Abstract
PagR is a transcriptional repressor in Bacillus anthracis that controls the chromosomal S-layer genes eag and sap, and downregulates the protective antigen pagA gene by direct binding to their promoter regions. The PagR protein sequence is similar to those of members of the ArsR repressor family involved in the repression of arsenate-resistance genes in numerous bacteria. The crystal structure of PagR was solved using multi-wavelength anomalous diffraction (MAD) techniques and was refined with 1.8 Å resolution diffraction data. The PagR molecules form dimers, as observed in all SmtB/ArsR repressor family proteins. In the crystal lattice four PagR dimers pack together to form an inactive octamer. Model-building studies suggest that the dimer binds to a DNA duplex with a bend of around 4 °.
INTRODUCTION
Bacillus anthracis is a Gram-positive bacterium that primarily infects herbivores such as cattle and sheep but can also infect humans, causing the fatal disease anthrax. B. anthracis belongs to the Firmicutes and is genetically very closely related to some Bacillus cereus strains (Helgason et al., 2000). A major difference of B. anthracis from B. cereus is that it harbours two virulence plasmids, pXO1 and pXO2. The pXO2 plasmid codes for proteins responsible for capsule formation. The pXO1 plasmid encodes the structural genes required for the production of toxins: pagA, which codes for protective antigen PA, cya, which codes for edema factor (EF), and lef, which codes for lethal factor (LF). The pag operon consists of two genes, pagA and pagR, the latter being a weak autogenous repressor co-transcribed with pagA (Hoffmaster & Koehler, 1999).
Sequence comparisons suggest that PagR is closely related to the SmtB/ArsR family of metal-binding repressors involved in the repression of metal-resistance genes (Busenlehner et al., 2003; Xu et al., 1996). The best-characterized members of this family are SmtB from Synechococcus (Eicken et al., 2003), and CzrA (Eicken et al., 2003) and CadC (Ye et al., 2005) from Staphylococcus aureus. They bind to the DNA either at the promoter region or in the region upstream of the promoter, inhibiting transcription. The binding sites are generally imperfect inverted repeats of 12 bases separated by two bases. In general, these repressors regulate the expression of genes involved in metal resistance, such as those encoding efflux pumps.
All members of the SmtB/ArsR repressor family have helix–turn–helix motifs for DNA binding. The algorithm of Dodd & Egan (1990) predicted a helix–turn–helix motif for the PagR sequence between amino acid residues 42 and 64. PagR probably represses transcription by binding directly to the promoter regions of the controlled gene. DNase I footprinting experiments showed that PagR protects the pag promoter region from nucleotide −52 to nucleotide −3 with respect to the transcription start site of the P2 promoter (Mignot et al., 2003). The regions of the sap promoter protected by PagR extend from nucleotide −63 to nucleotide −15 with respect to the transcription start site of sap. Further, PagR protects the eag promoter region from nucleotide +45 to nucleotide +94 with respect to the σH-dependent eag promoter (Mignot et al., 2002). The PagR-protected region in pagAR and the sap promoter overlaps with the σA recognition elements at the promoter −35 and −10 sites. These results suggest that PagR represses the pagAR and sap genes by preventing a σA-complexed RNA polymerase from binding to the sap and pag promoters (Hoffmaster & Koehler, 1999; Mignot et al., 2003). It was reported earlier that deletion of PagR increases the expression level of AtxA (Hoffmaster & Koehler, 1999), implying that PagR may also bind to the promoter region of atxA.
The role of PagR in virulence in animals is untested but there is little doubt of its role in the regulation of S-layer proteins. The S-layer proteins are required for the anchoring of the virulence-plasmid-encoded BslA, which is essential for B. anthracis to breach the blood–brain barrier (Kern & Schneewind, 2008). An examination of the sequences of the PagR-protected regions of the pag, sap and eag promoters revealed no specific DNA sequences for PagR binding. To gain a better understanding of PagR function, we crystallized the PagR protein and solved its crystal structure at a resolution of 1.8 Å.
METHODS
Cloning, expression and purification of PagR.
