Abstract
Auxin is an important plant hormone that plays significant roles in plant growth and development. Although numerous auxin-response mutants have been identified, auxin signal transduction pathways remain to be fully elucidated. We isolated ibr5 as an Arabidopsis indole-3-butyric acid–response mutant, but it also is less responsive to indole-3-acetic acid, synthetic auxins, auxin transport inhibitors, and the phytohormone abscisic acid. Like certain other auxin-response mutants, ibr5 has a long root and a short hypocotyl when grown in the light. In addition, ibr5 displays aberrant vascular patterning, increased leaf serration, and reduced accumulation of an auxin-inducible reporter. We used positional information to determine that the gene defective in ibr5 encodes an apparent dual-specificity phosphatase. Using immunoblot and promoter-reporter gene analyses, we found that IBR5 is expressed throughout the plant. The identification of IBR5 relatives in other flowering plants suggests that IBR5 function is conserved throughout angiosperms. Our results suggest that IBR5 is a phosphatase that modulates phytohormone signal transduction and support a link between auxin and abscisic acid signaling pathways.
INTRODUCTION
Auxins are an essential class of phytohormones that influence many aspects of plant growth and development. At the molecular level, auxins influence cell division, cell elongation, and cell differentiation (Davies, 1995). At the macroscopic level, auxins direct vascular development, promote apical dominance and lateral root formation, and mediate gravitropism and phototropism (Davies, 1995). Indole-3-acetic acid (IAA) and indole-3-butyric acid (IBA) are two endogenous auxins that can be interconverted (reviewed by Ludwig-Müller, 2000; Bartel et al., 2001).
Many auxin responses are mediated by changes in gene expression, and some of these are controlled by transcription from Auxin-Response Elements (AuxREs). Auxin-Response Factors (ARFs) were isolated initially because they bind AuxREs (Ulmasov et al., 1997a); they can either activate or repress transcription (Ulmasov et al., 1999). Aux/IAA proteins can heterodimerize with ARFs and repress the ability of activating ARFs to promote transcription (Ulmasov et al., 1997b; Tiwari et al., 2001). Increased auxin levels enhance the degradation rate of Aux/IAA proteins via ubiquitin-mediated degradation, allowing activating ARFs to increase the transcription of auxin-responsive genes (reviewed by Rogg and Bartel, 2001; Kepinski and Leyser, 2002).
Genetic analysis of auxin-response mutants in Arabidopsis has led to the identification of proteins necessary for IAA influx and efflux (reviewed by Muday and DeLong, 2001), several ARFs and Aux/IAA proteins (reviewed by Liscum and Reed, 2002), and proteins that mediate and regulate ubiquitin-dependent proteolysis (reviewed by Rogg and Bartel, 2001; Kepinski and Leyser, 2002). Moreover, biochemical experiments have revealed that the association of Aux/IAA proteins with a ubiquitin-protein ligase can be stimulated by auxin (Gray et al., 2001). Although it is likely that this binding leads to Aux/IAA protein degradation in vivo, the molecular signaling cascade by which auxin promotes Aux/IAA protein binding to the ubiquitin protein ligase is not understood, and the regulation of the components downstream of these initial events is only beginning to be elucidated (Xie et al., 2000, 2002).
Abscisic acid (ABA) is another phytohormone that affects many aspects of growth and development, including shoot and root growth, stomatal closure, storage protein synthesis, and seed dormancy (Davies, 1995). Gaps remain in our understanding of ABA signaling, although the transcription factors necessary for ABA sensitivity and the phosphatases that modulate ABA responses have been identified (reviewed by Finkelstein et al., 2002). Like auxin signaling, ABA responses depend on regulated ubiquitin-dependent proteolysis of a key transcription factor (Lopez-Molina et al., 2001, 2003). Analysis of Arabidopsis mutants has revealed interactions between ABA and other signaling pathways, including those for sugar, ethylene, and auxin (reviewed by Fedoroff, 2002; Gazzarrini and McCourt, 2003). Indeed, an emerging theme of phytohormone responses is that many such responses cannot be reduced to simple linear pathways that connect inputs and outputs but may more closely resemble interaction webs (Møller and Chua, 1999; Gazzarrini and McCourt, 2003).
Mutants that respond aberrantly to multiple phytohormones may be used to elucidate the connections between interacting phytohormone-response pathways. Here, we report the characterization of ibr5, an Arabidopsis mutant defective in specific auxin and ABA responses. ibr5 is less sensitive to inhibitory concentrations of exogenous auxins and ABA and appears to be less sensitive to endogenous auxin. IBR5 encodes a 257–amino acid protein with ∼35% identity to known dual-specificity mitogen-activated protein kinase (MAPK) phosphatases. Dual-specificity phosphatases often dephosphorylate signaling components; therefore, IBR5 may modulate auxin and ABA signal transduction pathways.
RESULTS
ibr5 Displays Attenuated Responses to Auxin and ABA
In an attempt to isolate auxin signaling components, we performed a screen for mutants resistant to the inhibitory effects of exogenous auxin on root elongation, focusing on mutants with slightly weaker auxin phenotypes than seen in previously isolated auxin-response mutants. ibr5-1 was isolated from a pool of ethyl methanesulfonate–mutagenized seeds as an IBA-response mutant (Zolman et al., 2000). Analysis of ibr5 after backcrossing revealed general auxin resistance. For example, ibr5 was less sensitive than the wild type to primary root elongation inhibition caused by the natural auxins IAA and IBA (Figure 1A); the synthetic auxins 2,4-D (Figure 1A), 2,4-dichlorophenoxybutyric acid, and 1-naphthaleneacetic acid; and auxin transport inhibitors, including 1-napthylphthalamic acid and 2,3,5-triiodobenzoic acid (data not shown). A strong allele of the auxin-response mutant axr1 (Estelle and Somerville, 1987; Lincoln et al., 1990) generally was more resistant than ibr5 to the root elongation inhibition caused by these auxins (Figure 1A) and by auxin transport inhibitors (data not shown), consistent with the idea that ibr5 is a weak auxin-response mutant. The ibr5 defects were recessive (data not shown), suggesting that the mutant lesion results in a loss of function.
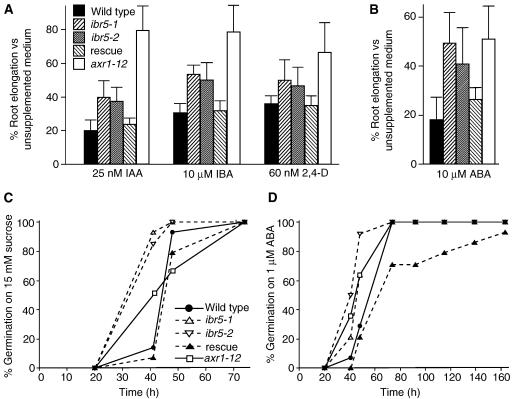
Figure 1.
ibr5 Responses to Phytohormones.
(A) Root elongation on auxin. After 8 days of growth on medium supplemented with the indicated concentrations of IAA, IBA, or 2,4-D, Col-0 (wild type), ibr5-1, ibr5-2, ibr5-1 transformed with pBINIBR5 (rescue), and axr1-12 seedlings were removed from the agar, and the length of the primary root was measured. Results were standardized against growth on unsupplemented medium.
(B) Root elongation on ABA. Seedlings were grown for 4 days on PNS and transferred to medium supplemented with 10 μM ABA for 4 days, after which the entire primary root was measured. Results were standardized against values from plants transferred to unsupplemented medium.
(C) and (D) Germination on medium lacking (C) or containing (D) ABA. Seedlings were scored for radical emergence at the times indicated on medium containing 15 mM sucrose (C) or lacking sucrose but containing 1 μM ABA (D).
Error bars represent standard deviations of the means (n ≥ 12). ibr5-1 was backcrossed four times before analysis; ibr5-2 was backcrossed once (A) or not backcrossed ([B] to [D]).
To examine the specificity of ibr5 auxin-response defects, we examined root elongation inhibition by other phytohormones. ibr5 showed decreased sensitivity to ABA (Figure 1B) but displayed wild-type sensitivity to the ethylene precursor 1-aminocyclopropane-1-carboxylic acid, the cytokinin 6-benzylaminopurine, the brassinosteroid epibrassinolide, the growth hormone gibberellic acid, and the defense hormones methyl jasmonate and salicylic acid (data not shown). Moreover, we observed no differences between ibr5 and the wild type in root elongation inhibition caused by 100 mM NaCl and in stress responses caused by 4% glucose, 0.5% mannitol, or 25% polyethylene glycol (data not shown). Thus, ibr5 response defects appear specific to the phytohormones auxin and ABA.
