Abstract
Background
Actin polymerization by Arp2/3 complex must be tightly regulated to promote endocytosis. While many Arp2/3 complex activators have been identified, mechanisms for its negative regulation have remained more elusive. We analyzed the yeast arp2-7 allele, which is biochemically unique in causing unregulated actin assembly in vitro in the absence of Arp2/3 activators.
Results
We examined endocytosis in arp2-7 mutants by live cell imaging of Sla1-GFP, a coat marker, and Abp1-RFP, which marks the later actin phase of endocytosis. Sla1-GFP and Abp1-RFP lifetimes were accelerated in arp2-7 mutants, which is opposite to actin nucleation-impaired arp2 alleles or deletions of Arp2/3 activators. We performed a screen for multi-copy suppressors of arp2-7 and identified SYP1, a FCHO1 homologue, which contains F-BAR and AP-2μ homology domains. Over-expression of SYP1 in arp2-7 cells slowed Sla1-GFP lifetimes closer to wild type cells. Further, purified Syp1 directly inhibited Las17/WASp stimulation of Arp2/3 complex-mediated actin assembly. This activity was mapped to a fragment of Syp1 located between its F-BAR and AP-2μ homology domains, and depends on non-VCA sequences in Las17/WASp.
Conclusions
Together, these data identify Syp1 as a novel negative regulator of WASp-Arp2/3 complex that helps choreograph the precise timing of actin assembly during endocytosis.
Keywords: Arp2/3 Complex, actin cytoskeleton, endocytosis, WASp, Nucleation Promoting Factors, endocytic coat factors, endocytic dynamics
INTRODUCTION
Clathrin mediated endocytosis (CME) is a complex process involving a large number of protein factors that collect membrane cargo, provide directional force to invaginate a vesicle, pinch it off, and ultimately move the vesicle into the cytoplasm. This process is highly conserved throughout evolution, and its efficiency depends on the cooperative interactions of many low affinity protein-protein and protein-lipid binding domains and highly tuned regulatory mechanisms [1]. Actin polymerization has emerged as critical for providing force during endocytosis [2-4]. This was first uncovered in yeast, where sites of endocytosis are marked by characteristic cortical actin patches [5, 6], and later extended to CME in animal cells [7, 8]. However, the underlying mechanisms that coordinate the precisely timed actin polymerization during endocytosis are still not fully understood.
Budding yeast has been an ideal system in which to dissect the specific roles and complex spatiotemporal relationships of proteins involved in endocytosis and actin assembly. This is largely because of the advanced molecular genetic approaches that can be combined with biochemical amenability and recent advances in imaging of endocytic progression using multi-color live-cell fluorescence microscopy. These studies have revealed discrete stages of vesicle development and a characteristic assembly/disassembly pathway of individual endocytic factors [9, 10]. Initially there is a prolonged immobile phase of 1-2 minutes where coat module components collect membrane cargo into an incipient vesicle bud site. Factors such as clathrin and Ede1, an Eps15 homology (EH) domain protein, arrive early during this ordered phase [10-12]. Sla2, related to mammalian Hip1/R, appears fairly late in the immobile stage and is the first actin binding protein seen at the patch [9, 11]. Finally, late immobile phase factors assemble ~15-20 seconds prior to actin assembly, which include a SH3 domain-containing protein (Sla1), two EH domain proteins (Pan1, End3), yeast Epsins (Ent1/Ent2) and Las17, the yeast WASp homologue [9, 10, 13]. Las17 and Pan1 are Arp2/3 activators [9, 10, 14], and Sla1 serves as both a cargo adaptor [15] and an inhibitor of Las17 activity [16]. It is hypothesized that Las17, possibly together with Pan1, primes actin assembly for membrane invagination. The fast mobile stage of endocytosis is marked by the arrival of Abp1, Arp2/3 complex, type 1 myosins (Myo3/5) that further activate Arp2/3 complex, and a host of proteins that decorate actin filaments [9, 10, 17-22]. Actin-mediated invagination takes only ~10-15 sec, at which time vesicle scission occurs, facilitated by Rvs161/167 (amphiphysin homologues) [9, 10]. The coat module factors move inward about 200 nm with the invaginating membrane bud (e.g. clathrin, Sla1, Sla2, Pan1, End3, Ent1/2) and then, immediately after or around the time of scission, disassemble, while the released vesicle moves rapidly into the cell, along actin cables [9, 10, 12, 23]. Some studies suggest that Las17 and type 1 myosins dissipate from the surface rather than internalizing with the nascent vesicle [9, 19, 24], suggesting that actin assembly takes place at the cell surface. However, other studies suggest these proteins may, in fact, move inward as the endocytic bud neck elongates but dissipate upon vesicle scission [18, 25].
The Arp2/3 complex is the primary actin nucleator controlling the polymerization of actin filaments at yeast endocytic sites [26]. It promotes formation of a densely branched actin network, which provides the tensile strength needed for invagination and polarized movement. Alone, purified wild type yeast Arp2/3 complex fails to nucleate actin assembly efficiently [27]. However, yeast express five different nucleation-promoting factors (NPFs) that, when combined with Arp2/3 complex, stimulate actin nucleation: Las17/WASp, Abp1, Pan1, and the type 1 myosins Myo3 and Myo5 [14, 17, 20, 21, 27, 28]. The potency of their effects on Arp2/3 complex ranges greatly, with Las17/WASp appearing to be the strongest [16, 24]. Little is known about how the activities of these NPFs on Arp2/3 complex are spatially or temporally controlled. Since some of the NPFs arrive at sites of endocytosis well before Arp2/3 complex-mediated actin assembly occurs, presumably they must be negatively regulated until the appropriate stage of patch development. While two Las17/WASp inhibitors, Bbc1 and Sla1, have been identified in yeast [16], the potency of their activities has suggested that other factors may be involved in this negative regulation. Furthermore, little is known about how the other NPFs are down regulated, and to date, only one direct negative regulator of yeast Arp2/3 complex has been identified, Crn1/coronin [29], which arrives much later than most NPFs [18]. To obtain a full understanding of yeast Arp2/3 complex regulation, and therefore better elucidate the rules governing development of endocytic sites, it will be necessary to identify and characterize the full cast of regulators.
One set of genetic tools that has proven to be highly useful in dissecting yeast Arp2/3 complex function and regulation is a collection of seven arp2 temperature sensitive alleles that were generated through random mutagenesis [30, 31]. While all of the alleles show defects in actin organization and restricted cell growth at elevated temperatures, differences in their genetic interactions and biochemical activities suggest that they may uncouple different regulatory inputs controlling Arp2/3 complex [32]. Purification of the mutant Arp2/3 complexes revealed that some of the alleles (e.g. arp2-1 and arp2-2) are actin nucleation-impaired, while others (e.g. arp2-7) show unregulated (or ‘leaky’) nucleation in the absence of NPFs [32]. Previously, a dosage suppression screen of arp2-1 temperature sensitive growth identified the strong yeast NPF, LAS17/WASp [32, 33]. In this study, we asked conversely whether a dosage suppression screen of arp2-7 might identify negative regulators of Arp2/3 complex. We identify SYP1 (suppressor of yeast profilin), which encodes a FCH01/F-BAR-related protein that inhibits Las17/WASp activation of Arp2/3 complex-mediated actin assembly and localizes to cortical sites of endocytosis.
