Abstract
We report the identification and biotechnological utility of a plant gene encoding the tocopherol (vitamin E) biosynthetic enzyme 2-methyl-6-phytylbenzoquinol methyltransferase. This gene was identified by map-based cloning of the Arabidopsis mutation vitamin E pathway gene3-1 (vte3-1), which causes increased accumulation of δ-tocopherol and decreased γ-tocopherol in the seed. Enzyme assays of recombinant protein supported the hypothesis that At-VTE3 encodes a 2-methyl-6-phytylbenzoquinol methyltransferase. Seed-specific expression of At-VTE3 in transgenic soybean reduced seed δ-tocopherol from 20 to 2%. These results confirm that At-VTE3 protein catalyzes the methylation of 2-methyl-6-phytylbenzoquinol in planta and show the utility of this gene in altering soybean tocopherol composition. When At-VTE3 was coexpressed with At-VTE4 (γ-tocopherol methyltransferase) in soybean, the seed accumulated to >95% α-tocopherol, a dramatic change from the normal 10%, resulting in a greater than eightfold increase of α-tocopherol and an up to fivefold increase in seed vitamin E activity. These findings demonstrate the utility of a gene identified in Arabidopsis to alter the tocopherol composition of commercial seed oils, a result with both nutritional and food quality implications.
INTRODUCTION
The nutritional value of vitamin E in the human diet was recognized >75 years ago (Evans and Bishop, 1922). Vitamin E is made up of a group of structurally related compounds (vitamers), eight major forms of which are known to occur in nature: α-, β-, γ-, and δ-tocopherol and four corresponding unsaturated derivatives, α-, β-, γ-, and δ-tocotrienol (Figure 1A). The biological activities of these eight naturally occurring tocol isoforms vary according to the number and position of methyl groups on the chroman ring and by the configuration of the asymmetric carbons in the side chain (Table 1) (Sheppard et al., 1993; Bramley et al., 2000). The highest vitamin E activity in animals is seen with trimethylated RRR-α-tocopherol, which may be attributable in part to the fact that α-tocopherol is more readily retained by humans (Traber and Sies, 1996). During the past 20 years, a large body of medical evidence has indicated that daily vitamin E supplementation of 400 IU (250 mg of RRR-α-tocopherol) results in decreased risk for cardiovascular disease and cancer, aids in immune function, and prevents or slows a number of degenerative diseases (Buring and Hennekens, 1997; Tangney, 1997; Bramley et al., 2000). Tocotrienols also have been found to have hypocholesterolemic properties (Qureshi and Qureshi, 1993; Qureshi et al., 1995), leading to the suggestion that tocotrienols should be considered nutrients independent of the tocopherols (Hendrich et al., 1994).
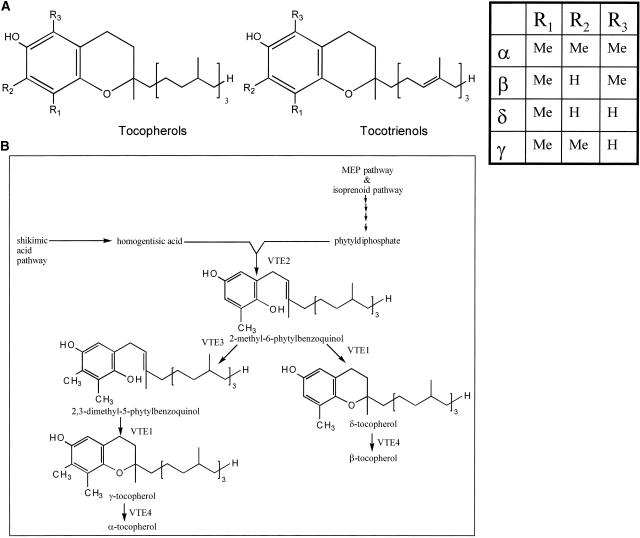
Figure 1.
Tocopherol and Tocotrienol Structures and Biosynthesis.
(A) Detailed chemical structures of tocopherol and tocotrienol. The canonical structures are shown, with R-group modifications illustrated in the gridded box at top right.
(B) Final steps of the tocopherol biosynthetic pathway (Bramley et al., 2000). MEP, methylerythritol phosphate; VTE1, tocopherol cyclase; VTE2, homogentisic acid prenyltransferase; VTE3, 2-methyl-6-phytylbenzoquinol (MPBQ) methyltransferase; VTE4, γ-tocopherol methyltransferase.
Table 1.
Vitamin E Activity of Tocopherols and Tocotrienols (Sheppard et al., 1993)
| Tocopherol | Activity as α-Tocopherol Equivalents (mg/mg Compound) |
|---|---|
| RRR-α-tocopherol | 1.0 |
| RRR-β-tocopherol | 0.5 |
| RRR-γ-tocopherol | 0.1 |
| RRR-δ-tocopherol | 0.03 |
| RRR-α-tocotrienol | 0.3 |
| RRR-β-tocotrienol | 0.05 |
| Synthetic α-tocopheryl acetate | 0.74 |
Tocopherols and tocotrienols are thought to have a number of vital functions in plants, including the protection of chloroplasts from photooxidative damage (Munné-Bosch and Alegre, 2002). Two recent findings underscore the fact that we do not understand all of the functions of tocopherols in plants. First, the altered plasmodesmata mutant sucrose export defective1 of maize appears to be defective in a tocopherol cyclase ortholog (Porfirova et al., 2002). This mutant has dramatic morphological alterations, presumably caused by altered photoassimilate export (Provencher et al., 2001). By contrast, the Arabidopsis loss-of-function cyclase mutant vitamin E pathway gene1 (vte1) is normal under standard growth conditions and shows only modest changes in photosynthesis and pigmentation under growth conditions that cause photooxidative stress (Porfirova et al., 2002).
Tocopherols are synthesized in photosynthetic microorganisms and plants, with the highest concentrations being found in seeds. The seeds of most plants have significant amounts of γ-tocopherol, whereas leaves have predominately α-tocopherol. Unfortunately, γ-tocopherol has only one-tenth the vitamin E activity of α-tocopherol (Table 1). Twenty to 30% of the vitamin E in the U.S. diet comes from oil-based products such as margarines, dressings, and mayonnaise (Sheppard et al., 1993; Eitenmiller, 1997). Soybean oil accounts for ∼70% of the edible oil consumed by humans in the United States and 30% of the worldwide oil consumption (USDA Foreign Agriculture Service, 2003). It has on average 100 mg of tocopherols per 100 g of oil (Eitenmiller, 1997), but the vitamin E activity of these tocopherols is relatively low, because only ∼10% is the highly active α-tocopherol. The major tocopherols are the highly abundant γ-tocopherol (60 to 65% of the total) and δ-tocopherol (20 to 26% of the total) (Tan, 1989). Manipulating the seed tocopherol biosynthetic pathway in soybean to convert the less active tocopherols to α-tocopherol could have significant human health benefits and make this crop an attractive target for the improvement of tocopherol composition.
Tocopherols are synthesized from precursors derived from two pathways. The methylerythritol phosphate pathway contributes to the side chain of tocopherol through the synthesis of phytyldiphosphate (Figure 1B) (Bramley et al., 2000). The shikimate pathway produces homogentisic acid, which contributes the aromatic ring of tocopherol. The first committed step in the tocopherol biosynthetic pathway is the prenylation of homogentisic acid with phytyldiphosphate to form 2-methyl-6-phytylbenzoquinol (MPBQ). The overall tocopherol composition is determined by the combined activities and substrate specificities of the homogentisate prenyltransferase (VTE2), tocopherol cyclase (VTE1), and two methyltransferase enzymes (VTE3 and VTE4) present in a given tissue (Grusak and DellaPenna, 1999). The action of the cyclase directly on MPBQ leads to the formation of δ-tocopherol. By contrast, ring methylation at position R2 by the first methyltransferase, MPBQ methyltransferase (VTE3), leads to 2,3-dimethyl-5-phytylbenzoquinol and the subsequent action of tocopherol cyclase leads to the formation of γ-tocopherol. The second methyltransferase, γ-tocopherol methyltransferase (VTE4), performs ring methylation at R3, converting the γ and δ isoforms to the α and β isoforms, respectively. Tocotrienols are formed when geranylgeranyldiphosphate is used in place of phytyldiphosphate as the side chain precursor.