Because ibr5 mutants display ABA-resistant root elongation and because exogenous and endogenous ABA inhibit germination (Finkelstein et al., 2002), we also examined ibr5 germination kinetics with and without supplied ABA. We found that ibr5 germinated a few hours earlier than the wild type on sucrose-supplemented medium and was slightly less sensitive to germination inhibition by exogenous ABA (Figures 1C and 1D).
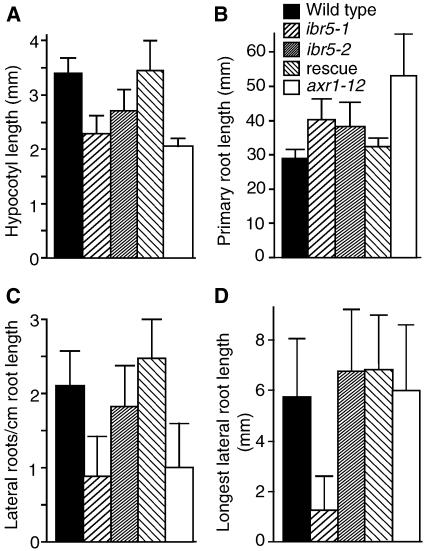
ibr5 seedlings also displayed defects in the absence of exogenous phytohormones. When grown in the light on unsupplemented medium, ibr5 mutants had shorter hypocotyls than wild-type plants (Figure 2A). In addition, light-grown ibr5 seedlings had longer primary roots with slightly fewer lateral roots than wild-type seedlings on unsupplemented medium (Figures 2B and 2C). Moreover, the few lateral roots in ibr5-1 elongated less than those in the wild type (Figure 2D). These phenotypes also were seen in axr1 (Figures 2A to 2C) (Estelle and Somerville, 1987; Lincoln et al., 1990) and other auxin-response mutants, such as axr4 (Hobbie and Estelle, 1995). In contrast to these defects in light-grown seedlings, ibr5 hypocotyls and roots elongated normally in the dark (data not shown).
Figure 2.
ibr5 on Unsupplemented Medium.
(A) Hypocotyl length in the light. Hypocotyls of Col-0 (wild type), ibr5-1, ibr5-2, ibr5-1 transformed with pBINIBR5 (rescue), and axr1-12 seedlings were measured after 8 days of growth on PNS under yellow filters.
(B) Root length in the light. Primary roots were measured after 11 days of growth on PNS under yellow-filtered light.
(C) Lateral roots were counted with a dissecting microscope after 11 days of growth on PNS under yellow-filtered light. Primordia that emerged from the main root were counted as lateral roots.
(D) The lengths of the longest lateral roots counted in (C) were measured.
Error bars represent standard deviations of the means (n ≥ 12).
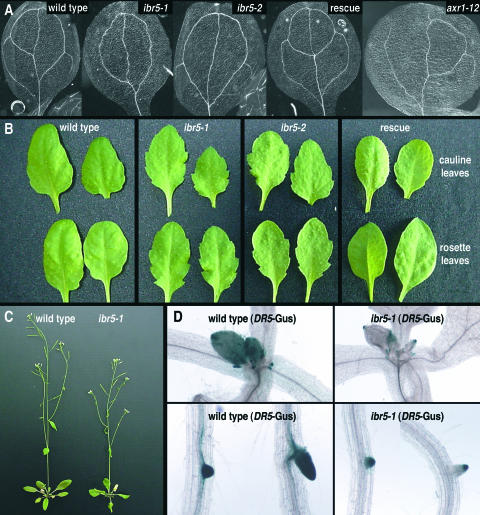
ibr5 mutants also displayed vascularization defects. Wild-type seedlings typically formed continuous veins in cotyledons and leaves (Figure 3A), whereas ibr5, like axr1, often formed discontinued veins and spurs (Figure 3A). Adult ibr5 plants were slightly smaller than wild-type plants and had epinastic leaves (Figure 3C). In addition, ibr5 had serrated rosette (base) and cauline (inflorescence stem) leaves, in contrast to the smoother margins of wild-type leaves (Figure 3B).
Figure 3.
ibr5 Morphological and Auxin-Related Phenotypes.
(A) ibr5 displays aberrant vascular patterning. Cleared cotyledons of 10-day-old Col-0 (wild type), ibr5-1, ibr5-2, ibr5-1 transformed with pBINIBR5 (rescue), and axr1-12 seedlings are shown.
(B) ibr5 has increased leaf serration. Cauline (top) and rosette (bottom) leaves of 6-week-old Col-0 (wild type), ibr5-1, ibr5-2, and ibr5-1 transformed with 35S-IBR5 plants grown under short-day conditions are shown.
(C) Adult ibr5 plants are smaller than wild-type Col-0 plants. Five-week-old plants grown in continuous light are shown.
(D) ibr5 shows decreased levels of an auxin-inducible reporter. Shoot apex (top) and lateral root (bottom) staining of homozygous wild-type and ibr5-1 seedlings expressing DR5-GUS are shown. Seedlings were grown for 10 days in the light on unsupplemented medium before staining.
Gene Expression in ibr5
To examine the molecular consequences of IBR5 inactivation, we crossed ibr5-1 to a line expressing DR5-GUS, which contains the DR5 auxin-responsive element driving the expression of β-glucuronidase (Guilfoyle, 1999). Analysis of ibr5 DR5-GUS lines grown on unsupplemented medium revealed decreased GUS expression in the shoot apex, root tips, hydathodes, true leaves (Figure 3D), and sepals (data not shown), suggesting that IBR5 is necessary for full responsiveness to endogenous auxin.
In an attempt to identify specific genes misregulated in the ibr5 background, which might include targets of IBR5-modulated signaling, we performed whole-genome microarray analysis using RNA prepared from 7-day-old ibr5-1 and wild-type seedlings. Comparison of the results from three independent preparations each of ibr5-1 and wild-type RNA revealed no significant (>2.5-fold) and reproducible alterations in mRNA accumulation of the ∼22,000 genes analyzed (data not shown). Therefore, at 7 days, any gene expression changes in ibr5-1 seedlings may be local, as seen with the DR5-GUS reporter (Figure 3D), and not apparent in whole seedling RNA.
IBR5 Encodes a Putative Dual-Specificity Phosphatase
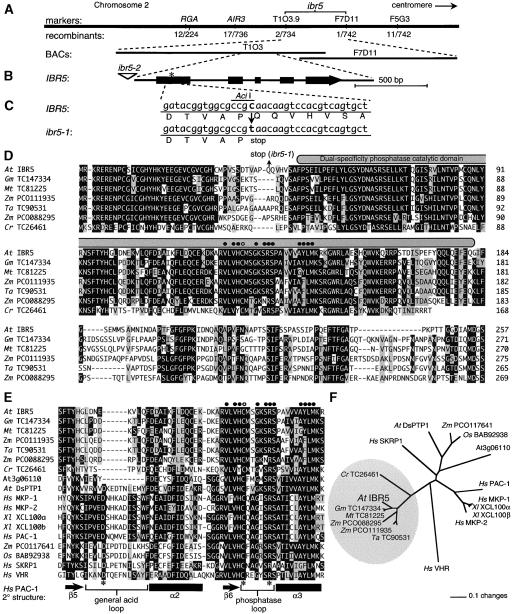
We identified the IBR5 gene using map-based positional cloning (see Methods). ibr5-1 was mapped to the top of chromosome 2, between the markers AIR3 and F5G3 (Zolman et al., 2000). We refined the ibr5-1 position to a 41-kb region containing most of BAC T1O3 and one end of BAC F7D11 (Figure 4A). We then used a candidate gene approach to identify the mutant gene. We sequenced a putative phosphatase (At2g04550) within this region and found a C-to-T mutation in the first exon, changing a Gln at position 42 to a premature stop codon in the ibr5-1 mutant and destroying an AciI restriction site (Figure 4C).
Figure 4.
Map-Based Positional Cloning of IBR5 Reveals Similarity to Dual-Specificity Phosphatases.