RESULTS
arp2 mutants display altered endocytic cortical patch dynamics
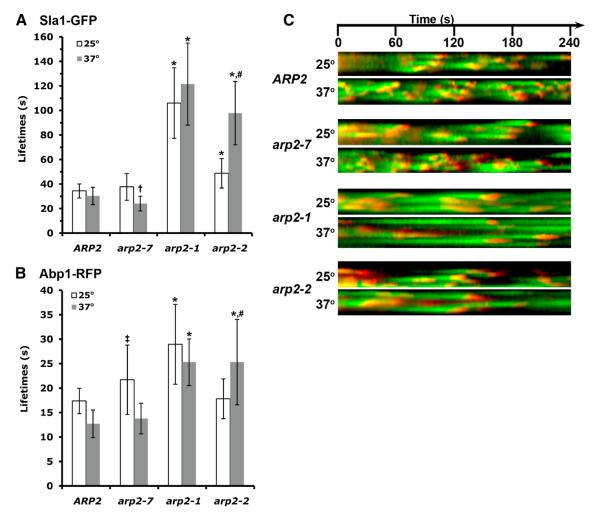
Previous reports characterizing seven temperature sensitive alleles of the Arp2 subunit of Arp2/3 complex suggested that arp2-1 and arp2-2 are loss of function (hypomorphic) mutations, while arp2-7 is unique, behaving as a hypermorphic mutation [30-33]. Each of these alleles leads to defects in actin organization [30-33]; however, their patterns of genetic interactions are distinct [32, 33]. The arp2-1 and arp2-2 mutations are synthetically lethal when combined with las17Δ or myo3Δ myo5Δ, and their temperature sensitivity is suppressed by overexpression of LAS17, indicating a dependence on elevated NPF activity. In contrast, arp2-7 demonstrates no additional defects when combined with these NPF null mutations, and overexpression of LAS17 fails to rescue this mutation [30-33]. Furthermore in vitro assays using purified mutant Arp2/3 complexes show that the arp2-7 mutation causes increased/unregulated actin assembly in the absence of NPFs, whereas arp2-1 or arp2-2 cause impaired actin nucleation [32]. To better understand how the biochemical properties caused by these different arp2 mutations translate into in vivo effects on endocytic vesicle formation, we used live cell imaging to examine the spatiotemporal patterns of Sla1-GFP (a component of the endocytic coat) and Abp1-RFP (a later-arriving actin binding protein that marks the mobile actin phase of internalization).
In wild type cells, Sla1-GFP persists at cortical patches for about 30-40 sec. After an immobile phase of 15-20 sec, actin assembly initiates and invagination begins. About 10 sec later, vesicle scission occurs and Sla1 and other coat factors rapidly disassemble, while the vesicle with associated actin is propelled into the cell away from the surface. At 25°C, the immobile phase, as assessed by the lifetime of Sla1-GFP, was prolonged significantly in arp2-1 and arp2-2 mutants, while arp2-7 was similar to wild type yeast (Figures 1A and 1C). Of the two affected alleles, arp2-1 caused the longest Sla1 lifetime (106 ± 29 sec, compared to 34 ± 6 sec for ARP2, p ≤ 0.0001), although the arp2-2 was also significantly prolonged (49 ± 12 sec, p ≤ 0.0001, compared with ARP2). After shift to 37°C for 15 minutes, the arp2-2 phenotype was further exacerbated, with Sla1-GFP showing an average lifetime of 98 seconds. In contrast, the Sla1-GFP lifetime was accelerated at the elevated temperature in both wild type and arp2-7, but it was significantly more rapid in arp2-7 than ARP2 cells (24 ± 6 sec for arp2-7 vs. 30 ± 7 sec for ARP2, p ≤ 0.0003) (Figures 1B and 1C; Supplemental Figure S1). The accelerated lifetime in arp2-7 can be seen clearly in kymographs (Figure 1C), which show many Sla1-GFP patches with shorter duration.
Figure 1. Endocytic dynamics in arp2 mutant cells.
(A) Patch lifetimes ± standard deviation of Sla1-GFP at 25°C (white) or 37°C (gray) for ARP2 (SL5770), arp2-7 (SL5775), arp2-1 (SL5792), and arp2-2 (SL5780), respectively. * indicates p ≤ 0.0001 compared with ARP2 at comparable temperature, † indicates p ≤ 0.0003 compared with ARP2 at comparable temperature, # indicates p ≤ 0.0001 compared to same strain at 25 °C. (B) Patch lifetimes ± standard deviation of Abp1-RFP at 25°C (white) or 37°C (gray) for ARP2, arp2-7, arp2-1, and arp2-2, respectively. * Indicates p ≤ 0.0001 compared with ARP2, ‡ indicates p ≤ 0.002 compared with ARP2 at comparable temperature, # indicates p ≤ 0.0001 compared to same strain at 25 °C. (C) Kymographs of Sla1-GFP and Abp1-RFP at either 25°C (upper) or 37°C (lower) illustrate the lifetimes and overlap of these factors over 240 sec. Time lapse movies were acquired with 2 sec intervals between frames, and using 250 msec exposures for both Sla1-GFP and Abp1-RFP.
The mobile actin assembly phase of endocytosis was also prolonged in the arp2-1 and arp2-2 mutants (Figures 1B and 1C). At 25°C Abp1-RFP timing in arp2-1 was nearly doubled compared to wild type (29 ± 8 sec vs. 17 ± 3 sec, p ≤ 0.0001). A similar delay in arp2-2 was seen, but only at 37°C (25 ± 9 sec, p< 0.0001 compared to ARP2). Abp1 timing was slightly delayed in arp2-7 at 25°C (22 ± 7 sec; p ≤ 0.002 vs. ARP2). However, at 37°C wild type (13 ± 3 sec) and arp2-7 (14 ± 3 sec) Abp1-RFP lifetimes were similar, and accelerated compared to 25°C, which likely represents a temperature effect. No aberrant cortical patch structures or actin comet tails were seen in any of these arp2 mutants. The number of Sla1-containing patches per cell was increased in arp2-1 and arp2-2, consistent with the long delay in the immobile phase; however patch numbers seemed unaffected in the arp2-7 mutant (not shown). Therefore, the pronounced elongation of Sla1 and Abp1 lifetimes in arp2-1 and arp2-2 reflects their hypomorphic properties; while the accelerated lifetime of Sla1 in arp2-7 at 37°C is consistent with its hypermorphic character, corroborating the prior genetic and biochemical analyses [32, 33].