Tocopherol biosynthesis occurs in the plastids of higher plants, and the biosynthetic enzymes are associated with the chloroplast envelope. It has proven difficult to isolate and purify the membrane-bound proteins of the tocopherol biosynthetic pathway using protein purification techniques (Soll et al., 1980, 1985). However, the recent use of homology-based genomic database searching techniques has led to the identification of several tocopherol biosynthetic genes (Shintani and DellaPenna, 1998; Collakova and DellaPenna, 2001; Schledz et al., 2001; Subramaniam et al., 2001; Savidge et al., 2002). Using this approach, Shintani et al. (2002) identified a gene from Synechocystis sp PCC 6803 capable of methylating MPBQ to produce 2,3-dimethyl-5-phytylbenzoquinol based on its homology with VTE4. Proteins that are similar to this Synechocystis MPBQ methyltransferase (Syn-VTE3) have been identified from other cyanobacterial species (Van Eenennaam et al., 2003). There are no obvious orthologs to these cyanobacterial genes present in sequence databases derived from higher plants, including the sequenced Arabidopsis genome. Because the comparative genomics approach did not yield good candidates for this gene in higher plants, other approaches were necessary.
A screen was used to identify mutants with altered seed tocopherols in Arabidopsis. We hypothesized that mutants with increased δ-tocopherol might be deficient in MPBQ methyltransferase activity. This screen identified 38 mutants with altered tocopherol profiles, 18 of which had increased accumulation of δ-tocopherol. Map-based cloning of one such δ-tocopherol accumulation locus resulted in the identification of a novel tocopherol methyltransferase gene, At-VTE3 (Arabidopsis gene locus At3g63410.1). This gene has no significant similarity to the Synechocystis MPBQ (Syn-VTE3), but it is similar to a ubiquinone methyltransferase. Enzyme assay results supported the hypothesis that At-VTE3 encodes a methyltransferase that is responsible for the R2 methylation of MPBQ. Data from transgenic Arabidopsis and soybean plants demonstrated that the At-VTE3 protein is capable of performing the methylation of MPBQ in planta. Soybean seed-specific expression of At-VTE3 resulted in a 90% decrease in the percentage of δ-tocopherol. When At-VTE3 was coproduced with At-VTE4 in soybean, seed with >95% α-tocopherol was obtained. These results demonstrate the significant utility that map-based cloning in Arabidopsis can have for identifying genes that lead to crop improvement.
RESULTS
Isolation of High δ-Tocopherol Arabidopsis Lines in a Mutant Screen
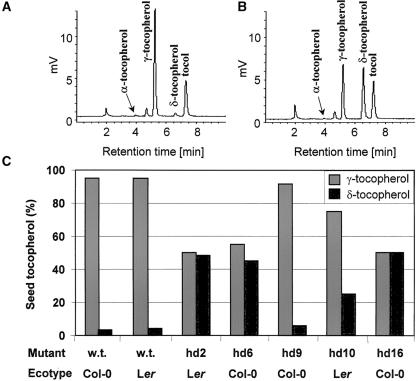
Normally, the tocopherol composition in Arabidopsis seed is predominately γ-tocopherol, with small amounts of δ-tocopherol, as shown in Figure 2A. A screen was designed to detect mutants with altered tocopherol seed composition. Tocopherols were extracted and analyzed from the M3 seed of each of the ∼8000 plant lines. Individual plant lines with altered tocopherol composition were selected as putative mutants. To both confirm the mutant phenotype and attempt to isolate true breeding lines, tocopherol levels were analyzed in M4 seeds. We confirmed 18 high δ (hd) tocopherol mutants. An HPLC trace of the representative mutant hd2 is shown in Figure 2B, and relative seed γ- and δ-tocopherol levels for five of the hd mutants are shown in Figure 2C. The total tocopherol accumulation in these mutants was not substantially different from wild-type levels; rather, the ratio of γ-tocopherol to δ-tocopherol was shifted. As a result of their heavy mutational load, any nonseed composition phenotypes noted could not be attributed definitively to the mutation responsible for the tocopherol phenotype. However, it was noted that there were no consistent phenotypes other than increased seed δ-tocopherol observed among the 18 hd mutants isolated.
Figure 2.
Tocopherol Content and Composition of Seeds from Wild-Type and High Delta (hd) Arabidopsis Mutants.
(A) HPLC scan of wild-type seed, with tocol standard.
(B) HPLC scan of hd2 mutant seed, with tocol standard.
(C) Percentage of γ-tocopherol and δ-tocopherol in Arabidopsis wild type and high δ-tocopherol mutants defective in VTE3. Col, Columbia-0; Ler, Landsberg erecta.
Identification of an Allelic Series of 2-Methyl-6-Phytylbenzoquinol Methyltransferase–Deficient High δ-Tocopherol Mutants
The recessive hd2 mutant was chosen for map-based cloning based on its strong high δ-tocopherol phenotype. This M4 line, Landsberg erecta ecotype, was outcrossed to the Columbia-0 wild type, segregating F2 populations were generated, F3 seeds were harvested from 384 individual F2 lines, tocopherol levels were measured in the F3 seeds, and 101 lines with a mutant phenotype were identified. Eighty-eight homozygous mutant lines were selected for genotyping with 25 molecular markers throughout the genome (Jander et al., 2002).
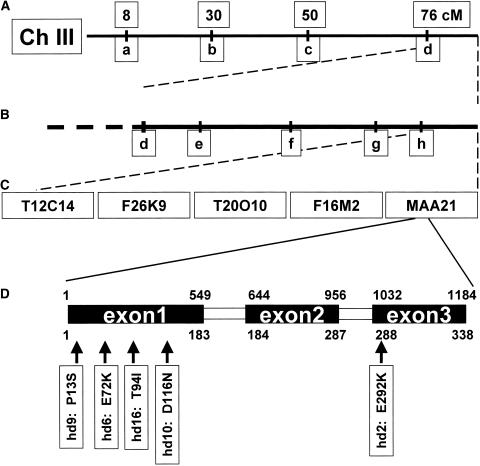
The mutation responsible for the hd2 phenotype showed linkage to a marker on BAC F28P10 at position 21,630 kb on chromosome III (Figure 3, marker d). The location of the mutation was narrowed further to between a marker on BAC T12C14 (Figure 3B, marker h) and the end of chromosome III, defining a region of five sequenced BACs (Figure 3B). Review of the existing GenBank annotations for this region revealed no likely candidate genes. By contrast, a search of Pfam Hidden Markov models (Bateman et al., 2002) of this region found homology between gene MAA21_40 and a ubiquinone methyltransferase. This was chosen as a candidate gene for further study. Sequencing of this gene from the hd2 mutant showed a single base pair change resulting in a Glu-to-Lys substitution at amino acid 292 (Figure 3D). Four other mutants (hd6, hd9, hd10, and hd16) with increased δ-tocopherol also showed mutations in this gene (Figure 3D). The presence of distinct alleles in multiple independently isolated mutants provided strong evidence that we had identified the gene, hereafter called At-VTE3, responsible for the high δ-tocopherol accumulation, and these five mutations appear to be allelic. No mutations in this gene were detected in the remaining 13 hd mutants, consistent with them defining one or more other complementation groups.
Figure 3.
Map-Based Cloning of a Gene Associated with the hd2 Mutant Phenotype.