(A) Recombination mapping of ibr5-1. Mapping with the PCR-based markers RGA1 (Silverstone et al., 1998), AIR3 (Silverstone et al., 1998), T1O3.9, F7D11, and F5G3 (see Methods) localized the defect to a region spanned by BACs T1O3 and F7D11.
(B) IBR5 contains five exons (thick boxes) separated by four introns (lines). The position of the stop codon in ibr5-1 is indicated with an asterisk, and the position of the T-DNA insertion in ibr5-2 is indicated with a triangle.
(C) ibr5-1 has a C-to-T mutation at position 123 (where 1 is the A of the initiator ATG) that converts a Gln (Q) residue at position 42 to a premature stop codon and destroys an AciI restriction site.
(D) Alignment of Arabidopsis (At) IBR5 and its plant homologs. Sequences were aligned with the MegAlign program (DNAStar, Madison, WI) using the CLUSTAL W method. Residues identical in at least four sequences are shaded in black, similar residues are shaded in gray, and dashes indicate gaps introduced to maximize alignment. The gray oval overlies the dual-specificity phosphatase catalytic domain, circles mark residues defining the dual-specificity phosphatase active-site motif, and the open circle marks the catalytic Cys nucleophile. Sequences from soybean (Gm), Medicago truncatula (Mt), wheat (Ta), and Chlamydomonas reinhardtii (Cr) are from plant genome projects assembled in the TIGR Gene Index Database (http://www.tigr.org/tdb/tgi) (Quackenbush et al., 2001), and the maize (Zm) sequences are from GenBank accession numbers AY105390 and AY108971. The Chlamydomonas cDNA sequence was truncated at the 3′ end, so only the N-terminal region is shown.
(E) Alignment, as in (D), of part of the phosphatase catalytic domains of the proteins shown in (D) with the corresponding catalytic domains of dual-specificity phosphatases from rice (Os), human (Hs), and Xenopus laevis (Xl). α-Helices (black rectangles), β-sheets (arrows), and loops (brackets) observed in the PAC-1 phosphatase domain (Farooq et al., 2003) are shown below the alignment. Asterisks indicate the PAC-1 catalytic residues (D226, C257, and R263).
(F) Phylogenetic tree of IBR5 and its relatives. The tree reconstructs the evolutionary relationship between the characterized and putative phosphatases shown in (E). The portions of the protein corresponding to the catalytic domain (shaded oval in [D]; residues 49 to 182 in IBR5) were aligned as described for (D), and the unrooted phylogram was generated using PAUP 4.05b (Swofford, 2001). The bootstrap method was performed for 100 replicates with a distance optimality criterion, and all characters were weighted equally.
To confirm that this gene was responsible for the ibr5-1 phenotypes, we transformed the wild-type IBR5 gene, driven by its own promoter, into ibr5-1 plants and assayed for rescue of the mutant phenotypes. The resulting transgenic plants displayed wild-type root elongation on the auxins IAA, IBA, 2,4-D, and 1-naphthaleneacetic acid; on the auxin transport inhibitors 1-napthylphthalamic acid and 2,3,5-triiodobenzoic acid; and on the phytohormone ABA (Figure 1). In addition, the delayed germination, lateral root defects, aberrant vascularization, and serrated leaves were rescued by wild-type IBR5 supplied from a transgene (Figures 1C, 1D, 2C, 2D, 3A, and 3B). Together, these analyses confirm that we have identified the IBR5 gene and that this single lesion is responsible for the pleiotropic ibr5 phenotypes.
To identify a second ibr5 allele, we obtained a mutant with a T-DNA insertion 161 bp upstream of the IBR5 start codon (27 bp upstream of the 5′ untranslated region; Figure 4B) from the Salk Institute Genomic Analysis Laboratory collection (Alonso et al., 2003). ibr5-2 has a similar but slightly weaker phenotype than ibr5-1 (Figures 1, 2, and 3), consistent with the potentially leaky nature of the ibr5-2 mutation (see below).
To determine the splicing pattern of the IBR5 gene and the amino acid sequence of the encoded protein, we cloned and sequenced a full-length IBR5 cDNA (see Methods). This analysis revealed that IBR5 has five exons separated by four introns (Figure 4B). Comparison of this cDNA with At2g04550 revealed that the computer-generated annotation failed to recognize the fourth intron, resulting in 26 extra amino acids predicted to include a transmembrane domain in the computer-annotated version.
IBR5 encodes a 257–amino acid protein similar to characterized dual-specificity phosphatases from Arabidopsis, human, mouse, and rat (Figures 4E and 4F). The IBR5 dual-specificity phosphatase catalytic domain (amino acids 49 to 182; Figure 4D) is ∼35% identical to the catalytic domain of known dual-specificity MAPK phosphatases, including human MKP-1 and PAC-1 (Sun et al., 1993; Farooq et al., 2003). Within this catalytic domain is the highly conserved dual-specificity phosphatase active-site motif VxVHCx2GxSRSx5AYLM (Figures 4D and 4E). The premature stop codon in ibr5-1 allows translation of only the first 41 amino acids, which would lack this catalytic domain (Figure 4D). Therefore, the ibr5-1 mutation is expected to be null.
Because IBR5 encodes a dual-specificity phosphatase-like protein, we sought to determine if IBR5 possesses phosphatase activity. Although we could express soluble IBR5 fused to glutathione S-transferase (GST) in E. coli and purify the recombinant protein (see Methods), we did not detect the dephosphorylation of the generic substrate p-nitrophenyl phosphate by this recombinant protein, even after cleavage of the GST tag (data not shown). The only Arabidopsis dual-specificity phosphatase with demonstrated in vitro activity after heterologous expression is AtDsPTP1, which nonetheless has very low specific activity with p-nitrophenyl phosphate (Gupta et al., 1998). Interestingly, certain mammalian MAPK phosphatases require the binding of their substrate MAPK for in vitro activity (Camps et al., 1998; Hutter et al., 2000). It may be necessary to identify the in vivo substrates of IBR5 before enzymatic characterization can be completed.
IBR5 Accumulates throughout the Plant
IBR5 transcript levels are not altered in 4-week-old wild-type plants treated with ABA (Hoth et al., 2002) or in 7-day-old wild-type plants treated with auxin (data not shown). Comparison of IBR5 mRNA levels in ibr5-1 and the wild type revealed an ∼2.5-fold decrease in ibr5 message in the ibr5-1 mutant (data not shown), consistent with the nonsense-mediated decay of the mutant mRNA (Wagner and Lykke-Andersen, 2002).
To determine in which tissues IBR5 is expressed, we transformed wild-type plants with an IBR5 promoter–GUS fusion. We found that IBR5-GUS was expressed in root tips, root vasculature, cotyledons, and true leaves, including the vasculature and hydathodes; in the sepals, anther filaments, and carpels of flowers; and in siliques (Figures 5A to 5E and 5G). In addition, IBR5-GUS was expressed in dark-grown seedlings in both hypocotyls and cotyledons (Figure 5F). Thus, IBR5-GUS was expressed widely throughout development, including in most of the tissues in which the ibr5 mutant displayed phenotypes. However, it is possible that this reporter is not sufficiently sensitive to detect all tissues in which IBR5 plays important roles. For example, we detected no IBR5-GUS expression in lateral root primordia or newly emerged lateral roots (Figure 5B), but we did detect reduced expression of the auxin-responsive DR5-GUS promoter in these tissues in the ibr5 mutant (Figure 3D).
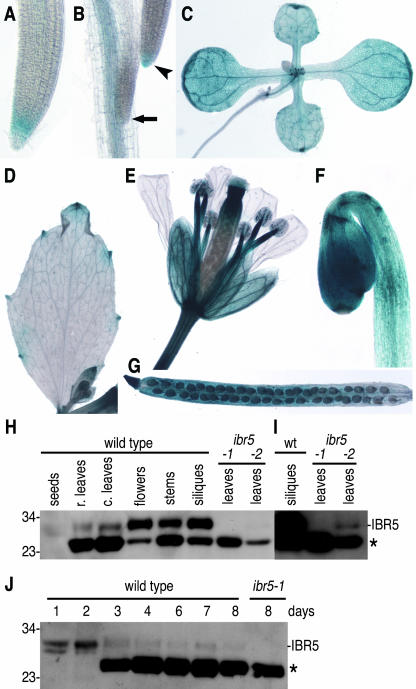
Figure 5.