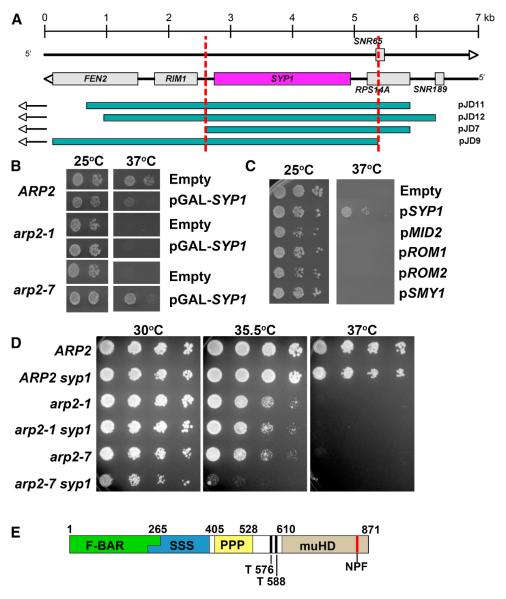
A genetic screen for high copy suppressors of arp2-7
To identify negative regulators of Arp2/3 complex, we screened for dosage suppressors of the temperature sensitive, hyperactive arp2-7 allele. A high-copy plasmid (2μ) S. cerevisiae genomic DNA library was transformed into the arp2-7 strain (BGY142). Four plasmids that suppressed temperature sensitivity (pJD7, pJD9, pJD11 and pJD12) contained overlapping regions of the right arm of chromosome III. One intact ORF was common to these four plasmids, SYP1 (Figure 2A) [34]. To confirm the suppression was by SYP1, we constructed a plasmid with SYP1 under the control of the GAL1 promoter and found that galactose-inducible overexpression of the SYP1 open reading frame could suppress the conditional growth defects of arp2-7 (Figure 2B). Moreover, SYP1 suppression was specific to arp2-7, as it failed to suppress the temperature sensitive growth defects of other arp2 alleles such as arp2-1 (Figure 2B) and arp2-2 (not shown). Noteworthy is that GAL1 driven overexpression of SYP1 negatively affected the growth of wild type yeast at 37°C.
Figure 2. Identification of SYP1 as a dosage suppressor of arp2-7.
(A) Schematic of S. cerevisiae genomic inserts contained in library plasmids pJD11, pJD12, pJD7 and pJD9, which were isolated as high copy suppressors of arp2-7 temperature sensitive growth. (B) ARP2 (BGY134), arp2-1 (BGY136), and arp2-7 (BGY142) strains transformed with empty vector (pBG006) or pGAL1-SYP1 (pBG287) were 10-fold serially diluted, plated on selective galactose-containing medium, and grown at 25 and 37°C. (C) Previously identified high copy suppressors of pfy1Δ [34] were transformed into the arp2-7 (BGY142) strain (BGY142). Transformants were 10-fold serially diluted, plated on selective medium, and grown at 25°C and 37°C. (D) ARP2, arp2-1 and arp2-7 strains containing SYP1 or syp1Δ were 5-fold serially diluted and grown at 30°C, 35.5°C or 37°C for 48 hours. (E) Schematic of Syp1 demonstrating regions of homology to F-BAR (Green) or AP μ (Brown) domains. Also highlighted are serine (Blue) and proline (Yellow) rich regions, possible Ark/Prk phosphorylation sites (T576, T588) and an NPF motif (Orange).
SYP1 was originally identified, along with four other genes (MID2, ROM1, ROM2 and SMY1), in a genetic screen for dosage suppressors of a yeast profilin null mutant [34]. Interestingly, all of the suppressors restored the defects in actin patch polarization caused by pfy1Δ, but not the loss of actin cables. However, of these five genes that suppress pfy1Δ, only SYP1 could suppress arp2-7 (Figure 2C), which stresses the specificity of the genetic interaction.
Deletion of SYP1 alone causes no discernable alterations in cell growth, morphology, or cytoskeletal organization [34]. However, when combined with arp2-7, syp1Δ exacerbated the temperature-sensitive phenotype (Figure 2D). This result is consistent with our observation above that elevated SYP1 expression suppresses arp2-7. Further, in accordance with the suppression data, the syp1Δ synthetic growth defect was only seen in arp2-7, not arp2-1 (Figure 2D).
Syp1 is an early component of cortical endocytic/actin patches
By structural modeling in silico using PHYRE [35], we found that Syp1 contains an N-terminal F-BAR domain (aa 1-265) and a C-terminal μ homology region (aa 609-870) similar to the tyrosine sorting motif binding region of the clathrin heterotetrameric adaptor medium chain μ subunits (Figure 2E) [36]. This has recently been corroborated by X-ray crystallographic analysis [36]. F-BAR domains are thought to sense and/or induce membrane curvature [37]. Additional elements in Syp1 include serine-rich (aa 251-405) and proline-rich (aa 417-528) sequences, two putative Ark1/Prk1 phosphorylation sites (aa T576/T588) [38, 39], and a C-terminal Asp-Pro-Phe (NPF) motif (aa 846-848) that is a potential Eps15 homology (EH) domain binding partner [40-42]. Previous global protein interaction studies have suggested physical interactions between Syp1 and Ede1, an early endocytic coat factor that contains three EH domains, as well as Las17 and Sla1 [43-46].
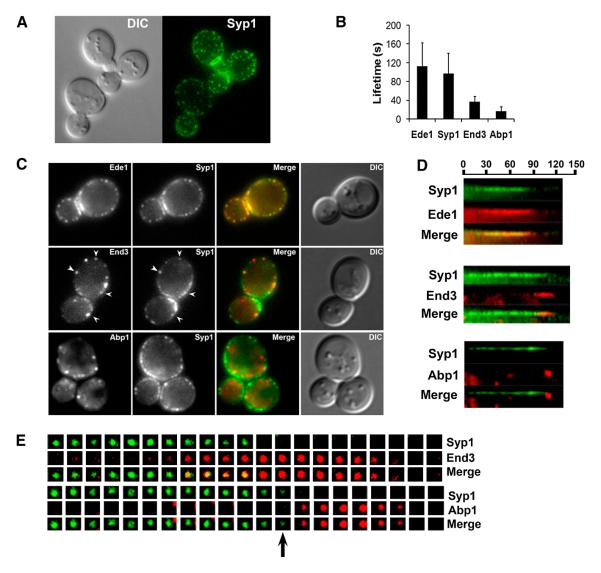
Since we identified SYP1 as a suppressor of arp2-7, we investigated whether Syp1 also localizes to sites of actin assembly and endocytosis by live cell imaging. Previously, Syp1 was shown to interact with septins and localize to the bud neck throughout most of the cell cycle [47]. Also, faint cortical Syp1 was reported by Marcoux and colleagues [34]. With more sensitive imaging, we observed distinct cortical patches of Syp1-GFP, in addition to the bud neck localization (Figure 3A). To determine whether the cortical puncta were sites of endocytosis we co-localized Syp1-GFP with other endocytic markers (Figure 3C). Syp1-GFP did not overlap with Abp1-RFP, a marker of the actin–mediated mobile internalization phase of endocytosis. However, Syp1-GFP closely co-localized with Ede1 (yellow in the merged image), which is one of the earliest known factors to arrive during the immobile phase of endocytosis. We also observed partial co-localization with End3, a later-arriving coat-module factor that appears near the end of the immobile phase along with Sla1 and Pan1 (Figure 3C). Similar to other coat module endocytic factors, Syp1 was still recruited to cortical patches in the presence of Latrunculin A, which prevents actin assembly (Supplemental Figure S2A).