Lowercase letters a to h refer to genetic markers on BACs with the nomenclature as follows: BAC number_nucleotide position on BAC. a, T12J13_32820; b, MCB17_75129; c, K11J14_6841; d, F28P10_21630; e, T5P19_12659; f, F15B8_57589; g, F17J16_24643; h, T12C14_1563. cM, centimorgan.
(A) First-pass mapping of hd2 showed linkage to genetic markers near the end of chromosome III.
(B) Narrowing with additional markers to five BACs at the end of chromosome III.
(C) BACs in the region of the hd2 mutation.
(D) Scheme of the predicted exon-intron structure of MAA21_40 (At-VTE3) with sites of vte3-1 through vte3-5. Numbers above the schematic of the gene model refer to nucleotides from the first codon, while numbers below represent amino acids.
Transgenic Restoration of Normal δ-Tocopherol Levels in hd2
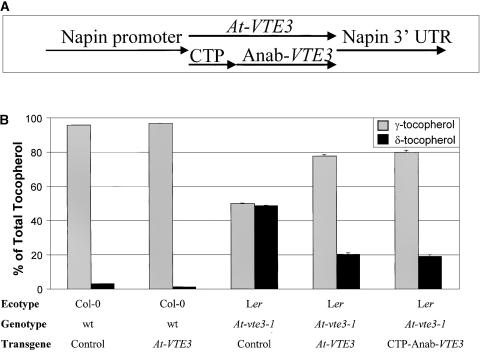
To confirm that mutation of the wild-type At-VTE3 was responsible for the hd2 phenotype, we transformed the mutant with the wild-type gene under the control of the seed-specific napin promoter, as shown in Figure 4A. As a control, transgenic plants also were produced with the Anabaena-VTE3 (Anab-VTE3) gene fused to the N-terminal CTP1 chloroplast-targeting sequence and placed under the control of the same napin promoter. The results of tocopherol analysis of T2 segregating seeds are shown in Figure 4B. There was a substantial decrease in δ-tocopherol and an accompanying increase in γ-tocopherol in the seeds from these transgenic plants, as expected if At-VTE3 complemented the mutation in hd2. The chloroplast targeted Anab-VTE3 gave a similar phenotype. Together, the genetic mapping, sequencing of five independently isolated mutant alleles, and genetic complementation provide extremely strong evidence that mutation of At-VTE3 causes a high δ-tocopherol phenotype.
Figure 4.
Percentage of γ-Tocopherol and δ-Tocopherol in Arabidopsis T2 Seeds from Wild-Type and hd2 Mutant Plants Expressing At-VTE3 or CTP-Anab-VTE3 Proteins under the Control of Napin.
(A) Scheme of the constructs that were used to transform Arabidopsis.
(B) Transgenic (n = 16) and control (n = 4) Arabidopsis T2 seed data. Error bars represent standard errors. Ler, Landsberg erecta; wt, wild type.
Demonstration That At-VTE3 Has 2-Methyl-6-Phytylbenzoquinol Methyltransferase Activity in Escherichia coli
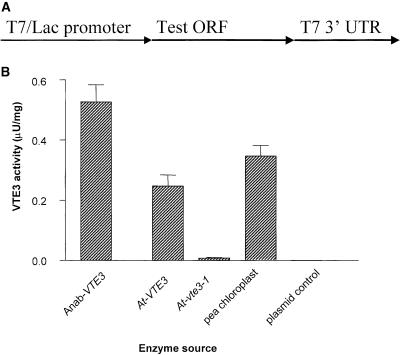
Because of the sequence similarity of At-VTE3 to ubiquinone methyltransferase and the accumulation of δ-tocopherol in the loss-of-function mutants, we hypothesized that At-VTE3 encodes a plant MPBQ methyltransferase (Figure 1B). This hypothesis was tested by expressing the mature protein from both wild-type Arabidopsis and the hd2 mutant in E. coli (Figure 5A) and determining whether the recombinant proteins could methylate MPBQ to produce 2,3-dimethyl-5-phytylbenzoquinol (Figure 5B). Consistent with our hypothesis, activity was seen in both the Anab-VTE3 and At-VTE3 E. coli extracts, in addition to a positive control extract from pea chloroplast (Figure 5). By contrast, extracts from E. coli expressing At-vte3-1 mutant protein had greatly decreased activity compared with that of the wild-type At-VTE3 protein. As expected, no activity was seen in E. coli extracts transformed with the empty vector control. These results support the hypothesis that At-VTE3 protein has MPBQ methylation activity in planta. Based on the mutant phenotype, gene sequence, and enzymatic activity, we conclude that At-VTE3 encodes a MPBQ methyltransferase, and we have renamed the hd2, hd6, hd9, hd10, and hd16 alleles At-vte3-1 through At-vte3-5 to reflect this fact.
Figure 5.
2-Methyl-6-Phytylbenzoquinol Methyltransferase Activity of Recombinant Proteins.
(A) Scheme of the constructs that were transformed into E. coli.
(B) Activity data for E. coli extracts expressing Anab-VTE3, At-VTE3, or At-vte3-1 protein, empty vector negative control, or positive control extract from pea chloroplasts. MPBQ transferase activity is given in units. One unit of MPBQ activity is defined as 1 μmol of 2,3-dimethyl-5-phytylbenzoquinol formation per minute per milligram of protein.
Demonstration That At-VTE3 Expression Significantly Alters the Tocopherol Composition in Transgenic Soybean
To determine if a multigenic approach could convert the majority of tocopherols to α-tocopherol in soybean seed, At-VTE3 and At-VTE4 were expressed either independently or simultaneously in transgenic soy seed. Numerous plants were generated, and plants that tested positive for the presence of the CP4 selectable marker were analyzed for tocopherol composition. A representative analysis of pools of ten seed from the first generation is shown for eight representative lines (Table 2). Because this is the first generation, the sample represents a mixture of transgenic and null seeds. Seed populations from plants transformed with the P7Sα′-At-VTE3 construct showed a dramatic decrease in δ- and β-tocopherol levels in all lines, with a proportionate increase in γ- and α-tocopherol levels. To examine this phenotype further, single seed analysis was performed on the progeny of several transgenic insertion events. The results for event 27930 are shown in Figure 6A. Because this analysis was performed on a population of R1 seeds segregating for the transgene, each individual seed may be a null or may carry one or more copies of the gene. For the eight single seeds tested, one seed was a null and the other seven seed showed the trait from At-VTE3 expression. In the transgenic seeds, the majority of the tocopherol accumulated as γ-tocopherol (75 to 85%) with increased α-tocopherol as well. By contrast, these seeds had very low levels of β- and δ-tocopherol, with these representing only 0.5 to 1.5% of total tocopherols. These data demonstrate that the At-VTE3 MPBQ methyltransferase enzyme converts δ- to γ-tocopherol in soybean. Soybean lines transformed with a P7Sα′-VTE4 expression construct had substantially decreased levels of γ- and δ-tocopherol, with a proportional increase in α- and β-tocopherol, respectively (Table 2). This finding is consistent with results reported previously in Arabidopsis (Shintani and DellaPenna, 1998).
Table 2.