IBR5 Expression and Protein Accumulation in Wild-Type Plants throughout Development.
(A) to (C) Staining of 10-day-old seedlings revealed IBR5-GUS expression in the primary root tip (A); in lateral root tips of longer (arrowhead) but not shorter (arrow) lateral roots and primary root vasculature (B); and in the shoot (C).
(D) to (G) Staining of mature plants revealed IBR5-GUS expression in the hydathodes of cauline leaves (D); in the sepals, anther filaments, and carpels of flowers (E); and in green siliques (G). Staining of 6-day-old dark-grown seedlings revealed expression throughout the hypocotyl and cotyledons (F).
(H) Immunoblot analysis with an anti-IBR5 antibody of protein prepared from seeds, rosette (r.) and cauline (c.) leaves, flowers, stems, and siliques of wild-type plants and from rosette leaves of ibr5-1 and ibr5-2 mutants.
(I) Overexposure of the last three lanes of (H). No IBR5 protein is detected in rosette leaves of ibr5-1, and reduced amounts of IBR5 protein are detected in rosette leaves of ibr5-2. wt, wild type.
(J) Immunoblot analysis with an anti-IBR5 antibody of protein prepared from seedlings at 1 to 8 days after imbibition.
The positions of IBR5 and an unidentified cross-reacting protein (asterisk) are indicated at right in (H) to (J), and the positions of molecular mass markers (in kD) are indicated at left.
We developed a polyclonal IBR5 antibody to examine IBR5 levels in our ibr5 alleles and to determine the tissues in which IBR5 protein accumulates. This antibody detects two proteins (∼30 and ∼25 kD) in wild-type Arabidopsis; the predicted size for the IBR5 open reading frame is 28.7 kD. The ∼30-kD protein was undetectable in the ibr5-1 mutant, which was disrupted early by a nonsense mutation, suggesting that the ∼30-kD protein is IBR5 (Figures 5H to 5J). The ibr5-2 mutant, which contains a T-DNA insertion upstream of the IBR5 5′ untranslated region, accumulated a reduced but detectable amount of IBR5 protein (Figure 5I). The presence of some residual IBR5 protein in this second allele probably accounts for the generally weaker phenotype of ibr5-2 compared with the ibr5-1 presumed null allele (Figures 1 to 3).
Consistent with the IBR5 promoter-reporter gene expression patterns, we found that IBR5 protein accumulated throughout wild-type plants, including in rosette and cauline leaves, flowers, stems, siliques, and seeds (Figure 5H). During germination, IBR5 levels declined slightly at 2 days after imbibition (Figure 5J).
Overexpression of IBR5
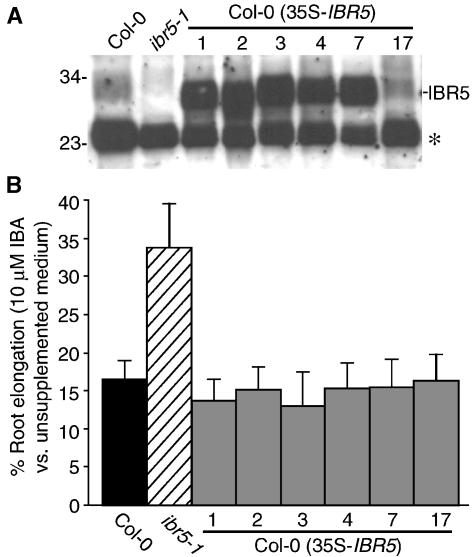
To determine whether IBR5 levels are normally limiting for auxin responsiveness, we transformed wild-type plants with a vector expressing the IBR5 cDNA driven by the strong 35S promoter of Cauliflower mosaic virus (35S-IBR5). We isolated several lines with increased IBR5 protein accumulation, as judged by immunoblot analysis (Figure 6A), and we assayed auxin responsiveness in these lines. We found that IBR5 overexpression did not dramatically alter auxin sensitivity (Figure 6B). Moreover, these lines appeared morphologically similar to the wild-type line (data not shown). The observation that these overexpressing lines did not have significantly shorter roots or longer hypocotyls than wild-type plants (data not shown) suggests that IBR5 is not normally limiting for auxin responsiveness.
Figure 6.
IBR5 Overexpression.
(A) Immunoblot analysis with an anti-IBR5 antibody of protein prepared from 8-day-old wild type (Col-0), ibr5-1, and six lines of Col-0 transformed with 35S-IBR5 grown on 10 μM IBA. The positions of IBR5 and an unidentified cross-reacting protein (asterisk) are indicated at right, and the positions of molecular mass markers (in kD) are indicated at left.
(B) Wild type (Col-0), ibr5-1, and six lines of Col-0 transformed with 35S-IBR5 were analyzed for root elongation inhibition by IBA as described for Figure 1A. Results were standardized against growth on unsupplemented medium. Error bars represent standard deviations of the means (n ≥ 12).
DISCUSSION
IBR5 Encodes an Apparent Dual-Specificity Phosphatase
IBR5 encodes an apparent phosphatase containing the highly conserved dual-specificity phosphatase active-site motif VxVHCx2GxSRSx5AYLM. Phosphatases have been most studied in animal and fungal systems, in which dual-specificity phosphatases often regulate MAPK pathways. MAPK kinases have dual activity, phosphorylating both Tyr and Thr residues on target MAPKs. MAPKs require this dual phosphorylation for full activation. Conversely, phosphatases can dephosphorylate MAPKs at either or both residues. Protein Tyr phosphatases dephosphorylate phosphotyrosine residues, whereas Ser/Thr protein phosphatases dephosphorylate phosphothreonine residues. In addition, a number of phosphatases that dephosphorylate both phosphotyrosine and phosphothreonine residues of MAPKs, termed dual-specificity or MAPK phosphatases, have been identified (reviewed by Keyse, 1995; Camps et al., 2000). Dual-specificity phosphatases have a highly conserved active-site motif, and in mammals, these phosphatases often are highly specific for their substrate (Keyse, 1995; Camps et al., 2000).
The Arabidopsis genome encodes 20 likely MAPKs (Ichimura et al., 2002) and 18 genes in the dual-specificity phosphatase family (Kerk et al., 2002). Of these 18, only 5 (IBR5/At2g04550, AtDsPTP1/At3g23610, At5g23720, At3g06110, and AtMKP1/At3g55270) have the complete dual-specificity phosphatase active-site motif VxVHCx2GxSRSx5AYLM. Two of these five apparent phosphatases have been characterized to date: AtDsPTP1 (Gupta et al., 1998) and AtMKP1 (Ulm et al., 2001, 2002). AtDsPTP1 (26% identical to IBR5) and the uncharacterized At3g06110 protein (29% identical) are the closest IBR5 relatives in the Arabidopsis genome (Figures 4E and 4F). A dsptp1 mutant has not been reported; however, AtDsPTP1 can dephosphorylate and inactivate the MAPK AtMPK4 in vitro (Gupta et al., 1998). AtMKP1 has not been characterized in vitro, but genetic analysis indicates that AtMKP1 negatively regulates an ∼49-kD MAPK. This MAPK has increased activity in mkp1 plants exposed to UV light, suggesting a role for AtMKP1 in UV resistance (Ulm et al., 2001). Although IBR5 has ∼35% sequence identity to AtDsPTP1 within the catalytic domain (Figure 4E; 26% overall), the N and C termini are quite dissimilar. IBR5 also is diverged from AtMKP1 (19% identical), which has an extended C-terminal region.