Figure 3. Temporal pattern of Syp1 localization to cortical patches.
(A) Syp1-GFP localization in JDY175. A Z-stack of 0.25 uM sections (600 msec exposures) was deconvolved and projected onto a single plane. (B) Patch lifetimes ± standard deviation of indicated endocytic factors were obtained from 4 min time lapse movies taken at 600 msec exposures, 2 sec between frames. Data are from SL5863 (Ede1-Cherry), SL5806 (Syp1-GFP and Abp1-RFP), SL5862 (End3-Cherry). (C) Localization of Syp1-GFP combined with Ede1-Cherry (SL5863), End3-Cherry (SL5862), or Abp1-RFP (SL5806) in a single cross-section plane. Exposures were 1 sec for GFP and Cherry fusions; 800 msec for Abp1-RFP. Note yellow in the merge shows nearly complete overlap of Ede1 and Syp1, with only occasional overlap for Syp1-GFP and End3-Cherry (arrowheads). There is essentially no overlap between Syp1-GFP and Abp1-RFP. (D) Kymographs from time lapse movies of Syp1-GFP combined with Ede1-Cherry (SL5863), End3-Cherry (SL5862), or Abp1-RFP (SL5806). Exposures were 1 sec for GFP and Cherry fusions; 800 msec for Abp1-RFP, with 3 sec between frames. Note the nearly complete overlap of Syp1-GFP and Ede1-Cherry, whereas End3-Cherry appears near the end of the fluorescent lifetime of Syp1. Syp1-GFP disappears just prior to Abp1-RFP appearance. (E) Tile views showing timing of appearance and disappearance of Syp1-GFP with End3-Cherry (SL5862) or Syp1-GFP with Abp1-RFP (SL5806). Exposures are as indicated in panel D. Beginning frames showing the early phase of Syp1 have been trimmed. Tile views were aligned based on the known timing of End3 and Abp1 [10] The black arrow in the lower tile view indicates the transition point at which Syp1 disappears but Abp1 has not yet appeared.
Time-lapse movies pairing Syp1-GFP with red fluorescent-tagged Ede1, End3 and Abp1 were consistent with the co-localization images in Figure 3C. Syp1 arrived early in the immobile phase and overlapped nearly completely with Ede1; both proteins showed long lifetimes of ~1-2 minutes (Figures 3B and 3D). Syp1’s cortical signal was relatively faint and it flickered on and off during its lifetime (see kymographs panel D). Furthermore, like Ede1, Syp1 did not internalize, and instead disappeared near the onset of the actin/mobile phase (Figures 3D and 3E). End3 arrived around 20 seconds before the end of the lifetime of Syp1-GFP, but as shown previously [10], persisted during invagination with Abp1/actin until vesicle scission and coat disassembly (Figures 3D and 3E).
Although Syp1 is an early endocytic factor, deletion of SYP1 had no detectable effect on the lifetimes of Sla2, Sla1, Las17 and Abp1 (Supplemental Figure S3), consistent with previous findings [10]. As expected for an early endocytic factor, deletion of genes encoding later-arriving endocytic factors had no effect on the cortical recruitment of Syp1-GFP (not shown). Syp1-GFP was efficiently recruited in ede1Δ; however, deletion of clathrin heavy chain or light chain genes reduced the patch localization of Syp1-GFP at the cell surface (Supplemental Figure S2B, not shown). This suggests that clathrin increases the efficiency of recruitment or retention of Syp1 at the cortex. Together these results define Syp1 as an early component of endocytic sites, arriving around the time of Ede1, and disappearing just before the actin mediated invagination phase of endocytosis.
Syp1 over-expression slows endocytic dynamics in arp2-7 mutant cells
Since SYP1 overexpression suppresses the ts growth of arp2-7, we compared endocytic dynamics in wild type and arp2 mutant strains bearing Sla1-GFP and Abp1-RFP markers with or without pSYP1, 2μ (pBG287). At 25°C, SYP1 overexpression caused a slight increase in Sla1-GFP lifetime in ARP2 (41 ± 8 sec vs. 34 ± 6 sec, p ≤ 0.0005, +SYP1 vs. −SYP1 respectively), and SYP1 had no significant effect on the lifetime of Sla1-GFP in arp2-7 or lifetimes of Abp1-RFP in arp2-7 and ARP2 (Supplemental Figure S4). On the other hand, in arp2-1 and arp2-2 strains at 25°C, SYP1 overexpression increased the lifetime of Sla1 from 106 sec to 149 sec and from 49 sec to 67 sec, respectively (p<0.0001, Supplemental Figure S4). Further, Abp1-RFP progression was delayed even further by SYP1 overexpression in arp2-1 (43 ± 14 sec vs. 29 ± 8 sec, p ≤ 0.0001, +SYP1 vs. −SYP1 respectively) and arp2-2 (28 ± 6 sec vs. 18 ± 4 sec, p ≤ 0.0001, +SYP1 vs. −SYP1 respectively) (Supplemental Figure S4).
At 37°C, overexpression of SYP1 in wild type cells caused no detectable shift in either Sla1-GFP or Abp1-RFP dynamics; however, it did attenuate the accelerated Sla1 lifetime of arp2-7 cells (38 ± 12 sec vs. 24 ± 6 sec, p ≤ 0.0001, +SYP1 vs. −SYP1 respectively). In addition, Abp1 timing was delayed at 37 °C when SYP1 was over-expressed in arp2-7, leading to significantly longer lifetimes (22 ± 8 sec vs. 13 ± 3 sec, +SYP1 vs. −SYP1 respectively, p ≤ 0.0001). Overall, the effect of SYP1 overexpression on the arp2 mutants is consistent with its possible role as a negative regulator of Arp2/3 complex-stimulated actin assembly during endocytosis.