The pagR gene was PCR amplified from chromosomal DNA of B. anthracis (34F2 strain) using oligonucleotides BaPagR5′BspH (5′-AGAATTATCATGACAGTATTTGTAGATC-3′) and BaPagR3′Bam (5′-CGCGGATCCCTATTGGATAGGGTTTAAC-3′). After digestion of PCR-amplified DNA with BspHI and BamHI, it was ligated with plasmid pET28a (Novagen), generating an ORF of the gene that codes for the PagR protein without a histidine tag. PagR was expressed in Escherichia coli BL21(DE3) grown at 30 °C in LB medium. Selenomethionine-containing PagR was expressed in the methionine-auxotrophic strain E. coli B834(DE3). The cells were grown in defined medium containing 40 mg seleno-dl-methionine ml−1. The protein purification was carried out in 50 mM HEPES, 50 mM NaCl, pH 8.0 buffer (buffer A), using a two-step chromatographic purification procedure. After sonication and centrifugation, the supernatant was loaded on a cation-exchange column that was washed with a buffer A and eluted with buffer B (buffer A containing an additional 1 M NaCl). The fractions including PagR were pooled, desalted and further loaded onto a heparin column, which was again washed with buffer A and eluted with buffer B. Purified PagR was stored in buffer containing 5 mM Bistris, 50 mM NaCl, pH 6.5. The protein was concentrated using a Centriprep YM-3 concentrator to 6–10 mg ml−1, and DTT was added to a final concentration of 5 mM. The incorporation of selenomethionine was confirmed by MALDI (data not shown).
Protein crystallization.
PagR crystals were obtained at 22 °C by the hanging-drop method using a reservoir solution containing 0.2 M sodium acetate, 1.6 M NaH2PO4 and 0.4 M K2HPO4. Crystals grew in 2 weeks to a maximal size of 0.08×0.08×0.08 mm in a tetragonal space group P41212, with unit cell dimensions of a=73.01 Å, c=151.12 Å. The volume of the unit cell suggested that there were four PagR molecules in the asymmetrical unit with a solvent content of 48 %. Crystals of PagR containing selenomethionine grew under similar conditions, but the crystal size improved to 0.2×0.2×0.2 mm after 2 weeks. The crystals were flash-frozen after transferring them into a cryosolution containing 20 % ethylene glycol.
Data collection.
Multi-wavelength anomalous diffraction (MAD) data were collected at the Advanced Light Source (ALS, Berkeley, CA) on beamline 8.3.1 at wavelengths corresponding to the peak (λ1) and low-energy remote (λ2) of a selenium MAD experiment. A native dataset (λ0) to 2.4 Å was also collected on beamline 8.3.1. The datasets were collected at 100 K using ADSC CCD detectors. The MAD data were integrated and reduced using mosflm and then scaled with scala from the ccp4 program (Leslie, 2006). The native data were integrated and reduced using Denzo and then scaled with the program Scalepack (Otwinowski & Minor, 1997). Data statistics are summarized in Table 1.
Table 1.
Crystallographic data
| SeMet λ1 peak inflection | SeMet λ2 remote | Native λ0 | |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 0.97969 | 1.00344 | 0.98012 |
| Resolution (Å) | 1.90 (1.90–1.97) | 1.75 (1.75–1.81) | 2.40 (2.40–2.49) |
| No. of total reflections | 23 1301 | 24 7423 | 121 266 |
| No. of unique reflections | 32 272 | 36 711 | 17 132 |
| Completeness (%) | 97.0 (82.2) | 86.8 (48.9) | 99.1 (99.3) |
| Rmerge (%) | 6.7 (31.0) | 3.7 (43.3) | 7.6 (39.7) |
| I/σ(I) | 35.4 (5.6) | 36.0 (2.4) | 18.8 (4.1) |
| Phasing statistics | |||
| Figure of merit | 0.49 | ||
| Refinement statistics | |||
| Model composition | 341 residues, 79 H2O | ||
| Resolution limits (Å) | 2.0 | ||
| Reflection in working/test sets | 92.4 %/4.7 % | ||
| R/Rfree (%) | 24.5 %/26.7 % | ||
| Bond (Å) angle (°) rms | 0.005 Å/1. ° | ||
| Ramachandran plot | |||
| Most favoured regions | 96 % | ||
| Additional allowed regions | 4 % |
Structure solution and refinement.