Although IBR5 lacks close relatives within the Arabidopsis genome, examination of assembled cDNAs from various plant genome projects (TIGR Gene Index Databases; http://www.tigr.org/tdb/tgi) (Quackenbush et al., 2001) revealed that IBR5 does have close relatives in monocots, dicots, and the unicellular green alga Chlamydomonas reinhardtii (Figures 4D to 4F). These proteins represent a unique clade of similarly sized proteins (257 to 275 amino acids; Figure 4F), of which IBR5 is the first characterized member. Unlike the similarity between IBR5 and other characterized phosphatases, which is limited to the catalytic domain, the similarity among these IBR5-like proteins extends throughout the proteins (Figure 4D). For example, IBR5 is 74% identical throughout its length to the wheat protein encoded by TC90531 (Figure 4D). Moreover, proteins in the IBR5 clade share a deletion of part of the general acid loop (Figure 4E) observed in structural studies of the human phosphatases VHR (Yuvaniyama et al., 1996) and PAC-1 (Farooq et al., 2003). Interestingly, the 30 N-terminal residues of the proteins in the IBR5 clade are almost identical among the flowering plants within this clade (Figure 4D). Certain mammalian MAPK phosphatases use N-terminal docking modules to bind their substrate MAPKs (Tanoue et al., 2002); it is possible that the IBR5-like proteins also use this region to dock substrates. If so, the extreme conservation of this region suggests that the substrate(s) of the IBR5-like phosphatases will be conserved as well. Unfortunately, this N-terminal region of IBR5 is responsible for the transcriptional activation seen when IBR5 is fused to a DNA binding domain in a yeast two-hybrid bait vector (data not shown), rendering the yeast two-hybrid system unsuitable for identifying IBR5 substrates.
Models for the IBR5 Regulation of Phytohormone Signaling
The phenotypes of plants lacking IBR5 suggest that IBR5 normally modulates auxin signaling. ibr5 has decreased sensitivity to root elongation inhibition by the application of natural auxins, synthetic auxins, and auxin transport inhibitors (Figure 1A and data not shown). Moreover, examination of the mutant on medium lacking hormones revealed that ibr5 may be less sensitive to endogenous auxin as well. Endogenous auxin promotes lateral root development (Casimiro et al., 2003) and hypocotyl elongation in the light (Romano et al., 1995; Gray et al., 1998; Jensen et al., 1998). Whereas auxin may be necessary for primary root elongation as well, the concentration of auxin in wild-type Arabidopsis roots is thought to be supraoptimal for elongation (Hobbie and Estelle, 1995). Consequently, mutants that overproduce auxin have longer hypocotyls, shorter primary roots, and more lateral roots than the wild type when grown in the light (Delarue et al., 1998; Zhao et al., 2001). Conversely, certain mutants with auxin-response defects, including axr1 and axr4, have short hypocotyls and long roots with fewer lateral roots compared with the wild type when grown on unsupplemented medium (Figure 2) (Estelle and Somerville, 1987; Lincoln et al., 1990; Hobbie and Estelle, 1995). Like these auxin-response mutants, both ibr5 alleles have shorter hypocotyls and longer primary roots with slightly fewer lateral roots on unsupplemented medium compared with the wild type (Figure 2), consistent with reduced responsiveness to endogenous auxin. Auxin also directs vascular patterning in leaves (reviewed by Berleth and Mattsson, 2000), and the discontinuous veins and spurs seen in ibr5 vasculature (Figure 3A) further suggest auxin-response defects. Because ibr5 appears to be less responsive to endogenous and exogenous auxin, and because ibr5-1 is a recessive loss-of-function mutation, these results indicate that IBR5 may normally promote auxin responsiveness, either directly or indirectly. In support of this idea, the auxin-responsive DR5-GUS reporter displays decreased expression in the shoot apex, roots, hydathodes, true leaves, and sepals in the ibr5-1 mutant (Figure 3D and data not shown).
Interestingly, IBR5 also appears to modulate certain aspects of ABA signaling. ibr5 is less sensitive than the wild type to the inhibitory effects of ABA on root elongation (Figure 1B) and germination (Figure 1D). In addition, ibr5 appears to germinate earlier than the wild type even on unsupplemented medium (Figure 1C), suggesting that ibr5 may be less sensitive to both endogenous and exogenous ABA. However, not all ABA responses are disrupted in ibr5. For example, ibr5 does not have a wilty phenotype, and detached ibr5 rosettes lose water at a similar rate as wild-type rosettes (data not shown), suggesting that ABA regulation of stomatal closure is functioning normally. In addition, ibr5 responds normally to high levels of exogenous sugar (data not shown), whereas several previously described ABA-response mutants were defective in both ABA and sugar signaling (reviewed by Fedoroff, 2002; Gazzarrini and McCourt, 2003).
Links between auxin and ABA have been noted previously. For example, auxin can enhance the inhibitory effects of ABA on germination (Brady et al., 2003), and certain mutants originally identified as auxin insensitive, such as axr1 and axr2, also display decreased ABA sensitivity during germination or in root elongation inhibition (Figure 1B) (Wilson et al., 1990; Tiryaki and Staswick, 2002). In addition, mutants defective in the RCN1 protein phosphatase 2A regulatory subunit, which influences auxin transport and gravity responses (Garbers et al., 1996; Rashotte et al., 2001), have decreased sensitivity to ABA (Kwak et al., 2002). Moreover, ABA inhibits DR5-GUS expression in emerging lateral roots (De Smet et al., 2003), and the abi3 mutant, isolated originally as ABA insensitive during germination (Koornneef et al., 1984), shows decreased sensitivity to the auxin induction of lateral roots (Brady et al., 2003). ABI3 expression is induced by auxin in roots (Brady et al., 2003), and when the maize ABI3 homolog VP1 is expressed in Arabidopsis, ABA is able to suppress auxin-induced lateral root formation (Suzuki et al., 2001), further supporting a role for ABI3 in auxin–ABA interactions.
Because ibr5 has decreased sensitivity to auxin and ABA and is defective in an apparent dual-specificity phosphatase, we can envision several possible roles for IBR5. One possibility is that IBR5 dephosphorylates a single MAPK acting in a signaling pathway. Because MAPK phosphatases inactivate MAPKs and the loss-of-function ibr5 mutants are less responsive to auxin and ABA, this putative MAPK may normally inhibit both auxin and ABA responses. The integration of auxin and ABA responses could be either a direct or an indirect result of this signaling. It also is possible that IBR5 has more than one MAPK substrate, explaining the pleiotropic nature of the ibr5 mutant phenotypes. Finally, IBR5 may dephosphorylate a protein or proteins not involved in a canonical signaling pathway, and the loss of this dephosphorylation reduces sensitivity to auxin and ABA.
Previous evidence has implicated MAPK pathways in both auxin and ABA signaling. Studies with the Arabidopsis MAPKKK ANP1 and the tobacco homolog NPK1, which are involved in oxidative stress pathways, show that transient expression in protoplasts of constitutively active ANP1 or NPK1 correlates with the activation of the AtMPK3 and AtMPK6 MAPKs and with a decrease in auxin-responsive gene transcription (Kovtun et al., 1998, 2000). AtMPK3 is activated by ABA, and AtMPK3 overexpression increases Arabidopsis ABA sensitivity (Lu et al., 2002). Coexpression of the mouse MAPK phosphatase MKP1 in protoplasts abolishes NPK1's effects on MAPK activation and restores auxin-responsive transcription (Kovtun et al., 1998). Therefore, it is unlikely that IBR5 functions to inactivate AtMPK3 or AtMPK6, because failure to inactivate these MAPKs is expected to increase, rather than decrease, ABA sensitivity.
In addition, Mockaitis and Howell (2000) demonstrated auxin-induced activation of an ∼44-kD MAPK in Arabidopsis. The activation of this MAPK correlates with the induction of auxin responses, and the reduced activation of this MAPK is seen in the axr4 auxin-response mutant (Hobbie, 1998; Mockaitis and Howell, 2000). Failure to inactivate this auxin-activated MAPK presumably would heighten rather than reduce auxin responsiveness, making it unlikely that IBR5 inactivates this MAPK. Thus, it is likely that IBR5 acts on a MAPK not currently known to influence ABA or auxin signaling or that IBR5 acts on substrates that are not MAPKs.