Tocopherol Composition in R1 Soy Seeds from P7Sα′-At-VTE3, P7Sα′-At-VTE4, and P7Sα′-At-VTE3/P7Sα′-At-VTE4 Lines
| Tocopherol (%)
|
Total Tocopherol (ng/mg seed) |
|||||
|---|---|---|---|---|---|---|
| Construct | Line | δ | β | γ | α | |
| Wild type | A3244 | 22.9 | 2.7 | 64.0 | 10.4 | 297 |
| Wild type | A3244 | 22.9 | 2.7 | 64.2 | 10.3 | 302 |
| Wild type | A3244 | 22.9 | 2.3 | 64.4 | 10.5 | 306 |
| Wild type | A3244 | 23.1 | 2.3 | 64.2 | 10.4 | 299 |
| Wild type | A3244 | 23.0 | 2.4 | 64.2 | 10.5 | 296 |
| VTE3 | A28081 | 11.1 | 1.2 | 76.9 | 10.8 | 324 |
| VTE3 | A27627 | 8.3 | 1.5 | 75.9 | 14.2 | 324 |
| VTE3 | A27857 | 7.3 | 1.0 | 78.5 | 13.3 | 316 |
| VTE3 | A27995 | 6.1 | 0.9 | 78.1 | 14.9 | 329 |
| VTE3 | A28075 | 5.6 | 1.7 | 73.6 | 19.1 | 303 |
| VTE3 | A28063 | 1.2 | 0.6 | 74.6 | 23.6 | 327 |
| VTE3 | A28072 | 1.0 | 0.3 | 76.7 | 22.1 | 317 |
| VTE3 | A27930 | 0.7 | 0.0 | 79.1 | 20.3 | 296 |
| VTE4 | A18288 | 28.4 | 4.5 | 60.1 | 7.0 | 307 |
| VTE4 | A18446 | 19.6 | 2.7 | 69.0 | 8.8 | 356 |
| VTE4 | A18571 | 16.7 | 3.7 | 67.1 | 12.6 | 350 |
| VTE4 | A18416 | 5.9 | 27.6 | 15.6 | 50.8 | 288 |
| VTE4 | A18411 | 6.3 | 17.6 | 24.7 | 51.4 | 310 |
| VTE4 | A18335 | 5.1 | 23.5 | 17.5 | 53.9 | 340 |
| VTE4 | A18533 | nd | 28.2 | nd | 71.8 | 294 |
| VTE4 | A18764 | nd | 24.8 | nd | 75.3 | 321 |
| VTE3 + VTE4 | A27712 | 10.2 | 2.5 | 26.9 | 60.4 | 283 |
| VTE3 + VTE4 | A27936 | 5.2 | 20.4 | 13.8 | 60.6 | 269 |
| VTE3 + VTE4 | A28093 | 8.3 | 3.3 | 21.2 | 67.2 | 302 |
| VTE3 + VTE4 | A27934 | 5.0 | 3.1 | 16.5 | 75.4 | 260 |
| VTE3 + VTE4 | A28096 | 5.0 | 3.4 | 12.5 | 79.1 | 320 |
| VTE3 + VTE4 | A27935 | 1.9 | 2.7 | 6.9 | 88.5 | 261 |
| VTE3 + VTE4 | A27998 | 1.9 | 2.5 | 6.0 | 89.6 | 318 |
| VTE3 + VTE4 | A27711 | 1.0 | 3.3 | 4.3 | 91.4 | 302 |
Data are from pools of 10 seeds each. nd, below the detection limit.
Figure 6.
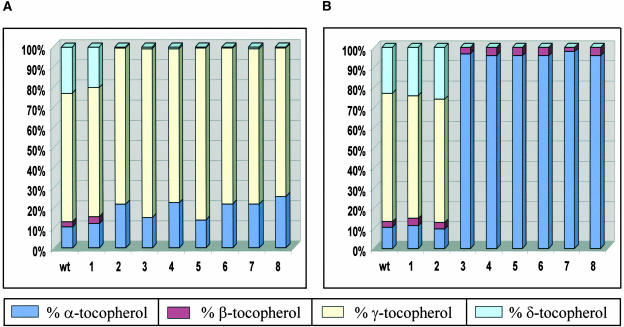
Tocopherol Composition Analysis of Segregating R1 Single Soybean Seed.
(A) Data for wild-type control and eight seeds of transgenic line 27930, containing P7Sα′-At-VTE3.
(B) Data for wild-type control and eight seeds of transgenic line 28906, containing both P7Sα′-At-VTE3 and P7Sα′-At-VTE4.
Simultaneous expression of At-VTE3 and At-VTE4 in soybean seeds resulted in a dramatic increase in α-tocopherol and decrease in δ-tocopherol levels (Table 2). This finding is consistent with the hypothesis that the At-VTE4 enzyme efficiently converts γ-tocopherol produced by At-VTE3 to α-tocopherol. One line, 27936, did not fit this pattern and showed the phenotype seen if only At-VTE4 enzyme were produced: an increase in α- and β-tocopherol with a decrease in γ- and δ-tocopherol. Single seed analysis was performed on seeds from several lines transformed with the double gene construct. Analysis of seeds from a representative line (A28096) revealed the presence of two null seeds and six seeds showing the At-VTE3 plus At-VTE4 expression phenotype (Figure 6B). These seeds contained substantial levels of α-tocopherol (96 to 98%) and β-tocopherol (2 to 4%), whereas δ- and γ-tocopherol levels were below the detection limit (<0.2%). Similar results were seen in single seed analysis of other lines (data not shown). Seed-specific expression of At-VTE3 and At-VTE4 as single genes or in combination did not cause a significant effect on total tocopherol levels in the seed (Table 2).
DISCUSSION
We report the use of Arabidopsis “forward” genetics to identify a gene that encodes a plant MPBQ methyltransferase, At-VTE3, and demonstrate its efficacy in the conversion of δ- and β-tocopherol to γ- and α-tocopherol in transgenic soybean seed. Eighteen mutants with increased levels of seed δ-tocopherol were identified in a screen of ∼8000 mutagenized Arabidopsis lines. The range of increase in δ-tocopherol was 4- to 25-fold (Figure 2C). This change in δ-tocopherol was accompanied by a reduction of γ-tocopherol in the seed, resulting in no net change in total tocopherol levels. It seemed likely that this change in composition could result from a mutation affecting the activity of the enzyme responsible for the methylation of MPBQ (Figure 1). A reduction in activity in this enzyme would cause more MPBQ to be converted to δ-tocopherol via the tocopherol cyclase enzyme, VTE1. Because VTE4 activity is limiting in seeds (Shintani and DellaPenna, 1998), efficient conversion of δ- to β-tocopherol is not expected, and this would result in the accumulation of abnormally large amounts of δ-tocopherol. We hypothesized three different molecular mechanisms that could lead to this recessive phenotype: a mutation in the methyltransferase enzyme structural gene, reduced synthesis of a cofactor required for the activity of this enzyme, or production of a defective regulator of the enzyme.
The vte3-1 (hd2) mutant was chosen for map-based cloning because it had a 25-fold increase in δ-tocopherol levels (Figure 2C). Genetic analysis narrowed the mutation-containing region to the area between a marker at position 1563 on BAC T12C14 and the end of the chromosome (Figure 3C). A Pfam Hidden Markov models analysis of this region found homology between MAA21_40 (At-VTE3) and a ubiquinone methyltransferase. For this reason, At-VTE3 was considered a strong candidate gene because tocopherols and ubiquinone are both isoprenoids with similar biosynthetic pathways. The homology of genes that encode proteins in these two isoprenoid pathways recently led to the identification of another tocopherol gene, homogentisate phytyltransferase, VTE2 (Collakova and DellaPenna, 2001; Schledz et al., 2001; Savidge et al., 2002).
Sequencing of the At-VTE3 open reading frame from five independently isolated hd mutants showed unique single base pair changes, all predicted to cause amino acid substitutions. Interestingly, four of the five alleles have mutations in the first exon (Figure 3D), suggesting that this protein region may be crucial for enzyme activity or that mutations in this area are less detrimental. It is noteworthy that none of the five mutations was predicted to create a null allele, such as a splice site change or protein truncation. This finding, coupled with the observation that significant amounts of γ-tocopherol accumulate as a result of even the strongest vte3 mutant allele (Figure 2C), led us to hypothesize that complete loss-of-function mutations of At-VTE3 may be lethal or may have an unexpected effect on seed tocopherol. Consistent with this hypothesis, the At-vte3-1 mutant protein showed some residual ability to perform the methyltransferase reaction (Figure 5B). Another line of evidence that VTE3 is an essential gene comes from a recent report on the apg1 mutation (Motohashi et al., 2003). This vte3 Ds insertion mutant did not progress beyond the seedling stage when grown on soil and presumably has a complete loss-of-function allele. The data presented by Motohashi et al. (2003) suggest a role for At-VTE3 in the methylation of 2-methyl-6-solanylbenzoquinol, the plastoquinone 9 precursor. This observation is consistent with the 2-methyl-6-solanylbenzoquinol methyltransferase activity reported with Syn-VTE3 by Shintani et al. (2002).