PagR is a 99-residue protein containing five methionines. The crystals of selenomethionine-incorporated protein diffracted to a resolution of 1.8 Å. Structure solution was carried out by multi-wavelength anomalous dispersion (MAD) techniques. The initial phase were determined with the 1.8 Å selenium MAD data (λ1, λ2) using the program solve (Terwilliger, 2003). Automatic model building was performed with resolve (Terwilliger, 2003) and the model was completed using the program O (Jones et al., 1991); refinement of the coordinates was carried out using the diffraction data at the wavelength λ2 (Table 1) using program cns1.0 (Brunger et al., 1998) . Refinement statistics are summarized in Table 1. The final model consists of four molecules of PagR and 79 water molecules in the asymmetrical unit. No electron density was observed for residues 1–13 and 97–99 in Mol.A, 1–10 and 97–99 in Mol.B, 1–10 and 97–99 in Mol.C, and 1–12, 73–79 and 99 in Mol.D.
RESULTS AND DISCUSSION
Description of the overall structure
The PagR molecules exist as an octamer in the crystal lattice and the asymmetrical unit contains four molecules (Mol.A, Mol.B, Mol.C and Mol.D). Mol.A is related to Mol.B by non-crystallographic twofold symmetry. Mol.A is additionally related to Mol.D by another non-crystallographic diad axis displaying a different mode of association. Mol.B is related to Mol.C also by a similar non-crystallographic twofold symmetry.
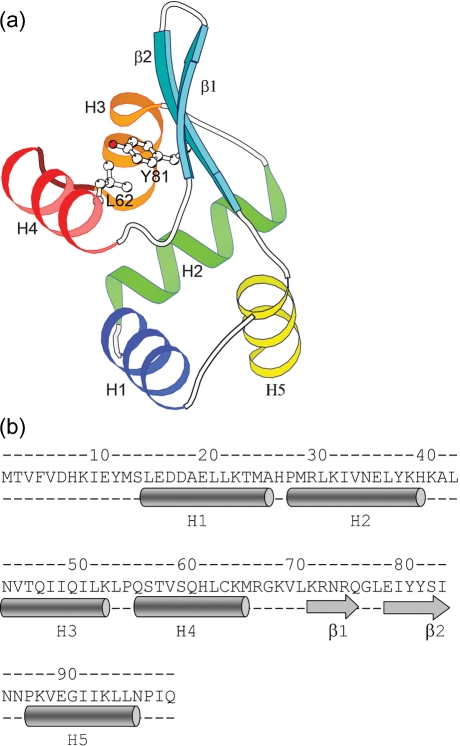
PagR is an α/β protein with the same fold as the members of SmtB/ArsR family of metal-binding repressors. These proteins belong to the class of winged helix–turn–helix (HTH) motif-containing transcription factors. They have a unique topology of five α-helices and a β-hairpin in the following order: ααααββα. The structure of PagR comprises α-helices H1 (residues 14–25), H2 (27–39), H3 (43–52), H4 (55–65) and H5 (86–96) and a two-stranded β sheet with β1 (residues 71–75) and β2 (78–83) (Fig. 1a, b). Two of the helices, H3 and H4, along with the connecting residues form the putative HTH motif seen in many prokaryotic and eukaryotic DNA-binding proteins. The β-strands, β1 and β2, are arranged in an anti-parallel manner to form a hairpin structure that is anchored by hydrophobic and hydrogen-bonding interactions to the rest of the protein. The conserved residues from helices H2 and H3, the termini of the β hairpins and the H5 helix form the hydrophobic core. The classic inverse γ-turn between helices H1 and H2 is a common feature in the SmtB/ArsR family and it is also present in PagR. A GXX motif (where X is an amino acid with a large hydrophobic side chain) seen towards the C-terminal end of the recognition helix in most transcription factors is absent in PagR. The type II′ β-turn in the β-hairpin with the Gly in the second position is also a common feature observed in most members of the SmtB/ArsR family of repressors. The residue Y82, located on strand β2, has been predicted to be a potential tyrosine kinase phosphorylation site (Hoffmaster & Koehler, 1999) and its side chain points towards the recognition helix (Fig. 1a). The residues Y81 and L62 are part of the hydrophobic core between the helical part of the protein and the β-strands (Fig. 1a). The conserved nature of the residues in the interface between the β-strands and helices indicates that these residues are important for the structural integrity of the protein. The above-mentioned structural features in this fold play an important role in understanding the allosteric mechanism of DNA binding in this family of repressors (Eicken et al., 2003). These conserved features in PagR structure suggest that it might have a mechanism similar to that of the members of the SmtB/ArsR family.