The identification of IBR5 provides a new tool with which to dissect auxin responsiveness and the interaction between auxin and ABA signaling pathways. The presence of IBR5 homologs throughout the plant kingdom implies that this subfamily of phosphatase-like proteins may play plant-specific signaling roles. Moreover, because IBR5 is related to dual-specificity phosphatases, this work will likely complement efforts to clarify the roles of MAPK pathways in plants. Ongoing efforts to identify IBR5 substrates by identifying genetic suppressors of ibr5-1 mutant phenotypes may further define the roles of IBR5 in plant growth and development.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana accessions Columbia (Col-0) and Wassilewskija (Ws) were used. The ibr5-1 mutant was described previously as B31, an ethyl methanesulfonate–induced IBA-response mutant in the Col-0 background (Zolman et al., 2000). The ibr5-2 allele was isolated from SALK_032185 seeds (also in the Col-0 accession) obtained from the ABRC (Columbus, OH). Plants were grown in soil (Metromix 200; Scotts, Marysville, OH) at 22 to 25°C under continuous illumination by cool-white fluorescent bulbs (Sylvania, Danvers, MA). Plants grown aseptically were plated on PNS (plant nutrient medium with 0.5% [w/v] sucrose) (Haughn and Somerville, 1986) solidified with 0.6% (w/v) agar, either alone or supplemented with hormones (from 0.1-, 1.0-, or 100-mM stocks in ethanol), BASTA (glufosinate ammonium; Crescent Chemical, Augsburg, Germany; from a 50-mg/mL stock in 25% [v/v] ethanol), or kanamycin (from a 25-mg/mL stock). Plates were wrapped with gas-permeable surgical tape (LecTec Corp., Minnetonka, MN) and grown at 22°C under continuous light. Plates were incubated under yellow long-pass filters to slow the breakdown of indolic compounds (Stasinopoulos and Hangarter, 1990) unless indicated otherwise.
Phenotypic Analyses
The ibr5-1 mutant was backcrossed to Col-0 at least four times before analysis. All assays were conducted at least twice with similar results. Seeds were surface-sterilized (Last and Fink, 1988) and plated on PNS with the hormone concentrations indicated in the figure legends. In root-elongation assays on phytohormones other than ABA, seedlings were grown for 8 days and removed from the agar, and the length of the primary root was measured. To measure root elongation inhibition by ABA, seedlings were grown for 4 days on PNS, transferred to PNS supplemented with 10 μM ABA, and grown for an additional 4 days, after which the entire primary root was measured. In lateral root assays, seedlings were grown on PNS for 11 days, and lateral roots were examined with a dissecting microscope, counting primordia emerged from the main root as a lateral root. The lengths of the primary root and the longest lateral root were measured. For hypocotyl elongation assays, seeds were plated on PNS and incubated for 8 days, when seedlings were removed from the agar and hypocotyl length was measured.
To observe vascular patterning, seedlings were grown for 10 days on PNS, removed from the agar, and placed in a chloral hydrate solution (80 g of chloral hydrate, 20 mL of glycerol, and 10 mL of water) for 3 weeks at room temperature, mounted, and photographed. In the adult leaf-shape analysis, wild-type and mutant plants were grown on PNS for 12 days under white light before being transferred to soil. Plants were grown under short-day conditions (8 h of white light, 16 h of dark), and rosette and cauline leaves were removed from 6-week-old plants and photographed.
Positional Cloning and Mutant Complementation
ibr5-1 was outcrossed to Ws for mapping and DNA was isolated (Celenza et al., 1995) from IBA-resistant F2 plants. The mutant was mapped using published (Konieczny and Ausubel, 1993; Bell and Ecker, 1994) and newly developed PCR-based markers. For the marker T1O3.9, PCR amplification with the primers 5′-CACAACCATAAACTCACTCTACGAGTTGAC-3′ and 5′-CACGCTGGAAGATGAGACTCTACTTTGCTC-3′ yielded a 1.3-kb product with two TaiI sites in Col-0 and one site in Ws. For the marker F7D11, PCR amplification with the primers 5′-GAGCATGACCATCATCATCATTCCCATAGC-3′ and 5′-GGATATGGATGTTATGGACAAAGTTGTTG-3′ yielded a 1.1-kb product with two EcoRV sites in Col-0 and no sites in Ws. For the marker F5G3, PCR amplification with the primers 5′-CGGACACACTAGCTAAAACAGTTAGAC-3′ and 5′-GCATATTAGATCCTAATCCGCAC-3′ yielded a 976-bp product with one DpnII site in Col-0 and no sites in Ws.
A candidate gene (IBR5; At2g04550) within the mapping interval was examined for changes in the mutant. Genomic DNA extracted from ibr5-1 mutant plants was amplified using two pairs of oligonucleotides (T1O3.4-1 [5′-CCTAATTTCCTCCGTCTGTGAAATCAAGGG-3′] and T1O3.4-2 [5′-GCAGCCGCAAGTTTAGGATGAGAGTAAGAG-3′]; T1O3.4-3 [5′-CAGACGGTTCCTATGTGCCAGAATCTCTAC-3′] and T1O3.4-4 [5′-CAAACTCTCTCAAGCAATGCACCAATCTCC-3′]) with a program of 40 cycles of 94°C for 15 s, 55°C for 15 s, and 72°C for 30 s. The resulting overlapping fragments were ∼900 bp each and covered the gene from 90 bp upstream of the putative translation start site to 115 bp downstream of the stop codon. Amplification products were purified by ethanol precipitation followed by gel purification using a Matrix Gel Extraction kit (Marligen Biosciences, Ijamsville, MD) and sequenced directly using an automated DNA sequencer (Seqwright, Houston, TX; LoneStar Labs, Houston, TX) with the primers used for amplification. Subsequent ibr5-1 genotyping was performed by amplifying genomic DNA with the primers T1O3.4-1 and T1O3.4-6 (5′-CAAGGCAAAACCCTAACTAAACAAACCG-3′), which yielded a 463-bp product with one AciI site in Col-0 and no sites in ibr5-1.
ibr5-2 (SALK_032185) contains a T-DNA insertion generated by the Salk Institute Genomic Analysis Laboratory, La Jolla, CA (Alonso et al., 2003). Segregating seeds from the ABRC were plated on 12 μg/mL kanamycin, and resistant seedlings were transferred to soil after 12 days. Plants were genotyped by amplifying genomic DNA from these plants with the primers IBR5-2.1 (5′-GGCATTTTGCCATCTATTTGGACATGCGGCC-3′) and LB1-SALK (5′-CAAACCAGCGTGGACCGCTTGCTGCAACTC-3′), which yielded an ∼600-bp fragment from ibr5-2. This fragment was sequenced directly with the primers used for amplification to determine the precise location of the T-DNA insert in the ibr5-2 allele.
To create a genomic rescue clone, DNA from BAC T1O3 was isolated and digested with HindIII. The resulting genomic fragment (containing the IBR5 gene plus 4.5-kb 5′ and 0.3-kb 3′ flanking sequences) was subcloned into pBluescript KS+ (Stratagene) to give pKSIBR5g. The HindIII fragment was excised and subcloned into the plant transformation vector pBIN19 (Bevan, 1984) to give pBINIBR5. This plasmid was electroporated (Ausubel et al., 1999) into Agrobacterium tumefaciens strain GV3101 (Koncz et al., 1992) and transformed into ibr5-1 mutant plants using the floral-dip method (Clough and Bent, 1998). Transformants were identified on PNS medium supplemented with 12 μg/mL kanamycin after 10 days under white light. Rescue assays were performed using seeds from homozygous progeny of kanamycin-resistant transformants.
cDNA Isolation
An IBR5 cDNA was isolated by hybridizing a cDNA library (Minet et al., 1992) with a 32P-labeled probe made by PCR amplifying genomic DNA with T1O3.4-3 and T1O3.4-5 (5′-CAAGTACGCTACAACAACCGCTGGTG-3′). The 1138-bp NotI insert from the hybridizing clone was subcloned into pBluescript KS+ to give pKSIBR5c, which was sequenced.
35S-IBR5 was made by ligating the NotI insert of the IBR5 cDNA into NotI-cut 35SpBARN (LeClere and Bartel, 2001), a plant transformation vector with the 35S promoter of Cauliflower mosaic virus and the nos terminator. The 35S-IBR5 plasmid was electroporated (Ausubel et al., 1999) into A. tumefaciens, which was used to transform wild-type Col-0 and ibr5-1. Transformants were identified on PNS medium supplemented with 7.5 μg/mL BASTA after 10 days under white light. Phenotypic analyses were performed using seeds from homozygous progeny of BASTA-resistant transformants.
Reporter Gene Analysis
A 2.0-kb IBR5 promoter fragment (−2005 to −30 from the IBR5 initiator ATG) was excised from pKSIBR5g with BglII and ligated into BamHI-cut pBI101.1 (Jefferson et al., 1987), forming an IBR5 promoter–GUS fusion (IBR5-GUS), which was transformed into Col-0 plants. β-Glucuronidase activity was localized histochemically by staining for 2 days with 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide as described (Bartel and Fink, 1994). Three independent lines showed similar staining patterns with variable intensity.