Mutations in multiple genes can lead to the high δ-tocopherol phenotype, because 13 of the original 18 hd mutants did not have lesions in At-VTE3. Genetic mapping studies with the remaining hd mutants demonstrated the existence of at least two other loci (K. Lincoln and S. Norris, unpublished data). It will be interesting to define the biochemical mechanisms that lead to high seed δ-tocopherol accumulation in these mutants.
Enzyme assays and transgenic Arabidopsis and soybean data support the hypothesis that At-VTE3 encodes a tocopherol methyltransferase responsible for methylating MPBQ. The transgenic Arabidopsis data indicate that the expression of At-VTE3 under the control of the napin seed-specific promoter substantially restores the wild-type phenotype to vte3-1 seed (Figure 4B). Interestingly, the napin–At-VTE3 construct further decreased the already low levels of δ-tocopherol that normally are present in wild-type Arabidopsis seed (Figure 4B). The decrease in δ-tocopherol in these transgenic lines was accompanied consistently by a corresponding increase in γ-tocopherol.
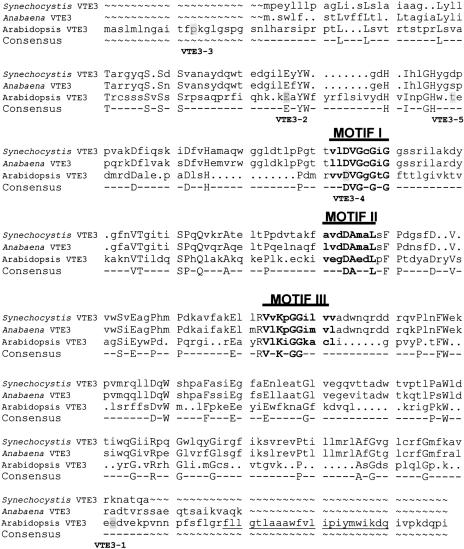
The inferred At-VTE3 protein-coding region has structural features that are consistent with its identity as an S-adenosylmethionine–dependent methyltransferase, as shown in Figure 7. Three regions of sequence similarity—called motifs I, II, and III—are seen in a broad group of methyltransferases (Kagan and Clarke, 1994). Motif I consists of a nine-residue consensus sequence, (V/I/L)(L/V)(D/E)(V/I)G(G/C)G(T/P)G, whereas motif II comprises an eight-residue conserved region, (P/G)(Q/T)(F/Y/A)DA(I/V/Y)(F/I)(C/V/L), located 36 to 90 (average of 57) amino acids after the terminal Gly of motif I. Finally motif III consists of 10 amino acids, LL(R/K)PGG(R/I/L)(L/I)(L/F/I/V)(I/L), and is located 12 to 38 (average of 22) residues after motif II. The At-VTE3 protein sequence contains matches to all 9 amino acids of motif I and is followed 40 residues later by a sequence that matches 3 of the 8 residues typically found in motif II. Thirty-four amino acids downstream from the end of motif II is a sequence that matches 5 of the 10 residues typically found in motif III. In contrast to the findings of Shintani et al. (2002) and Motohashi et al. (2003), motif II was assigned to a sequence 11 and 12 amino acids upstream of the sequences chosen by these groups as motif II for Syn-VTE3 and At-VTE3, respectively. This choice was made to better reflect the highly conserved AspAla amino acid motif. Two of the five vte3 mutant alleles identified in the screen had changes in amino acids that are conserved between At-VTE3 and the two cyanobacterial VTE3 proteins (Figure 7).
Figure 7.
Protein Sequence Alignment of VTE3 from Arabidopsis and Two Cyanobacteria.
The protein sequence of Arabidopsis VTE3 was aligned with the Anabaena VTE3 and Synechocystis VTE3 sequences. Motifs I to III are as described (Kagan and Clarke, 1994). Labels under the sequence correspond to the location of the amino acid change in each of the five hd mutants as described in Figure 3, and the affected amino acid residues are indicated by shading. A putative C-terminal membrane anchor proposed by Motohashi et al. (2003) is underlined.
At-VTE3 has a surprisingly low level of sequence similarity (∼35% amino acid identity over an area of 68 amino acids) with the cyanobacterial VTE3 genes (Figure 7), which is why homology searching of full Arabidopsis genomic and EST sequences did not reveal its presence. Furthermore, a transmembrane domain identified by Motohashi et al. (2003) at the C-terminal end of At-VTE3 is located in a C-terminal extension of the Arabidopsis sequence and cannot be defined clearly in the Syn-VTE3 sequence (Figure 7). One possible reason for this difference in sequence homology could relate to substrate specificity. Syn-VTE3 was found to be active on both MPBQ and 2-methylsolanylbenzoquinol (Shintani et al., 2002), suggesting that prenyl side chain length is not a factor in substrate preference for this enzyme. However, characterization of the Arabidopsis apg1 mutant suggests that At-VTE3 is involved in the methylation of 2-methylsolanylbenzoquinol as well (Motohashi et al., 2003). It would be interesting to test the activity of At-VTE3 on 2-methylsolanylbenzoquinol to determine whether the sequence differences between the plant and cyanobacterial VTE3 proteins can be explained partially by a divergence in substrate specificity. It also would be interesting to examine whether either VTE3 can use the geranylgeranyl-substituted quinols that produce tocotrienol, because biochemical assays found that geranylgeranyl-substituted quinols were methylated to a greater extent than phytyl-substituted quinols in spinach chloroplasts (Soll and Schultz, 1979).
The transgenic soy seed data clearly demonstrate that At-VTE3 will be useful for modifying the composition and thus the quality of the tocopherols in plants. The expression of At-VTE3 protein caused a significant decrease in seed δ- and β-tocopherol, with a corresponding increase in α- and γ-tocopherol. The increase in α-tocopherol likely is the result of a larger γ-tocopherol pool available as a substrate for VTE4. Expression of the combination of At-VTE4 and At-VTE3 led to the accumulation of >95% α-tocopherol in the seed, an eightfold increase in α-tocopherol, and a fivefold increase in vitamin E equivalents. These data show that simultaneous overexpression of At-VTE3 and At-VTE4 can lead to methylation at both the R2 and R3 sites in the tocopherol pool, resulting in nearly complete conversion of all tocopherols present in the seed to α-tocopherol. These findings indicate that by modulating the expression of both VTE3 and VTE4, we may be able to tailor the tocopherol composition of a variety of different crops for both nutritional and food uses. Because oil accounts for 20 to 30% of the tocopherols in the U.S. diet and the majority of the oil consumed in the United States is from soybean, an eightfold increase in α-tocopherol has the potential to significantly alter the vitamin E intake of the population.
Soybean is not the only crop in which increased VTE3 activity could have a beneficial impact. Several other important oilseed crop species have a relatively high percentage of seed δ-tocopherol: for example, safflower and maize seed oil contain 30 and 7% δ-tocopherol, respectively (Hess, 1993). Our results with soybean suggest that transgenic expression of VTE3 under the control of a seed-specific promoter could have the beneficial effect of reducing δ-tocopherol in favor of γ-tocopherol in these crops. With the complete sequencing of the Arabidopsis genome as well as significant progress in the genome sequencing of crops such as rice, an important next step for crop science is to use this information to positively affect crop productivity and human health. Our results show the power of model organism functional genomics in this process and its potential application to an important crop plant. The plant VTE3 gene is one of the first of many potentially useful genes for crops obtained using these techniques.
