., Meister, A., Gernand, D., Turner, B.M., and Schubert, I. (2003). Histone modifications in Arabidopsis: High methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J. 33, 471–480. [DOI] [PubMed] [Google Scholar]
- Jeddeloh, J.A., Stokes, T.L., and Richards, E.J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22, 94–97. [DOI] [PubMed] [Google Scholar]
- Johnson, L., Cao, X., and Jacobsen, S. (2002). Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12, 1360–1367. [DOI] [PubMed] [Google Scholar]
- Jones, P.A. (1985). Altering gene expression with 5-azacytidine. Cell 40, 485–486. [DOI] [PubMed] [Google Scholar]
- Kouzarides, T. (2002). Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12, 198–209. [DOI] [PubMed] [Google Scholar]
- Lachner, M., and Jenuwein, T. (2002). The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 14, 286–298. [DOI] [PubMed] [Google Scholar]
- Logemann, J., Schell, J., and Willmitzer, L. (1987). Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 163, 16–20. [DOI] [PubMed] [Google Scholar]
- Mathieu, O., Picard, G., and Tourmente, S. (2002. a). Methylation of a euchromatin-heterochromatin transition region in Arabidopsis thaliana chromosome 5 left arm. Chromosome Res. 10, 455–466. [DOI] [PubMed] [Google Scholar]
- Mathieu, O., Yukawa, Y., Prieto, J.L., Vaillant, I., Sugiura, M., and Tourmente, S. (2003). Identification and characterization of transcription factor IIIA and ribosomal protein L5 from Arabidopsis thaliana. Nucleic Acids Res. 31, 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu, O., Yukawa, Y., Sugiura, M., Picard, G., and Tourmente, S. (2002. b). 5S rRNA gene expression is not inhibited by DNA methylation in Arabidopsis. Plant J. 29, 313–323. [DOI] [PubMed] [Google Scholar]
- Moffatt, B.A., McWhinnie, E.A., Agarwal, S.K., and Schaff, D.A. (1994). The adenine phosphoribosyltransferase-encoding gene of Arabidopsis thaliana. Gene 143, 211–216. [DOI] [PubMed] [Google Scholar]
- Murata, M., Heslop-Harrison, J.S., and Motoyoshi, F. (1997). Physical mapping of the 5S ribosomal RNA genes in Arabidopsis thaliana by multi-color fluorescence in situ hybridization with cosmid clones. Plant J. 12, 31–37. [DOI] [PubMed] [Google Scholar]
- Probst, A.V., Fransz, P.F., Paszkowski, J., and Scheid, O.M. (2003). Two means of transcriptional reactivation within heterochromatin. Plant J. 33, 743–749. [DOI] [PubMed] [Google Scholar]
- Rangasamy, D., Berven, L., Ridgway, P., and Tremethick, D.J. (2003). Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 22, 1599–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro, R., and Grummt, I. (2001). Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol. Cell 8, 719–725. [DOI] [PubMed] [Google Scholar]
- Schubert, I., Fransz, P.F., Fuchs, J., and de Jong, J.H. (2001). Chromosome painting in plants. Methods Cell Sci. 23, 57–69. [PubMed] [Google Scholar]
- Soppe, W.J., Jasencakova, Z., Houben, A., Kakutani, T., Meister, A., Huang, M.S., Jacobsen, S.E., Schubert, I., and Fransz, P.F. (2002). DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21, 6549–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, B.M. (2000). Histone acetylation and an epigenetic code. Bioessays 22, 836–845. [DOI] [PubMed] [Google Scholar]
- van Tassel, D. (1981). BASIC-Pack Statistics for Small Computers. (Englewood Cliffs, NJ: Prentice-Hall), pp. 146–151.
- Vicient, C.M., and Delseny, M. (1999). Isolation of total RNA from Arabidopsis thaliana seeds. Anal. Biochem. 268, 412–413. [DOI] [PubMed] [Google Scholar]
- Vongs, A., Kakutani, T., Martienssen, R.A., and Richards, E.J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260, 1926–1928. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., and Reinberg, D. (2001). Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes Dev. 15, 2343–2360. [DOI] [PubMed] [Google Scholar]