Backcrossed ibr5-1 was crossed to Col-0 carrying DR5-GUS (Guilfoyle, 1999), and lines homozygous for both DR5-GUS and ibr5-1 were selected on 12 μg/mL kanamycin and by PCR genotyping the ibr5-1 mutation. For histochemical localization, homozygous seedlings or plant parts were stained for 2 days with 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide as described (Bartel and Fink, 1994).
IBR5 Antibody Production and Purification
A BglII-NotI fragment from pKSIBR5c was ligated into the protein expression vector pGEX-KTO (Davies et al., 1999) cut with BamHI and NotI to make pGEX-IBR5, which expresses a GST-IBR5 fusion protein in Escherichia coli. Protein expression and purification using glutathione-agarose were performed as described previously (Davies et al., 1999). After purification, the GST was removed from IBR5 using a Thrombin Cleavage kit (Novagen, Madison, WI) for 1 h at room temperature. The released IBR5 was separated from GST by SDS-PAGE, detected using Coomassie Brilliant Blue R250 staining (Ausubel et al., 1999), and excised from the gel.
To obtain an IBR5 polyclonal antibody, gel fragments containing IBR5 protein were used to immunize two rabbits at Cocalico Biologicals (Reamstown, PA) with a primary injection (100 μg) followed by three boosts (50 μg each). IBR5 antibody was selected from the resulting serum by incubating for 1 h on IBR5-GST–soaked nitrocellulose membrane, removing the unbound serum, rinsing the membrane two times for 5 min with TBS-T (Ausubel et al., 1999), and eluting bound IBR5 antibodies by incubating for 10 min with 100 mM glycine, pH 2.5. Eluted affinity-purified antibodies were neutralized immediately by adding 0.1 volume of 1 M Tris-HCl, pH 8.0, and stored at −80°C until use.
IBR5 Protein Analysis
Protein was extracted from ibr5-1 and Col-0 by grinding frozen tissue with a pestle, adding 1 volume of buffer (0.1 M Tris-HCl, pH 6.8, 20% glycerol, and 4% SDS), vortexing, removing debris by brief centrifugation, and heating to 100°C for 5 min. Protein extracts were separated by SDS-PAGE beside Cruz markers (Santa Cruz Biotechnology, Santa Cruz, CA) using NuPAGE 10% Bis-Tris gels (Invitrogen, Carlsbad, CA) and transferred for 40 min at 24 V to a Hybond enhanced chemiluminescence nitrocellulose membrane (Amersham Pharmacia Biotech, Piscataway, NJ) using NuPAGE transfer buffer. After blocking for 1 h in 5% milk in TBS-T, the membrane was incubated overnight at 4°C with affinity-purified IBR5 antibody diluted 1:100 or 1:25 in TBS-T, incubated with a 1:500 or 1:300 dilution of horseradish peroxidase–conjugated goat anti-rabbit IgG (Santa Cruz Biotechnology) for 1 h at room temperature, and then visualized using LumiGLO reagent (Cell Signaling, Beverly, MA).
Microarray Analysis
Total RNA was extracted using RNeasy Mini Kits (Qiagen, Valencia, CA) from triplicate samples of 7-day-old ibr5-1 and wild-type (Col-0) seedlings grown on filter paper overlaid on PNS under white light. Thirty to 40 μg of total RNA from each sample was sent to the laboratory of Thomas McKnight at Texas A&M University (College Station), where ibr5-1 and wild-type mRNA were converted to cDNA, amplified to yield biotin-labeled cRNA, and hybridized to Affymetrix (Santa Clara, CA) ATH1 Arabidopsis whole-genome microarray chips containing ∼22,000 genes per chip. The 13,169 mRNAs that were statistically present (Microarray Suite 5.0; Affymetrix) on all six chips were analyzed further. Fold changes were calculated for each ibr5-1 chip compared with each Col-0 chip individually. The fold changes from all nine comparisons were averaged. A >2.5-fold change in message level observed in all nine comparisons was defined as significant.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Bonnie Bartel, bartel@rice.edu.
Accession Number
The GenBank accession number for IBR5 is AY337455.
Acknowledgments
We thank Sherry LeClere for the 35SpBARN vector, Mark Estelle for axr1-12 seeds, Tom Guilfoyle and Jane Murfett for DR5-GUS seeds, and Mary Ellen Lane for microscope use and assistance with microscopy. The Salk Institute Genomic Analysis Laboratory generated the sequence-indexed T-DNA insertion mutant (ibr5-2), and the ABRC at Ohio State University supplied ibr5-2 seeds and BAC T1O3. We are grateful to Dereth Phillips, Rebekah Rampey, and Andrew Woodward for critical comments on the manuscript. This research was supported by the National Science Foundation (IBN-9982611 and IBN-0315596), by National Institutes of Health Training Grant T32-GM08362 (to M.M.-A.), and by Houston Livestock Show and Rodeo Fellowships (to M.M.-A.). Funding for the SIGnAL indexed insertion mutant collection was provided by the National Science Foundation.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.017046.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Ausubel, F., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1999). Current Protocols in Molecular Biology. (New York: Greene Publishing Associates and Wiley-Interscience).
- Bartel, B., and Fink, G.R. (1994). Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 91, 6649–6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, B., LeClere, S., Magidin, M., and Zolman, B.K. (2001). Inputs to the active indole-3-acetic acid pool: De novo synthesis, conjugate hydrolysis, and indole-3-butyric acid β-oxidation. J. Plant Growth Regul. 20, 198–216. [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Berleth, T., and Mattsson, J. (2000). Vascular development: Tracing signals along veins. Curr. Opin. Plant Biol. 3, 406–411. [DOI] [PubMed] [Google Scholar]
- Bevan, M. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12, 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady, S.M., Sarkar, S.F., Bonetta, D., and McCourt, P. (2003). The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 34, 67–75. [DOI] [PubMed] [Google Scholar]
- Camps, M., Nichols, A., and Arkinstall, S. (2000). Dual specificity phosphatases: A gene family for control of MAP kinase function. FASEB J. 14, 6–16. [PubMed] [Google Scholar]
- Camps, M., Nichols, A., Gillieron, C., Antonsson, B., Muda, M., Chabert, C., Boschert, U., and Arkinstall, S. (1998). Catalytic activation of the phosphatase MKP-3 by ERK2 mitogen-activated protein kinase. Science 280, 1262–1265. [DOI] [PubMed] [Google Scholar]
- Casimiro, I., Beeckman, T., Graham, N., Bhalerao, R., Zhang, H., Casero, P., Sandberg, G., and Bennett, M.J. (2003). Dissecting Arabidopsis lateral root development. Trends Plant Sci. 8, 165–171. [DOI] [PubMed] [Google Scholar]
- Celenza, J.L., Grisafi, P.L., and Fink, G.R. (1995). A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 9, 2131–2142. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Davies, P.J. (1995). Plant Hormones. (Dordrecht, The Netherlands: Kluwer Academic Publishers), p. 833.
- Davies, R.T., Goetz, D.H., Lasswell, J., Anderson, M.N., and Bartel, B. (1999). IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue, M., Prinsen, E., Van Onckelen, H., Caboche, M., and Bellini, C. (1998). Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 14, 603–611. [DOI] [PubMed] [Google Scholar]
- De Smet, I., Signora, L., Beeckman, T., Inze, D., Foyer, C.H., and Zhang, H. (2003). An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 33, 543–555. [DOI] [PubMed] [Google Scholar]
- Estelle, M.A., and Somerville, C. (1987). Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Gen. Genet. 206, 200–206. [Google Scholar]
- Farooq, A., Plotnikova, O., Chaturvedi, G., Yan, S., Zeng, L., Zhang, Q., and Zhou, M.M. (2003). Solution structure of the MAPK phosphatase PAC-1 catalytic domain: Insights into substrate-induced enzymatic activation of MKP. Structure 11, 155–164. [DOI] [PubMed] [Google Scholar]
- Fedoroff, N.V. (2002). Cross-talk in abscisic acid signaling. Science's STKE 2002, RE10.. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., Gampala, S.S.L., and Rock, C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14 (suppl.), S15.–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbers, C., DeLong, A., Deruére, J., Bernasconi, P., and Söll, D. (1996). A mutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis. EMBO J. 15, 2115–2124. [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini, S., and McCourt, P. (2003). Cross-talk in plant hormone signalling: What Arabidopsis mutants are telling us. Ann. Bot. 91, 605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of Aux/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., Östin, A., Sandberg, G., Romano, C.P., and Estelle, M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 7197–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle, T.J. (1999). Auxin-regulated genes and promoters. In Biochemistry and Molecular Biology of Plant Hormones, P.J.J. Hooykaas, M.A. Hall, and K.R. Libbenga, eds (Amsterdam: Elsevier), pp. 423–459.