Syp1 directly inhibits Las17/WASp activation of Arp2/3 complex
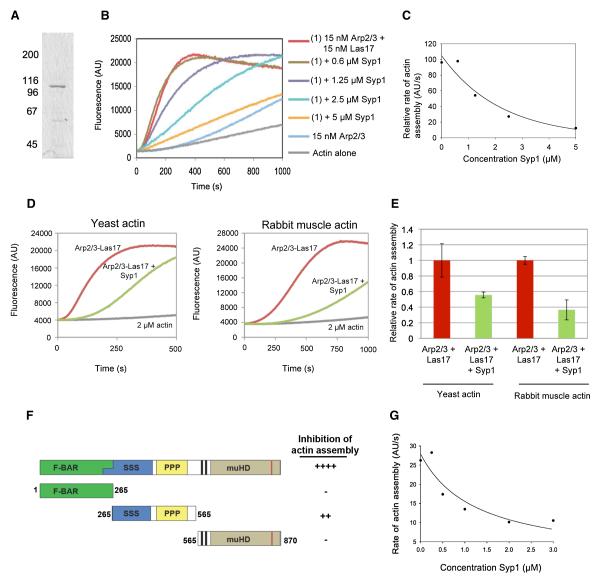
To better understand the mechanistic basis underlying SYP1 suppression of arp2-7, we purified full-length 6His-Syp1 from E. coli and tested its effects on Arp2/3 complex-mediated actin assembly in vitro. Purified 6His-Syp1 migrated on SDS gels with an apparent molecular weight of 105kDa, close to its predicted molecular weight of 96kDa (Figure 5A). In pyrene-actin assembly assays, Syp1 inhibited the actin assembly activity of Las17-Arp2/3 complex in a concentration-dependent manner, with half-maximal effects at 1.25 μM Syp1 (Figures 5B and 5C). The inhibition was specific, as Syp1 did not alter the rate of actin assembly in the absence of Arp2/3 complex (Supplemental Figure S5). Syp1 showed a similar ability to inhibit Las17-Arp2/3 complex activity in assays containing yeast actin and rabbit muscle actin (Figures 5D and 5E).
Figure 5. Purified Syp1 inhibits Las17/WASp-Arp2/3-complex mediated actin assembly.
(A) Coomassie stained gel of full-length 6His-Syp1 purified from E. coli. (B) 3 μM actin (5% pyrene-labeled) was polymerized in the presence of 15 nM yeast Arp2/3 complex, 15 nM yeast Las17/WASp, and a range of concentrations of 6His-Syp1. (C) Concentration-dependent effects of 6His-Syp1 on rate of actin assembly induced by 15nM Las17/WASp and 15nM Arp2/3 complex. (D) 2 μM yeast or rabbit muscle actin (5% pyrene labeled) was assembled alone and in the presence of 10nM Arp2/3 complex and 10 nM Las17/WASp with and without 2.5 μM Syp1. (E) Quantification of data in panel D. Rates of actin assembly for reactions containing yeast actin and rabbit muscle actin and lacking Syp1 were both normalized to 1 for comparative purposes. (F) Schematic of Syp1 fragments that were purified and tested for inhibition of Las17/WASp-Arp2/3 complex activity. Each Syp1 fragment (>1 μM) was tested for effects on 15nM Las17/WASp and 15nM Arp2/3 complex. (G) Concentration-dependent effects of Syp1 (aa 265-565) on rate of actin assembly induced by 15nM Las17/WASp and 15nM Arp2/3 complex.
The inhibitory activity of Syp1 was mapped further using three distinct purified fragments of the protein (Figure 5F). Inhibition was mapped to the central fragment of Syp1, whereas N- and C-terminal fragments of Syp1 lacked any detectable effects on Las17-Arp2/3-mediated actin assembly. Activity of the central fragment of Syp1 was titered (Figure 5G) and found to be somewhat reduced compared to full-length Syp1. The reason for this difference is not yet clear. One possibility is that sequences in the N- and/or C-terminal regions containing the F-BAR and μ2 homology regions, respectively, contribute to inhibition, but alone are not sufficient for inhibition. Another possibility is that Syp1 dimerization via its N-terminal F-BAR domain increases the activity of the central inhibitory region.
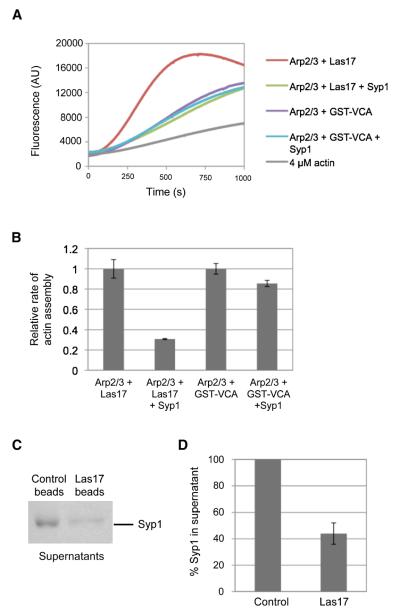
Next, we compared Syp1 inhibitory effects on Arp2/3 complex activated by full-length Las17/WASP and VCA domain of Las17/WASp (Figures 6A and 6B). Syp1 showed strong inhibition of Las17- but not VCA-stimulated Arp2/3 complex activity. This demonstrates that the inhibitory effects of Syp1 depend on sequences in Las17 outside of the VCA domain, emphasizing the specificity of inhibition. This also suggests that Syp1 directly inactivates Las17 rather than Arp2/3 complex. This model is further supported by direct binding interactions between purified Syp1 and Las17-coated beads but not control beads (Figures 6C and 6D), and by the failure of Syp1 to bind Arp2/3 complex (not shown).
Figure 6. Syp1 directly regulates Las17/WASp.
(A) 4 μM actin (5% pyrene-labeled) was polymerized in the presence of 10 nM yeast Arp2/3 complex and 10 nM yeast Las17/WASp or 20 nM yeast Arp2/3 complex and 200 nM yeast GST-VCA in the presence or absence of 2.5 μM Syp1. (B) Quantification of data from A. Rates of actin assembly for reactions containing Las17/WASp and GST-VCA domain but lacking Syp1 were normalized to 1 for comparative purposes. (C) Control beads or beads coated with Las17/WASp were incubated with soluble Syp1, then pelleted, and the supernatants were analyzed by SDS PAGE and Coomassie staining. (D) Quantification of data in C; levels of Syp1 in the supernatants were measured by densitometry.
DISCUSSION
Over ten years ago, a collection of seven ts arp2 mutant alleles was generated, which is still providing powerful tools for dissecting the function of Arp2/3 complex and the role of NPFs in actin assembly [30, 31, 33]. Here, we focused on three arp2 alleles with distinct biochemical activities, genetic interactions, and in vivo properties. Two alleles (arp2-1 and arp2-2) encode mutations that reside at the barbed end of Arp2, where actin polymerization is seeded. Hence, both of these alleles are partially-impaired for actin assembly in vitro [32] and their conditional growth can be rescued in vivo by elevated expression of Las17/WASp [33]. The third allele, arp2-7, carries two mutations, one (F203Y) located at the pointed end of Arp2, and another (F127S) located near the arp2-2 mutation. The first mutation (F203Y) may reverse a repulsive interaction between Arp2 and Arp3 that maintains the Arp2/3 complex in an inactive state, such that this mutation triggers unregulated actin nucleation in the absence of NPFs [32]. Thus arp2-7 behaves as a constitutively active or leaky allele. Consistent with these properties, here we have shown that endocytic dynamics in both arp2-1 and arp2-2 cells are slowed, leading to prolonged lifetimes for both Sla1 and Abp1. In contrast, shorter Sla1 lifetimes were observed in arp2-7 cells at the non-permissive temperature, suggesting an accelerated endocytic progression, which supports the view that the arp2-7 Arp2/3 complex is constitutively active.