Fig. 1.
3D structure and sequence of the PagR momomer. (a) Molecular architecture of PagR. The fold comprises five α-helices and a β-hairpin in the following order: H1 (blue), H2 (green), H3 (orange), H4 (red), β1 (cyan), β2 (cyan), H5 (yellow). Helices H3 and H4 and the connecting residues in between form the HTH motif. The side chains of Y81 and L62 are shown; they form part of the hydrophobic core between the β-hairpin and the α-helices. (b) Sequence and secondary structure of PagR. The α-helices are depicted as cylinders and the β-strands as arrows.
Two types of intermolecular associations
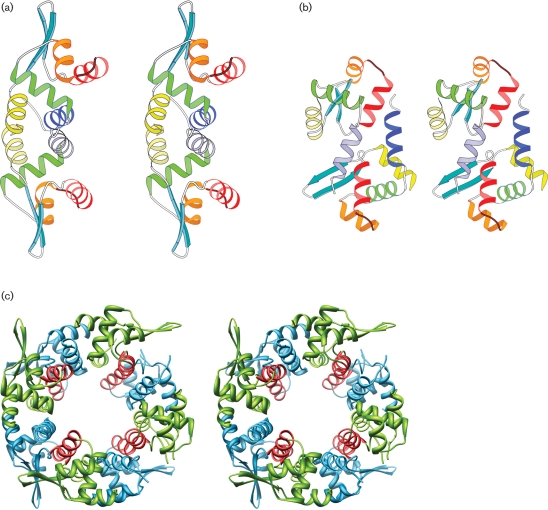
The metal-binding repressor proteins in the SmtB/ArsR family (Cook et al., 1998; Eicken et al., 2003) form elongated dimers, with the recognition helices separated by about 34 Å to interact with the DNA. In the crystal lattice, PagR molecules also form such dimers. Mol.A and Mol.B form a dimer and they are related by a non-crystallographic twofold axis (Fig. 2a), while Mol.C forms a dimer with its symmetry-related partner and so does Mol.D. The association of these two monomers buries a total of 17 % of the surface area of the two molecules at the interface. The core of this binding surface is a hydrophobic region formed by the residues of helices H1 and H5 from each monomer, which run antiparallel to each other. Dimerization of the molecule appears to be critical for its function, as it positions the recognition helices suitably for interactions with DNA.
Fig. 2.
Two modes of association of PagR molecules in the crystal lattice. (a) Stereo view of the dimer formed by Mol.A and Mol.B. They associate to form a biologically relevant dimer, related by a non-crystallographic twofold axis. The recognition helices (red) are in a suitable orientation for interaction with DNA. (b) Stereo view of the dimer formed by Mol.A and Mol.D. The molecules are related by a non-crystallographic diad axis, making the recognition helices inaccessible for DNA interactions. (c) Stereo view of the octamer formed in the crystal lattice. The recognition helices are shown in red.
The crystal lattice displays an additional mode of association (Fig. 2b). Mol.B and Mol.C (similarly Mol.A and Mol.D) are also related by a non-crystallographic twofold axis, and they associate through the interactions of helix H1 (Mol.B) with H4 of (Mol.C) and vice versa. The association buries a total of 9 % of the surface area of two molecules at the interface and it can be visualized as another mode of dimer formation (Fig. 2b). By using these two modes of associations alternately, the PagR molecules form cyclical octamers in the crystal lattice (Fig. 2c). The second mode of association (Mol.B with Mol.C and Mol.A with Mol.D) is stabilized by two salt bridges between each pair of monomers at their interfaces, indicating that the stability of the octamer may be dependent on the ionic strength of the medium.
While there is evidence for the presence of dimers in solution from mass spectroscopy (data not shown), there is no experimental evidence so far for the existence of higher-order oligomers in solution. Hydrophilic interactions play a major role in the second mode of dimer association, and therefore formation of stable tetramers or octamers would appear unlikely. In the crystal lattice, when the molecules pack themselves to form an octamer, the recognition helices are no longer accessible for interaction with DNA (Fig. 2c). Therefore the formation of the octamer could be thought of as an inactivation mechanism.