- Gupta, R., Huang, Y., Kieber, J., and Luan, S. (1998). Identification of a dual-specificity protein phosphatase that inactivates a MAP kinase from Arabidopsis. Plant J. 16, 581–589. [DOI] [PubMed] [Google Scholar]
- Haughn, G.W., and Somerville, C. (1986). Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204, 430–434. [Google Scholar]
- Hobbie, L., and Estelle, M. (1995). The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. Plant J. 7, 211–220. [DOI] [PubMed] [Google Scholar]
- Hobbie, L.J. (1998). Auxin: Molecular genetic approaches in Arabidopsis. Plant Physiol. Biochem. 36, 91–102. [Google Scholar]
- Hoth, S., Morgante, M., Sanchez, J.-P., Hanafey, M.K., Tingey, S.V., and Chua, N.-H. (2002). Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J. Cell Sci. 115, 4891–4900. [DOI] [PubMed] [Google Scholar]
- Hutter, D., Chen, P., Barnes, J., and Liu, Y. (2000). Catalytic activation of mitogen-activated protein (MAP) kinase phosphatase-1 by binding to p38 MAP kinase: Critical role of the p38 C-terminal domain in its negative regulation. Biochem. J. 352, 155–163. [PMC free article] [PubMed] [Google Scholar]
- Ichimura, K., et al. (2002). Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 7, 301–308. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, P.J., Hangarter, R.P., and Estelle, M. (1998). Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol. 116, 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski, S., and Leyser, O. (2002). Ubiquitination and auxin signaling: A degrading story. Plant Cell 14 (suppl.), S81.–S95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk, D., Bulgrien, J., Smith, D.W., Barsam, B., Veretnik, S., and Gribskov, M. (2002). The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 129, 908–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyse, S.M. (1995). An emerging family of dual specificity MAP kinase phosphatases. Biochim. Biophys. Acta 1265, 152–160. [DOI] [PubMed] [Google Scholar]
- Koncz, C., Schell, J., and Rédei, G.P. (1992). T-DNA transformation and insertion mutagenesis. In Methods in Arabidopsis Research, C. Koncz, N.-H. Chua, and J. Schell, eds (Singapore: World Scientific), pp. 224–273.
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Reuling, G., and Karssen, C.M. (1984). The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 61, 377–383. [Google Scholar]
- Kovtun, Y., Chiu, W.L., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.L., Zeng, W., and Sheen, J. (1998). Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature 395, 716–720. [DOI] [PubMed] [Google Scholar]
- Kwak, J.M., Moon, J.H., Murata, Y., Kuchitsu, K., Leonhardt, N., DeLong, A., and Schroeder, J.I. (2002). Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14, 2849–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last, R.L., and Fink, G.R. (1988). Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science 240, 305–310. [DOI] [PubMed] [Google Scholar]
- LeClere, S., and Bartel, B. (2001). A library of Arabidopsis 35S-cDNA lines for identifying novel mutants. Plant Mol. Biol. 46, 695–703. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum, E., and Reed, J.W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400. [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., and Chua, N.-H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98, 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., Kinoshita, N., and Chua, N.-H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 17, 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C., Han, M.H., Guevara-Garcia, A., and Fedoroff, N.V. (2002). Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc. Natl. Acad. Sci. USA 99, 15812–15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Müller, J. (2000). Indole-3-butyric acid in plant growth and development. Plant Growth Regul. 32, 219–230. [Google Scholar]
- Minet, M., Dufour, M.-E., and Lacroute, F. (1992). Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 2, 417–422. [DOI] [PubMed] [Google Scholar]
- Mockaitis, K., and Howell, S.H. (2000). Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant J. 24, 785–796. [DOI] [PubMed] [Google Scholar]
- Møller, S.G., and Chua, N.-H. (1999). Interactions and intersections of plant signaling pathways. J. Mol. Biol. 293, 219–234. [DOI] [PubMed] [Google Scholar]
- Muday, G.K., and DeLong, A. (2001). Polar auxin transport: Controlling where and how much. Trends Plant Sci. 6, 535–542. [DOI] [PubMed] [Google Scholar]
- Quackenbush, J., Cho, J., Lee, D., Liang, F., Holt, I., Karamycheva, S., Parvizi, B., Pertea, G., Sultana, R., and White, J. (2001). The TIGR Gene Indices: Analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 29, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte, A.M., DeLong, A., and Muday, G.K. (2001). Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. Plant Cell 13, 1683–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogg, L.E., and Bartel, B. (2001). Auxin signaling: Derepression through regulated proteolysis. Dev. Cell 1, 595–604. [DOI] [PubMed] [Google Scholar]
- Romano, C.P., Robson, P.R.H., Smith, H., Estelle, M., and Klee, M. (1995). Transgene-mediated auxin overproduction in Arabidopsis: Hypocotyl elongation phenotype and interactions with the hy6-1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol. Biol. 27, 1071–1083. [DOI] [PubMed] [Google Scholar]
- Silverstone, A.L., Ciampaglio, C.N., and Sun, T. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10, 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasinopoulos, T.C., and Hangarter, R.P. (1990). Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 93, 1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., Charles, C.H., Lau, L.F., and Tonks, N.K. (1993). MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell 75, 487–493. [DOI] [PubMed] [Google Scholar]
- Suzuki, M., Kao, C.Y., Cocciolone, S., and McCarty, D.R. (2001). Maize VP1 complements Arabidopsis abi3 and confers a novel ABA/auxin interaction in roots. Plant J. 28, 409–418. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (2001). PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods) (Sunderland, MA: Sinauer Associates).
- Tanoue, T., Yamamoto, T., and Nishida, E. (2002). Modular structure of a docking surface on MAPK phosphatases. J. Biol. Chem. 277, 22942–22949. [DOI] [PubMed] [Google Scholar]
- Tiryaki, I., and Staswick, P.E. (2002). An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol. 130, 887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, S.B., Wang, X.-J., Hagen, G., and Guilfoyle, T.J. (2001). Aux/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13, 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm, R., Ichimura, K., Mizoguchi, T., Peck, S.C., Zhu, T., Wang, X., Shinozaki, K., and Paszkowski, J. (2002). Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J. 21, 6483–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm, R., Revenkova, E., di Sansebastiano, G.P., Bechtold, N., and Paszkowski, J. (2001). Mitogen-activated protein kinase phosphatase is required for genotoxic stress relief in Arabidopsis. Genes Dev. 15, 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1997. a). ARF1, a transcription factor that binds auxin response elements. Science 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96, 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997. b). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, E., and Lykke-Andersen, J. (2002). mRNA surveillance: The perfect persist. J. Cell Sci. 115, 3033–3038. [DOI] [PubMed] [Google Scholar]
- Wilson, A.K., Pickett, F.B., Turner, J.C., and Estelle, M. (1990). A dominant mutation in Arabidopsis confers resistance to auxin, ethylene, and abscisic acid. Mol. Gen. Genet. 222, 377–383. [DOI] [PubMed] [Google Scholar]
- Xie, Q., Frugis, G., Colgan, D., and Chua, N.-H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14, 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q., Guo, H.-S., Dallman, G., Fang, S., Weissman, A.M., and Chua, N.-H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419, 167–170. [DOI] [PubMed] [Google Scholar]
- Yuvaniyama, J., Denu, J.M., Dixon, J.E., and Saper, M.A. (1996). Crystal structure of the dual specificity protein phosphatase VHR. Science 272, 1328–1331. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Christensen, S.K., Fankhauser, C., Cashman, J.R., Cohen, J.D., Weigel, D., and Chory, J. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309. [DOI] [PubMed] [Google Scholar]
- Zolman, B.K., Yoder, A., and Bartel, B. (2000). Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156, 1323–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]