We took advantage of the unique properties of arp2-7 to search for negative regulators of the Arp2/3 complex and identified SYP1 as an allele-specific, multi-copy suppressor. Further, syp1Δ showed synthetic growth interactions with arp2-7, but not arp2-1 or arp2-2. Consistent with a previously reported association between Syp1 and Las17/WASp [46], we found that purified Syp1 directly binds Las17/WASp and inhibits its stimulation of Arp2/3 complex-mediated actin assembly. The precise mechanism by which Syp1 inhibits Las17/WASp is not yet clear, but our data suggest that Syp1 binds to sequences in Las17/WASp outside of the VCA domain. As expected for a negative regulator, overexpression of SYP1 in arp2-7 cells attenuated the accelerated lifetimes of endocytic factors Sla1 and Abp1. Moreover, excess SYP1 further slowed the endocytic dynamics in arp2-1 and arp2-2.
Arp2/3 complex-mediated actin assembly during endocytosis must be tightly regulated to enable the endocytic patch to properly mature before the burst of actin assembly that initiates membrane invagination. Further, this assembly must be controlled spatially and temporally to tailor the invagination into a vesicle bud rather than a tubule, and then to promote vesicle scission and propel the vesicle into the cell. In addition to Syp1 studied here, two other endocytic factors, Sla1 and Bbc1, were previously shown to inhibit Las17-Arp2/3 complex activity [16]. Both of these proteins interact directly with Las17 via SH3 domains that bind to the proline-rich region of Las17 [16]. Since Syp1 lacks an SH3 domain, the mechanism of its inhibition is likely to be distinct. In addition, only SYP1, and not SLA1 or BBC1, was isolated as a suppressor of arp2-7. Further, our direct tests showed that neither SLA1 nor BBC1 overexpression suppress arp2-7 (our unpublished data).
Since there was relatively little effect of syp1Δ on cell growth or endocytic dynamics (our results, [10]), it is likely that Syp1 shares genetically redundant functions with Sla1, Bbc1 or other Las17/Arp2/3 complex inhibitors. In fact, the timing of appearance of these factors at cortical sites and the effects of their mutations, suggests that each protein may have unique yet genetically overlapping functions in negatively regulating actin assembly [10]. Bbc1 inhibits Las17 in vitro [16], but also interacts with the type 1 myosins, Myo3/5 [48]. Moreover, it arrives at and dissipates from the cell cortex with Myo3/5, during the 10 sec burst of actin assembly that promotes invagination [10]. In bbc1Δ cells, the speed and distance of inward movement of coat complex components and Abp1/actin are increased, suggesting that Bbc1’s role is to restrain, rather than completely inhibit, the potent NPF activity of myosins and Las17 during the mobile actin phase of endocytosis [10].
In contrast to Bbc1, Sla1 arrives with the later coat factors, around the time Las17 appears at the cortex [9, 10], and thus could negatively regulate Las17 prior to the mobile actin phase. Unlike Las17, Sla1 follows other coat factors once actin assembly occurs, decorating the tips of membrane invaginations and then disassembling shortly after vesicle scission [9, 25]. Possibly its inward movement with the vesicle relieves its inhibition of Las17. Consistent with Sla1 having roles as both a coat factor and as an inhibitor of actin assembly, sla1Δ causes major delays in progression of both the coat phase and the actin phase [9, 10]. Also, Sla1 is a known cargo adaptor, which is consistent with a coat function [15]. The capacity of Sla1 to negatively regulate actin assembly is dramatically unmasked when sla1Δ is combined with bbc1Δ [10]. sla1Δ bbc1Δ cells show exaggerated actin protrusions emanating from the surface, and Las17 is often found at the tips of these assemblies.
In contrast to Sla1 and Bbc1, Syp1 has a completely different temporal pattern of localization during endocytosis. Syp1 appears early, with Ede1, as the endocytic site is forming. Syp1 and Ede1 persist at the cortex until after the arrival of later immobile phase factors (such as End3, Pan1, and Sla1) and then both dissipate just prior to the appearance of the actin marker Abp1. The timing of Syp1 disappearance just prior to actin assembly is highly consistent with Syp1 acting as a negative regulator of Las17/WASp activity.
Like other proteins involved in CME, Syp1 likely has multiple roles during endocytosis. It has a novel modular structure found in a new protein family that includes mammalian FCHO1. These proteins have an N-terminal F-BAR domain and a heterotetrameric adaptor APμ-homology region at their C-termini. The μ-homology region in the Syp1/FCHO1 family is related to the μ chain-C-terminal segment that binds to tyrosine sorting motifs found in membrane cargo collected in clathrin coated pits [49-51]. Similar to Syp1, FCHO1 is also associated with clathrin-coated pits in animal cells [52]. The early arrival of Syp1 at the endocytic patch, diminished localization in the absence of clathrin, and presence of a μ-homology region, suggests that Syp1 could be a cargo specific adaptor [36]. Further studies will be needed to identify its putative cargo, and the specific sorting motif(s) it may recognize.
The F-BAR domain is a cousin of the Bin/amphiphysin/RVS family of BAR domains. These protein modules form crescent-shaped dimers that bind phospholipid bilayers and tubulate liposomes in vitro [53, 54]. They have different degrees of curvature and binding preferences for specific membrane arcs, depending on the BAR domain family. The F-BAR structure has a much shallower curvature (~600 Å) than the traditional amphiphysin-related BAR domains (~200-280 Å) [37, 55, 56]. It is hypothesized these different families of BAR domains bind to and promote membrane bending for different functional purposes, such as driving different stages of endocytosis; however they may also act as sensors of membrane curvature. In addition, many of the proteins in this family regulate localized actin assembly. One such example is Toca-1, a protein with an N-terminal F-BAR domain, which activates Arp2/3 mediated actin polymerization through a C-terminal SH3 domain interaction with WASp [57]. In vitro assays have shown that Toca1 induces actin polymerization on membranes in a curvature-dependent manner, providing maximal induction at the highest diameter curvatures [58]. Although Syp1 is an early endocytic factor, its ability to negatively regulate Las17/WASp activity suggests that Syp1 may also be functioning as a sensor. Assembly of coat module factors and maturation of the endocytic patch could begin to produce membrane curvature, especially in the late stages of the immobile phase. The Syp1 F-BAR domain itself may help induce the shallow membrane invaginations seen by EM at this stage [25]. Reaching a critical degree of curvature during this pre-actin phase might cause Syp1’s F-BAR domain to be released, leading to Syp1 dissociation from the incipient vesicle bud site, thus activating Las17-Arp2/3-complex-mediated actin assembly to promote full vesicle invagination. How this is orchestrated in the context of other endocytic factors is not clear at the moment. Thus, further studies are required to understand the multiple functions of Syp1 as a cargo adaptor, possible membrane curvature-sensor, and inhibitor of Arp2/3 complex activity.