METHODS
Plants and Growth Conditions
For the mutant screen, Arabidopsis thaliana was grown under continuous illumination at ∼120 μmol·m−2·s−1 and 21°C in Conviron MTPC432 chambers (Conviron, Winnipeg, Canada). The plants were grown in Metro Mix 200 soil (Scotts, Marysville, OH) in two three-eighth-inch pots at a density of 32 plants per flat, three flats to a group of 96. The three flats of a group were grown side by side at all times. A collection of Columbia and Landsberg erecta ethyl methanesulfonate–treated mutagenized plants was from two sources, Lehle Seeds (Red Rock, TX) and those generated at Cereon Genomics by the method described previously (Barczak et al., 1995). For Agrobacterium tumefaciens–mediated germline transformation, Arabidopsis plants were grown in a 21°C chamber under light at 200 μmol·m−2·s−1 for a 16-h photoperiod. After transformation, T0 plants recovering from transformation were grown at 19°C and T1 plants were grown at 21°C. Transgenic plants were selected by spraying with a 1:200 dilution of Finale (AgrEvo Environmental Health, Montvale, NJ) at both 7 and 14 days after seeding. Transformed hemizygous plants were grown to maturity, and the T2 segregating seeds were harvested for tocopherol analysis.
Tocopherol Analysis
Tocopherol analysis of Arabidopsis seed was performed as described (Savidge et al., 2002). For pooled soybean (Glycine max) seed analysis, 10 seed from each individual plant line were ground to a fine powder using a three-quarter-inch steel ball and vigorous shaking. For single seed analysis, one seed was treated as described above. For either pooled or single soy seed, 20 to 40 mg of ground seed was weighed into 2-mL polypropylene tubes and extracted as described for Arabidopsis seed by Savidge et al. (2002). Tocopherol analysis of ground Arabidopsis and soy seed was as described by Savidge et al. (2002).
Mutant Isolation and Gene Identification
For the mutant screen, ∼8000 M2 plants were grown in groups of 96, three flats of 32 wells each. M3 seed families were harvested from each of the M2 plants for tocopherol extraction and analysis. Individual plant lines with altered tocopherol composition were selected as putative mutants. To both confirm the mutant phenotype and attempt to isolate true breeding lines, tocopherol levels were analyzed in M4 seed. For map-based cloning, a homozygous mutant line with a 25-fold increase in seed δ-tocopherol, referred to as hd2, was outcrossed to wild-type Columbia-0 (Col-0) to generate a mapping population. F3 seed were analyzed for tocopherol composition, and lines presumed to be homozygous at the mutant locus were selected for genotyping. DNA for genotyping was isolated from pooled F3 seed of these lines using the Qiagen DNeasy 96 Plant Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Genotyping was performed using the TaqMan genotyping system (Applied Biosystems, Foster City, CA) with 25 single nucleotide polymorphism markers distributed throughout the genome (Jander et al., 2002) and additional markers near the telomere of chromosome III (Figures 3A and 3B).
For sequencing of the candidate Arabidopsis gene, PCR amplification was performed with DNA isolated from the mutant or wild-type lines using primer pairs 1 to 11 listed in Table 3. The reaction mixture for PCR was preincubated for 10 min at 94°C. The product then was amplified for 45 cycles of 94°C for 15s, 56°C for 15s, and 72°C for 1.5 min, followed by a 10-min hold at 72°C.
Table 3.
Primers Used for Gene Identification and Amplification
| No. | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) |
|---|---|---|
| 1 | TGTAAAACGACGGCCAGTTGCTGAAAGTTGAAAAGAGCAA | CAGGAAACAGCTATGACCCAATTTGATCAATGTTCCACGA |
| 2 | TGTAAAACGACGGCCAGTAGCTATGCGGATTGATGGTC | CAGGAAACAGCTATGACCTCCTCCTGGGAACTCTAGCA |
| 3 | TGTAAAACGACGGCCAGTTGCTGACTTGCGAGTTTTTG | CAGGAAACAGCTATGACCCCTGTCAACAACCCCTTCTC |
| 4 | TGTAAAACGACGGCCAGTCCACAAGAGGGGTTTACAATG | CAGGAAACAGCTATGACCACCCAACCTTCTGGCTCTCT |
| 5 | TGTAAAACGACGGCCAGTGGTCTTTGGGAACGATCTGA | CAGGAAACAGCTATGACCAGGGAAGCGTACAGGGTTCT |
| 6 | TGTAAAACGACGGCCAGTCCTCTTGAGCTGAACGTCCT | CAGGAAACAGCTATGACCGGCGGAACTGGTTTCACTAC |
| 7 | TGTAAAACGACGGCCAGTTGTCAGCATAATCGGTTGGA | CAGGAAACAGCTATGACCTCCCCAAAGGTTTAGGTTCC |
| 8 | TGTAAAACGACGGCCAGTAAGCCTCCTTCTTGTGCTGA | CAGGAAACAGCTATGACCCGACTTTTCCCTTCCATTTG |
| 9 | TGTAAAACGACGGCCAGTTGGAGGTTCGGGTAACTGAG | CAGGAAACAGCTATGACCCATCCTCTCGCTAGCAGGTC |
| 10 | TGTAAAACGACGGCCAGTGGAACCAGGGGAACCTAAAC | CAGGAAACAGCTATGACCGCCGTGAGAAACAGACTCCT |
| 11 | TGTAAAACGACGGCCAGTCAAATGGAAGGGAAAAGTCG | CAGGAAACAGCTATGACCGATCCAAAGAGAACCCAGCA |
| 12 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTAGAAGGAGA- TAGAACCATGAGTTGGTTGTTTTCTACACTGG |
GGGGACCACTTTGTACAAGAAAGCTGGGTCCTATTACTT- TTGAGCAACCTTGATCG |
| 13 | GGGGACAAGCAAAAAAGCAGGCTTAGAAGGAGATAGAAC- CATGAGTTGGTTGTTTTCTACACTGGTTTGTA |
GGGGACCACTTTGTACAAGAAAGCTGGGTCCTATTACTT- TTGAGCAACCTTGATCG |
| 14 | CATGCCATGGGAATGAAAGCAACTCTAGCAG | GTCAGAATTCTTATTAGAGTGGCTTCTGGCAAG |
| 15 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTAGAAGGAGA- TAGAACCATGGCTACTAGATGCAGCAGCAGCAGC |
GGGGACCACTTTGTACAAGAAAGCTGGGTCCTGCAGGTC- AGATGGGTTGGTCTTTGGGAACG |
Plant Vector Construction and Transformation
The At-VTE3 cDNA was amplified from an Arabidopsis accession Col-0 leaf cDNA library using the Expand High-Fidelity PCR System (Roche Molecular Biochemicals, Indianapolis, IN) and primer pair 12 (Table 3). The sequence of Syn-VTE3 was used to identify an Anab-VTE3 coding sequence (Kaneko et al., 2001). This Anab-VTE3 sequence was amplified from genomic DNA derived from 3-day-old Anabaena sp (ATCC 27893) cultures. Cultures were spun, and the pellet was washed with 1 mL of PBS to remove the medium. The suspension was centrifuged, and the supernatant was discarded. The pellet was resuspended in 1 mL of water and boiled for 10 min. Anabaena amplification reactions contained 10 μL of boiled Anabaena extract, the Expand High-Fidelity PCR System, and primer pair 13 (Table 3).
After amplification, PCR products were purified using a Qiagen PCR cleanup column and cloned into pDONR201 using the Gateway cloning system (Life Technologies, Rockville, MD). DNA sequences were confirmed and then recloned into a napin-driven cassette derived from pCGN3223 (Kridl et al., 1991) in a Gateway-compatible binary destination vector containing the BAR-selectable marker under the control of the Pe35S promoter. The Anab-VTE3 gene was cloned as a translational fusion with the plastid target peptide CTP1 from Arabidopsis SSU (Stark et al., 1992) to target this protein to the plastid. The resulting expression vectors (Figure 4A) were electroporated into A. tumefaciens strain ABI cells and grown under standard conditions (McBride et al., 1994), reconfirmed by restriction analysis, and transformed into Arabidopsis using the floral-dip method (Clough and Bent, 1998).