Comparison of PagR with metal-sensing proteins in the SmtB/ArsR family
In prokaryotes, there are two general classes of transcriptional regulatory proteins with the ability to respond to stress-induced metal toxicity, namely the MerR family and the SmtB/ArsR family (Busenlehner et al., 2003). MerR-like proteins function as repressors in the absence of metal ions, but function as activators upon metal binding. Conversely the SmtB/ArsR family of proteins act exclusively as prokaryotic transcriptional repressors which regulate the expression of genes associated with metal sequestration or efflux in both Gram-positive and Gram-negative bacteria (Busenlehner et al., 2003). PagR is closely related to theSmtB/ArsR family of proteins.
So far, six different crystal structures of the members of SmtB/ArsR family have been reported. They are SmtB in apo form (Cook et al., 1998), SmtB in one and two zinc-bound forms, CzrA in apo and zinc-bound form (Eicken et al., 2003), and CadC in zinc-bound form (Ye et al., 2005). PagR has 28 % sequence identity with CzrA and 21 % with CadC and SmtB. The structure of PagR has close resemblance to the structures of the proteins in this class. There are however two main differences: (i) PagR does not have metal-binding sites, and (ii) the PagR dimer has a more curved architecture compared to most of the molecules in this class.
While PagR does not possess metal-binding sites, certain members of the SmtB/ArsR family have two distinct metal-binding sites (Cook et al., 1998; Busenlehner et al., 2003). One of them is the α3N site formed by Cys residues of the third α-helix H3 along with Cys residues from the N-terminal region and it binds large, softer metal ions such as Cd(II), Pb(II) and Bi(III). The other is the α5 site; it is formed by carboxylate or imidazole side chains contributed by H5 from each monomer and it binds smaller, harder metal ions such as Co(II) and Zn(II) (Busenlehner et al., 2001, 2002). The two α3N sites are located on opposite ends of the dimer while the two α5 sites are at the dimeric interface (Fig. 3). PagR lacks the residues needed to form any of these binding sites.
Fig. 3.
Cartoon showing the two types of metal-binding sites in the SmtB/ArsR family depicted on the CadC structure. The α3N sites are located on opposite ends of the dimer and are formed by thiolate groups (orange circles) from helix H3 in one monomer and the N-terminal region of the other monomer. The α5 sites are located on the dimeric interface and are formed by carboxylate (red circles) and imidazole (blue circles) side chains contributed by the H5 helix from each monomer.
In CadC, only the α3N site has functional relevance. On the other hand, in SmtB and CzrA, the N-terminal site is not well conserved and is not known to be involved in any metal binding, but the C-terminal metal-binding site α5 has a regulatory role. As PagR does not have the metal-binding sites, its function must be regulated by some other as yet unknown means.
B. anthracis Sterne strain has four additional proteins related to the SmtB/ArsR family. Three of these appear to be cotranscribed with metal-associated functions: BAS0563, in an operon with a heavy-metal-transporting ATPase; BAS2974, in an operon with a protein known to reduce arsenate to arsenite, and BAS3305, in an operon with a zinc-requiring alcohol dehydrogenase. The fourth gene, BAS4321, appears to be transcribed alone. All of these maintain the metal-binding site.
The crystallographic data of repressors in the SmtB/ArsR family illustrate that the metal binding influences the conformation of the molecule and provides a basis for understanding the allosteric regulation of operator–promoter binding (Eicken et al., 2003). Metal binding at the dimeric interface causes a rigid body reorientation of one subunit with respect to the other. In SmtB, the metal binding causes a dramatic change, a movement of 2.4 Å for recognition helix in each of the subunits (Eicken et al., 2003). The altered conformation is less suitable for DNA binding, resulting in a drastic drop of the binding affinity and causing the release of repressor from the promoter sites and subsequent expression of metal-regulatory proteins.
Dimer architecture
Comparison of the PagR dimer with other structures of the SmtB/ArsR family shows that PagR has a more curved architecture. Fig. 4 shows the superposition of CzrA with PagR and it indicates that CzrA is more linear while PagR is more curved, making the recognition helices less parallel. It appears that the curvature of PagR and the concomitant deviated orientation of the recognition helices would correspond to a significant bend in the DNA.
Fig. 4.
Superposition of the PagR dimer (red and blue) with the CzrA dimer (grey). The CzrA molecules associate in a linear fashion to form the dimer while PagR molecules associate with a curved architecture.