Supplementary Material
Figure 4. Overexpression of SYP1 slows the accelerated endocytic dynamics of arp2-7 cells.

(A) Sla1-GFP lifetimes (± standard deviation) in ARP2 (SL5770) or arp2-7 (SL5775) ± SYP1 2μ shifted to 37°C. * indicates p ≤ 0.0001 comparing ± SYP1 over-expression. (B) ABP1-RFP lifetimes (± standard deviation) shifted to 37°C in the same strains as above. (C) Kymographs of Sla1-GFP and Abp1-RFP at 37°C from strains indicated above. Time lapse movies were taken with 2 sec intervals between frames, and using 250 msec exposures for both Sla1-GFP and Abp1-RFP.
ACKNOWLEDGMENTS
We are grateful for the yeast genomic DNA library provided by Angelica Amon, the arp2 strains from Barbara Winsor, and the pfy1 suppression plasmids from Dominick Pallotta. D.R.B. was supported by an NIH NRSA fellowship (F32-GM084677), and A.R. by an NIH Training Grant (T32-GM07231). This work was funded by NIH grants to B.W. (R01-GM060979), S.K.L. (R01-GM055796), and B.L.G. (R01-GM063691 and R01-GM083137).
Abbreviations
- NPF
Nucleation Promoting Factor
- ts
Temperature Sensitivity
- WASp
Wiskott-Aldrich Syndrome protein
- GFP
Green Fluorescent Protein
- RFP
Red Fluorescent Protein
- CME
Clathrin Mediated Endocytosis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Traub LM. Common principles in clathrin-mediated sorting at the Golgi and the plasma membrane. Biochim Biophys Acta. 2005;1744:415–437. doi: 10.1016/j.bbamcr.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Engqvist-Goldstein AE, Drubin DG. Actin assembly and endocytosis: from yeast to mammals. Annu Rev Cell Dev Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 3.Galletta BJ, Cooper JA. Actin and endocytosis: mechanisms and phylogeny. Curr Opin Cell Biol. 2009;21:20–27. doi: 10.1016/j.ceb.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Girao H, Geli MI, Idrissi FZ. Actin in the endocytic pathway: from yeast to mammals. FEBS Lett. 2008;582:2112–2119. doi: 10.1016/j.febslet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 5.Kubler E, Riezman H. Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J. 1993;12:2855–2862. doi: 10.1002/j.1460-2075.1993.tb05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munn AL. Molecular requirements for the internalisation step of endocytosis: insights from yeast. Biochim Biophys Acta. 2001;1535:236–257. doi: 10.1016/s0925-4439(01)00028-x. [DOI] [PubMed] [Google Scholar]
- 7.Merrifield CJ. Seeing is believing: imaging actin dynamics at single sites of endocytosis. Trends Cell Biol. 2004;14:352–358. doi: 10.1016/j.tcb.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Yarar D, Waterman-Storer CM, Schmid SL. A dynamic actin cytoskeleton functions at multiple stages of clathrin-mediated endocytosis. Mol Biol Cell. 2005;16:964–975. doi: 10.1091/mbc.E04-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 10.Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 11.Newpher TM, Smith RP, Lemmon V, Lemmon SK. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev Cell. 2005;9:87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Toshima JY, Toshima J, Kaksonen M, Martin AC, King DS, Drubin DG. Spatial dynamics of receptor-mediated endocytic trafficking in budding yeast revealed by using fluorescent alpha-factor derivatives. Proc Natl Acad Sci U S A. 2006;103:5793–5798. doi: 10.1073/pnas.0601042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toret CP, Lee L, Sekiya-Kawasaki M, Drubin DG. Multiple pathways regulate endocytic coat disassembly in Saccharomyces cerevisiae for optimal downstream trafficking. Traffic. 2008;9:848–859. doi: 10.1111/j.1600-0854.2008.00726.x. [DOI] [PubMed] [Google Scholar]
- 14.Duncan MC, Cope MJ, Goode BL, Wendland B, Drubin DG. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat Cell Biol. 2001;3:687–690. doi: 10.1038/35083087. [DOI] [PubMed] [Google Scholar]
- 15.Howard JP, Hutton JL, Olson JM, Payne GS. Sla1p serves as the targeting signal recognition factor for NPFX(1,2)D-mediated endocytosis. J Cell Biol. 2002;157:315–326. doi: 10.1083/jcb.200110027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodal AA, Manning AL, Goode BL, Drubin DG. Negative regulation of yeast WASp by two SH3 domain-containing proteins. Curr Biol. 2003;13:1000–1008. doi: 10.1016/s0960-9822(03)00383-x. [DOI] [PubMed] [Google Scholar]
- 17.Evangelista M, Klebl BM, Tong AH, Webb BA, Leeuw T, Leberer E, Whiteway M, Thomas DY, Boone C. A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J Cell Biol. 2000;148:353–362. doi: 10.1083/jcb.148.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galletta BJ, Chuang DY, Cooper JA. Distinct roles for Arp2/3 regulators in actin assembly and endocytosis. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.0060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jonsdottir GA, Li R. Dynamics of yeast Myosin I: evidence for a possible role in scission of endocytic vesicles. Curr Biol. 2004;14:1604–1609. doi: 10.1016/j.cub.2004.08.055. [DOI] [PubMed] [Google Scholar]
- 20.Lechler T, Shevchenko A, Li R. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J Cell Biol. 2000;148:363–373. doi: 10.1083/jcb.148.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winter D, Lechler T, Li R. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr Biol. 1999;9:501–504. doi: 10.1016/s0960-9822(99)80218-8. [DOI] [PubMed] [Google Scholar]
- 22.Barker SL, Lee L, Pierce BD, Maldonado-Baez L, Drubin DG, Wendland B. Interaction of the endocytic scaffold protein Pan1 with the type I myosins contributes to the late stages of endocytosis. Mol Biol Cell. 2007;18:2893–2903. doi: 10.1091/mbc.E07-05-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huckaba TM, Gay AC, Pantalena LF, Yang HC, Pon LA. Live cell imaging of the assembly, disassembly, and actin cable-dependent movement of endosomes and actin patches in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 2004;167:519–530. doi: 10.1083/jcb.200404173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y, Martin AC, Drubin DG. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev Cell. 2006;11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Idrissi FZ, Grotsch H, Fernandez-Golbano IM, Presciatto-Baschong C, Riezman H, Geli MI. Distinct acto/myosin-I structures associate with endocytic profiles at the plasma membrane. J Cell Biol. 2008;180:1219–1232. doi: 10.1083/jcb.200708060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goode BL, Rodal AA. Modular complexes that regulate actin assembly in budding yeast. Curr Opin Microbiol. 2001;4:703–712. doi: 10.1016/s1369-5274(01)00272-7. [DOI] [PubMed] [Google Scholar]