Three soy expression constructs were synthesized: two constructs containing either At-VTE3 or At-VTE4 under the control of the P7Sα′ promoter (Chen et al., 1986; Wang and Dubois, 2003) with a 3′ untranslated region from pea SSU, and a construct containing both of these expression cassettes. The At-VTE4 cDNA was obtained by PCR amplification of Arabidopsis Col-0 using primer pair 14. The PCR product was digested with EcoRI-NcoI, gel-purified, and sequenced. Sequencing of the fragment revealed that it exhibited two nucleotide changes compared with the published sequence (Shintani and DellaPenna, 1998). Although the first substitution (position 345, C to T) was translationally silent, the second nucleotide change (position 523, T to G) resulted in an amino acid change of Ser to Ala.
The At-VTE3 and At-VTE4 expression cassettes were cloned individually and jointly into a plant binary expression vector containing a Pe35S-CP4 gene as a selectable marker (Martinell et al., 2002). A. tumefaciens–mediated germline transformation of soybean was performed as described previously (Martinell et al., 2002) using freshly germinated soybean meristems that were induced to form shoots directly. Transgenic plants were grown in a greenhouse with a 10-h photoperiod, a daytime temperature of 29°C, and a nighttime temperature of 24°C. Plants were grown to maturity, and seed were harvested for analysis.
Expression of Wild-Type At-VTE3, Mutant At-vte3-1, and Anab-VTE3 Proteins in a Prokaryotic Expression System
The computer program ChloroP (Emanuelsson et al., 1999) (http://www.cbs.dtu.dk/services/ChloroP/) predicted the chloroplast-targeting peptide cleavage site of the plant At-VTE3 protein to be between amino acids 51 and 52. Based on this information, wild-type At-VTE3 from ecotype Col-0 and the vte3-1 mutant gene from ecotype Landsberg erecta were engineered to remove the predicted chloroplast target peptide and add an N-terminal Met to allow for the expression of these mature proteins in an E. coli expression system.
The mature At-VTE3 and vte3-1 sequences were amplified by PCR from cDNA using the Expand High-Fidelity PCR System as described previously and primer pair 15 (Table 3). PCR products were cloned into pDONR201 and sequenced and then At-VTE3, vte3-1, and Anab-VTE3 (as described previously) sequences were recombined behind the T7 promoter in the prokaryotic expression vector pET-DEST42 (Life Technologies) using the Gateway cloning system (Figure 5A). Overnight starter cultures of E. coli BL21 (DE3) cells transformed previously with one of these Anab-VTE3, At-VTE3, or vte3-1 prokaryotic expression constructs were inoculated into 100 mL of Luria-Bertani medium with appropriate antibiotics, and the cultures were grown at 25°C to a density of OD600 = 0.6, at which time isopropylthio-β-galactoside (0.4 mM) was added to induce protein expression. Cells were grown for an additional 3 h at 25°C, chilled on ice for 5 min, and spun down at 5000g for 10 min. The cell pellet was stored at −80°C overnight after thoroughly aspirating off the supernatant.
2-Methyl-6-Phytylbenzoquinol (MPBQ) Methyltransferase Enzyme Assay
The substrate MPBQ and a standard for the reaction product 2,3-dimethyl-5-phytylbenzoquinol for the methyltransferase assay were prepared as described previously (Soll and Schultz, 1980; Henry et al., 1987). Cell pellets expressing Anab-VTE3, At-VTE3, or vte3-1 protein were thawed on ice and resuspended in 4 mL of extraction buffer XB [10 mM Hepes-KOH, pH 7.8, 5 mM DTT, 1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride, 0.1 mM aprotinin, and 1 mg/mL leupeptin]. Cells were disrupted using a French press by making two passes through the pressure cell at 20,000 p.s.i. Triton X-100 was added to a final concentration of 1%, and then the cells were incubated on ice for 1 h. The cell homogenate was centrifuged at 5000g for 10 min at 4°C to separate cell debris. Enzyme assays, performed on the same day as cell extraction, were run in 10-mL polypropylene culture tubes with a final volume of 1 mL. Fifty microliters of the cell extract was added to a 950-μL reaction mixture consisting of 50 mM Tris-HCl, pH 8.0, 5 mM DTT, 100 μM MPBQ, and 0.5% Tween 80 (added directly to the phytylbenzoquinol after evaporating the solvent). The reaction was initiated by adding 1.7 μM 14C-S-adenosylmethionine (58 μCi/μmol) and incubated for 1 h at 30°C in the dark.
The reaction mixture was extracted with 4 mL of 2:1 CHCl3/methanol with 1 mg/mL butylated hydroxy toluene and vortexed for 30 s. The organic phase (bottom) was transferred to a fresh 15-mL glass tube after a 5-min low-speed centrifugation. The CHCl3 was evaporated off under a stream of nitrogen gas at 37°C for ∼15 min. The residue was dissolved in 200 μL of ethanol containing 1% pyrogallol, vortexed for 30s, filtered into a brown liquid chromatography vial equipped with an insert, and analyzed by HPLC using a normal phase column (4.6 × 250 mm Zorbax Sil; Agilent Technologies, Palo Alto, CA). Samples of 50 μL were injected onto the column, and quantitation of the 14C-labeled reaction products was performed using a flow scintillation counter (Packard 500TR). The elution program was an isocratic flow of 10% methyl-tert-butyl-ether in hexane at 1.5 mL/min for 12 min. As a positive control, a pea (Pisum sativum) chloroplast concentrate, which is known to have endogenous VTE3 activity, was prepared as described previously (Arango and Heise, 1998).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Henry E. Valentin, henry.e.valentin@monsanto.com.
Accession Numbers
The GenBank accession numbers for the genes mentioned in this article are as follows: CAB87794 (At-VTE3), BAA18485 (Syn-VTE3), BAA96953 (a ubiquinone methyltransferase), and NP_486161 (Anab-VTE3).
NOTE ADDED IN PROOF
While this manuscript was in press, a paper appeared describing cloning of the Arabidopsis VTE3 gene and a role for this enzyme in tocopherol and plastoquinone synthesis. The citation is Cheng, Z., Sattler., S., Maeda, H., Sakuragi, Y., Bryant, D.A., and DellaPenna, D. (2003). Highly divergent methyltransferases catalyze a conserved reaction in tocopherol and plastoquinone synthesis in cyanobacteria and photosynthetic eukaryotes. Plant Cell 15, 2343–2356.
Acknowledgments
This work was a team effort between four Monsanto research sites: Davis, California; Madison, Wisconsin; St. Louis, Missouri; and the former Cereon site in Cambridge, Massachusetts. We thank our current and past colleagues at all of these sites, particularly those in plant transformation, the plant growth facilities, the greenhouses, and leadership roles for their expert assistance and support. We also thank our colleagues at Renessen (Bannockburn, IL) for their staunch support and advice. Any views, opinions, or conclusions expressed in this article are those of the author (R.L.L.) and do not represent the official views, opinions, or policy of the National Science Foundation.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.015875.