Putative DNA-binding residues
PagR is the transcriptional repressor that regulates the pagAR operon and the S-layer genes eag and sap in B. anthracis by binding to the promoter regions of these genes (Mignot et al., 2003). As the molecule possesses the HTH motif, characteristic of DNA-binding proteins, it appears that helix H4 would function as the recognition helix and interact with the major groove of DNA.
In order to envision the mode of DNA binding, we examined the geometry of the binding of protein dimers with a twofold symmetry like the lambda Cro protein to palindromic DNA (Anderson et al., 1981). The two recognition helices bind to the major groove of DNA separated by 34 Å, corresponding to the pitch of the DNA helix. While some dimers bind to straight B-form DNA (Beamer & Pabo, 1992), certain others bind to bent forms of DNA (Schultz et al., 1991). For example, NarL binding to DNA causes the DNA to bend by 4 ° (Maris et al., 2002). A comparison of the orientation of the recognition helices in NarL and PagR shows that PagR could have reasonably good interactions with the DNA conformation observed in the NarL–DNA complex. Therefore, we propose that PagR dimer may bind to DNA with a similar bend of around 4 ° (Fig. 5). The side chains of the residues Lys64, Gln60 and Ser56 of the recognition helix are oriented favourably to interact with the DNA major groove. A recent study (Hadjifrangiskou & Koehler, 2008) on the pag operon using in vitro analysis and in silico modelling shows that there is intrinsic curvature associated with anthrax toxin promoter genes, in agreement with our proposed model.
Fig. 5.
DNA binding: a model of PagR dimer binding to DNA with a bend of about 4 °. Mol.A is shown in red and Mol.B in blue. The side chains of the residues Lys64, Gln60 and Ser56 (blue) of the recognition helix point towards the major groove. The residues His26, Arg29 and His61 cluster together (green), indicating possible interactions with the DNA backbone.
Three basic residues, His26, Arg29 and His61, cluster together and are favourably oriented for interactions with the DNA backbone. One of these residues, Arg29, is highly conserved in all the SmtB/ArsR family of proteins (Busenlehner et al., 2003). Modelling of the DNA complex of PagR indicates that Arg29, His26 and His61 could interact with the backbone and major groove of DNA. Sequence alignment (Supplementary Fig. S1, available with the online version of this paper) shows that residues S59, L62 and Y81 are also highly conserved in all the SmtB/ArsR family of proteins. Residue S59 lies on the recognition helix and could have interactions with DNA. As shown in Fig. 1(a), L62 and Y81 lie between the β-hairpin and the α-helices.
DNA footprinting data (Mignot et al., 2003) show that PagR binding protects the region of the DNA from nucleotides −52 to −3, a length representing nearly five helical turns. Additionally, the footprinting data suggest that PagR binds only one operator site at each promoter. The model of DNA–PagR binding (Fig. 5) shows that binding of PagR dimer accounts only for about 2.5 helical turns, suggesting the existence of two tandem binding sites for the promoter.
Conclusions
The crystal structure of PagR reveals a fold similar to that of members of SmtB/ArsR family of repressors and shows that it forms a dimer in a similar manner. In the crystal lattice four dimers are packed together to form an octamer. While the members of the SmtB/ArsR family are regulated by metals, the lack of metal-binding sites in PagR points to other modes of regulation. The structure provides a basis for further studies on the role of environmental and host factors in transcription regulation in B. anthracis.
Acknowledgments
This research was supported, in part, by grants GM54246 and GM19416 from the National Institute of General Medical Sciences, grants AI055860 and AI052289 from National Institutes of Allergy and Infectious Diseases, the National Institutes of Health, USPHS, and the Arkansas Biosciences Institute, the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000. This is paper number 19166 from The Scripps Research Institute. We thank the ALS Synchrotron Source at Berkeley for providing X-ray diffraction facilities.
Abbreviations
MAD, multi-wavelength anomalous diffraction
Footnotes
The atomic coordinates and structure factors for PagR (PDB ID 2ZKZ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org).
A supplementary sequence alignment is available with the online version of this paper.