- 27.Goode BL, Rodal AA, Barnes G, Drubin DG. Activation of the Arp2/3 complex by the actin filament binding protein Abp1p. J Cell Biol. 2001;153:627–634. doi: 10.1083/jcb.153.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toshima J, Toshima JY, Martin AC, Drubin DG. Phosphoregulation of Arp2/3-dependent actin assembly during receptor-mediated endocytosis. Nat Cell Biol. 2005;7:246–254. doi: 10.1038/ncb1229. [DOI] [PubMed] [Google Scholar]
- 29.Humphries CL, Balcer HI, D’Agostino JL, Winsor B, Drubin DG, Barnes G, Andrews BJ, Goode BL. Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J Cell Biol. 2002;159:993–1004. doi: 10.1083/jcb.200206113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreau V, Galan JM, Devilliers G, Haguenauer-Tsapis R, Winsor B. The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol Biol Cell. 1997;8:1361–1375. doi: 10.1091/mbc.8.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreau V, Madania A, Martin RP, Winson B. The Saccharomyces cerevisiae actin-related protein Arp2 is involved in the actin cytoskeleton. J Cell Biol. 1996;134:117–132. doi: 10.1083/jcb.134.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D’Agostino JL, Goode BL. Dissection of Arp2/3 complex actin nucleation mechanism and distinct roles for its nucleation-promoting factors in Saccharomyces cerevisiae. Genetics. 2005;171:35–47. doi: 10.1534/genetics.105.040634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madania A, Dumoulin P, Grava S, Kitamoto H, Scharer-Brodbeck C, Soulard A, Moreau V, Winsor B. The Saccharomyces cerevisiae homologue of human Wiskott-Aldrich syndrome protein Las17p interacts with the Arp2/3 complex. Mol Biol Cell. 1999;10:3521–3538. doi: 10.1091/mbc.10.10.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcoux N, Cloutier S, Zakrzewska E, Charest PM, Bourbonnais Y, Pallotta D. Suppression of the profilin-deficient phenotype by the RHO2 signaling pathway in Saccharomyces cerevisiae. Genetics. 2000;156:579–592. doi: 10.1093/genetics/156.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 36.Reider A, Barker SL, Mishra SK, Im YJ, Maldonado-Baez L, Hurley JH, Traub LM, Wendland B. Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J. 2009 doi: 10.1038/emboj.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 38.Huang B, Zeng G, Ng AY, Cai M. Identification of novel recognition motifs and regulatory targets for the yeast actin-regulating kinase Prk1p. Mol Biol Cell. 2003;14:4871–4884. doi: 10.1091/mbc.E03-06-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henry KR, D’Hondt K, Chang JS, Nix DA, Cope MJ, Chan CS, Drubin DG, Lemmon SK. The actin-regulating kinase Prk1p negatively regulates Scd5p, a suppressor of clathrin deficiency, in actin organization and endocytosis. Curr Biol. 2003;13:1564–1569. doi: 10.1016/s0960-9822(03)00579-7. [DOI] [PubMed] [Google Scholar]
- 40.Miliaras NB, Wendland B. EH proteins: multivalent regulators of endocytosis (and other pathways) Cell Biochem Biophys. 2004;41:295–318. doi: 10.1385/CBB:41:2:295. [DOI] [PubMed] [Google Scholar]
- 41.Confalonieri S, Di Fiore PP. The Eps15 homology (EH) domain. FEBS Lett. 2002;513:24–29. doi: 10.1016/s0014-5793(01)03241-0. [DOI] [PubMed] [Google Scholar]
- 42.de Beer T, Hoofnagle AN, Enmon JL, Bowers RC, Yamabhai M, Kay BK, Overduin M. Molecular mechanism of NPF recognition by EH domains. Nat Struct Biol. 2000;7:1018–1022. doi: 10.1038/80924. [DOI] [PubMed] [Google Scholar]
- 43.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 44.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 45.Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Tarassov K, Messier V, Landry CR, Radinovic S, Molina M.M. Serna, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 47.Qiu W, Neo SP, Yu X, Cai M. A novel septin-associated protein, Syp1p, is required for normal cell cycle-dependent septin cytoskeleton dynamics in yeast. Genetics. 2008;180:1445–1457. doi: 10.1534/genetics.108.091900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mochida J, Yamamoto T, Fujimura-Kamada K, Tanaka K. The novel adaptor protein, Mti1p, and Vrp1p, a homolog of Wiskott-Aldrich syndrome protein-interacting protein (WIP), may antagonistically regulate type I myosins in Saccharomyces cerevisiae. Genetics. 2002;160:923–934. doi: 10.1093/genetics/160.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–535. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- 50.Nesterov A, Carter RE, Sorkina T, Gill GN, Sorkin A. Inhibition of the receptor-binding function of clathrin adaptor protein AP-2 by dominant-negative mutant mu2 subunit and its effects on endocytosis. EMBO J. 1999;18:2489–2499. doi: 10.1093/emboj/18.9.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Owen DJ, Evans PR. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science. 1998;282:1327–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakaushi S, Inoue K, Zushi H, Senda-Murata K, Fukada T, Oka S, Sugimoto K. Dynamic behavior of FCHO1 revealed by live-cell imaging microscopy: its possible involvement in clathrin-coated vesicle formation. Biosci Biotechnol Biochem. 2007;71:1764–1768. doi: 10.1271/bbb.60720. [DOI] [PubMed] [Google Scholar]
- 53.Razzaq A, Robinson IM, McMahon HT, Skepper JN, Su Y, Zelhof AC, Jackson AP, Gay NJ, O’Kane CJ. Amphiphysin is necessary for organization of the excitation-contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila. Genes Dev. 2001;15:2967–2979. doi: 10.1101/gad.207801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takei K, Slepnev VI, Haucke V, De Camilli P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol. 1999;1:33–39. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- 55.Henne WM, Kent HM, Ford MG, Hegde BG, Daumke O, Butler PJ, Mittal R, Langen R, Evans PR, McMahon HT. Structure and analysis of FCHo2 F-BAR domain: a dimerizing and membrane recruitment module that effects membrane curvature. Structure. 2007;15:839–852. doi: 10.1016/j.str.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen L, et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 57.Ho HY, Rohatgi R, Lebensohn AM, Le M, Li J, Gygi SP, Kirschner MW. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell. 2004;118:203–216. doi: 10.1016/j.cell.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 58.Takano K, Toyooka K, Suetsugu S. EFC/F-BAR proteins and the N-WASP-WIP complex induce membrane curvature-dependent actin polymerization. EMBO J. 2008;27:2817–2828. doi: 10.1038/emboj.2008.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.