References
- Anderson, W. F., Ohlendorf, D. H., Takeda, Y. & Matthews, B. W. (1981). Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature 290, 754–758. [DOI] [PubMed] [Google Scholar]
- Beamer, L. J. & Pabo, C. O. (1992). Refined 1.8 Å crystal structure of the lambda repressor-operator complex. J Mol Biol 227, 177–196. [DOI] [PubMed] [Google Scholar]
- Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M. & other authors (1998). Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Busenlehner, L. S., Cosper, N. J., Scott, R. A., Rosen, B. P., Wong, M. D. & Giedroc, D. P. (2001). Spectroscopic properties of the metalloregulatory Cd(II) and Pb(II) sites of S. aureus pI258 CadC. Biochemistry 40, 4426–4436. [DOI] [PubMed] [Google Scholar]
- Busenlehner, L. S., Apuy, J. L. & Giedroc, D. P. (2002). Characterization of a metalloregulatory bismuth(III) site in Staphylococcus aureus pI258 CadC repressor. J Biol Inorg Chem 7, 551–559. [DOI] [PubMed] [Google Scholar]
- Busenlehner, L. S., Pennella, M. A. & Giedroc, D. P. (2003). The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol Rev 27, 131–143. [DOI] [PubMed] [Google Scholar]
- Cook, W. J., Kar, S. R., Taylor, K. B. & Hall, L. M. (1998). Crystal structure of the cyanobacterial metallothionein repressor SmtB: a model for metalloregulatory proteins. J Mol Biol 275, 337–346. [DOI] [PubMed] [Google Scholar]
- Dodd, I. B. & Egan, J. B. (1990). Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res 18, 5019–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicken, C., Pennella, M. A., Chen, X., Koshlap, K. M., VanZile, M. L., Sacchettini, J. C. & Giedroc, D. P. (2003). A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. J Mol Biol 333, 683–695. [DOI] [PubMed] [Google Scholar]
- Hadjifrangiskou, M. & Koehler, T. M. (2008). Intrinsic curvature associated with the coordinately regulated anthrax toxin gene promoters. Microbiology 154, 2501–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason, E., Okstad, O. A., Caugant, D. A., Johansen, H. A., Fouet, A., Mock, M., Hegna, I. & Kolsto, A. B. (2000). Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis – one species on the basis of genetic evidence. Appl Environ Microbiol 66, 2627–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmaster, A. R. & Koehler, T. M. (1999). Autogenous regulation of the Bacillus anthracis pag operon. J Bacteriol 181, 4485–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991). Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kern, J. W. & Schneewind, O. (2008). BslA, a pXO1-encoded adhesin of Bacillus anthracis. Mol Microbiol 68, 504–515. [DOI] [PubMed] [Google Scholar]
- Leslie, A. G. (2006). The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr 62, 48–57. [DOI] [PubMed] [Google Scholar]
- Maris, A. E., Sawaya, M. R., Kaczor-Grzeskowiak, M., Jarvis, M. R., Bearson, S. M., Kopka, M. L., Schroder, I., Gunsalus, R. P. & Dickerson, R. E. (2002). Dimerization allows DNA target site recognition by NarL response regulator. Nat Struct Biol 9, 771–778. [DOI] [PubMed] [Google Scholar]
- Mignot, T., Mesnage, S., Couture-Tosi, E., Mock, M. & Fouet, A. (2002). Developmental switch of S-layer protein synthesis in Bacillus anthracis. Mol Microbiol 43, 1615–1627. [DOI] [PubMed] [Google Scholar]
- Mignot, T., Mock, M. & Fouet, A. (2003). A plasmid-encoded regulator couples the synthesis of toxins and surface structures in Bacillus anthracis. Mol Microbiol 47, 917–927. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Schultz, S. C., Shields, G. C. & Steitz, T. A. (1991). Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science 253, 1001–1007. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T. C. (2003). SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol 374, 22–37. [DOI] [PubMed] [Google Scholar]
- Xu, C., Shi, W. & Rosen, B. P. (1996). The chromosomal arsR gene of Escherichia coli encodes a trans-acting metalloregulatory protein. J Biol Chem 271, 2427–2432. [DOI] [PubMed] [Google Scholar]
- Ye, J., Kandegedara, A., Martin, P. & Rosen, B. P. (2005). Crystal structure of the Staphylococcus aureus pI258 CadC Cd(II)/Pb(II)/Zn(II)-responsive repressor. J Bacteriol 187, 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]