References
- Arango, Y., and Heise, K.-P. (1998). Tocopherol synthesis from homogentisate in Capsicum annuum L. (yellow pepper) chromoplast membranes: Evidence for tocopherol cyclase. Biochem. J. 336, 531–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barczak, A.J., Zhao, J., Pruitt, K.D., and Last, R.L. (1995). 5-Fluoroindole resistance identifies tryptophan synthase beta subunit mutants in Arabidopsis thaliana. Genetics 140, 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R., Griffiths-Jones, S., Howe, K.L., Marshall, M., and Sonnhammer, E.L.L. (2002). The Pfam protein families database. Nucleic Acids Res. 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramley, P.M., Elmadfa, I., Kafatos, A., Kelly, F.J., Manios, Y., Roxborough, H.E., Schuch, W., Sheehy, P.J.A., and Wagner, K.-H. (2000). Vitamin E. J. Sci. Food Agric. 80, 913–938. [Google Scholar]
- Buring, J.E., and Hennekens, C.H. (1997). Antioxidant vitamins and cardiovascular disease. Nutr. Rev. 55, S53–S60. [DOI] [PubMed] [Google Scholar]
- Chen, Z.L., Schuler, M.A., and Beachy, R.N. (1986). Functional analysis of regulatory elements in a plant embryo-specific gene. Proc. Natl. Acad. Sci. USA 83, 8560–8564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Collakova, E., and DellaPenna, D. (2001). Isolation and functional analysis of homogentisate phytyltransferase from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiol. 127, 1113–1124. [PMC free article] [PubMed] [Google Scholar]
- Eitenmiller, R.R. (1997). Vitamin E content of fats and oils: Nutritional implications. Food Technol. 51, 78–81. [Google Scholar]
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, H.M., and Bishop, K.S. (1922). On the existence of a hitherto unrecognized dietary factor essential for reproduction. Science 56, 650–651. [DOI] [PubMed] [Google Scholar]
- Grusak, M.A., and DellaPenna, D. (1999). Improving the nutrient composition of plants to enhance human nutrition and health. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 133–161. [DOI] [PubMed] [Google Scholar]
- Hendrich, S., Lee, K.W., Xu, X., Wang, H.J., and Murphy, P.A. (1994). Defining food components as new nutrients. J. Nutr. 124, 1789s–1792s. [DOI] [PubMed] [Google Scholar]
- Henry, A., Powls, R., and Pennock, J.F. (1987). Intermediates of tocopherol biosynthesis in the unicellular alga Scenedesmus obliquus: The presence of three isomeric methylphytylbenzoquinones. Biochem. J. 242, 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, J.L. (1993). Vitamin E, alpha tocopherol. In Antioxidants in Higher Plants, R. Alscher and J. Hess, eds (Boca Ranton, FL: CRC Press), pp. 111–134.
- Jander, G., Norris, S.R., Rounsley, S.D., Bush, D.F., Levin, I., and Last, R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129, 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan, R.M., and Clarke, S. (1994). Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch. Biochem. Biophys. 310, 417–427. [DOI] [PubMed] [Google Scholar]
- Kaneko, T., et al. (2001). Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8, 205–213. [DOI] [PubMed] [Google Scholar]
- Kridl, J., McCarter, D.W., Rose, R.E., Scherer, D.E., Knutzon, D.S., Radke, S.E., and Knauf, V.C. (1991). Isolation and characterization of an expressed napin gene from Brassica napus. Seed Sci. Res. 1, 209–219. [Google Scholar]
- Martinell, B.J., Julson, L.S., Emler, C.A., Huang, Y., McCabe, D.E., and Williams, E.J. (2002). Soybean Agrobacterium transformation method. U.S. Patent 6,384,301, Monsanto Technology, St. Louis, MO. [Google Scholar]
- McBride, K.E., Schaaf, D.J., Daley, M., and Stalker, D.M. (1994). Controlled expression of plastid transgenes in plants based on a nuclear DNA-encoded and plastid-targeted T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 91, 7301–7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi, R., Ito, T., Kobayashi, M., Taji, T., Nagata, N., Asami, T., Yoshida, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2003). Functional analysis of the 37 kDa inner envelope membrane polypeptide in chloroplast biogenesis using a Ds-tagged Arabidopsis pale-green mutant. Plant J. 34, 719–731. [DOI] [PubMed] [Google Scholar]
- Munné-Bosch, S., and Alegre, L. (2002). The function of tocopherols and tocotrienols in plants. Crit. Rev. Plant Sci. 21, 31–57. [Google Scholar]
- Porfirova, S., Bergmüller, E., Tropf, S., Lemke, R., and Dörmann, P. (2002). Isolation of an Arabidopsis mutant lacking vitamin E and identification of a cyclase essential for all tocopherol biosynthesis. Proc. Natl. Acad. Sci. USA 99, 12495–12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencher, L.M., Miao, L., Sinha, N., and Lucas, W.J. (2001). Sucrose Export Defective1 encodes a novel protein implicated in chloroplast-to-nucleus signaling. Plant Cell 13, 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi, A.A., Bradlow, B.A., Brace, L., Manganello, J., Peterson, D.M., Pearce, B.C., Wright, J.J.K., Gapor, A., and Elson, C.E. (1995). Response of hypercholesterolemic subjects to administration of tocotrienols. Lipids 30, 1171–1177. [DOI] [PubMed] [Google Scholar]
- Qureshi, N., and Qureshi, A.A. (1993). Tocotrienols: Novel hypocholesterolemic agents with antioxidant properties. In Vitamin E in Health and Disease, L. Packer and J. Fuchs, eds (New York: Marcel Dekker), pp. 247–267.
- Savidge, B., Weiss, J.D., Wong, Y.-H.H., Lassner, M.W., Mitsky, T.A., Shewmaker, C.K., Post-Beittenmiller, D., and Valentin, H.E. (2002). Isolation and characterization of homogentisate phytyltransferase genes from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiol. 129, 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schledz, M., Seidler, A., Beyer, P., and Neuhaus, G. (2001). A novel phytyltransferase from Synechocystis sp. PCC 6803 involved in tocopherol biosynthesis. FEBS Lett. 499, 15–20. [DOI] [PubMed] [Google Scholar]
- Sheppard, A.J., Pennington, J.A., and Weihrauch, J.L. (1993). Analysis and distribution of vitamin E in vegetable oils and foods. In Vitamin E in Health and Disease, L. Packer and J. Fuchs, eds (New York: Marcel Dekker), pp. 9–31.
- Shintani, D., and DellaPenna, D. (1998). Elevating the vitamin E content of plants through metabolic engineering. Science 282, 2098–2100. [DOI] [PubMed] [Google Scholar]
- Shintani, D.K., Cheng, Z., and DellaPenna, D. (2002). The role of 2-methyl-6-phytylbenzoquinone methyltransferase in determining tocopherol composition in Synechocystis sp. PCC 6803. FEBS Lett. 511, 1–5. [DOI] [PubMed] [Google Scholar]
- Soll, J., Kemmerling, M., and Schultz, G. (1980). Tocopherol and plastoquinone synthesis in spinach chloroplast subfractions. Arch. Biochem. Biophys. 204, 544–550. [DOI] [PubMed] [Google Scholar]
- Soll, J., and Schultz, G. (1979). Comparison of geranylgeranyl and phytyl substituted methylquinols in the tocopherol synthesis of spinach chloroplasts. Biochem. Biophys. Res. Commun. 91, 715–720. [DOI] [PubMed] [Google Scholar]
- Soll, J., and Schultz, G. (1980). 2-Methyl-6-phytylquinol and 2,3-dimethyl-5-phytylquinol as precursors of tocopherol synthesis in spinach chloroplasts. Phytochemistry 19, 215–218. [Google Scholar]
- Soll, J., Schultz, G., Joyard, J., Douce, R., and Block, M.A. (1985). Localization and synthesis of prenylquinones in isolated outer and inner envelope membranes from spinach chloroplasts. Arch. Biochem. Biophys. 238, 290–299. [DOI] [PubMed] [Google Scholar]
- Stark, D.M., Timmerman, K.P., Barry, G.F., Preiss, J., and Kishore, G.M. (1992). Regulation of the amount of starch in plant tissues by ADP glucose pyrophosphorylase. Science 258, 287–292. [DOI] [PubMed] [Google Scholar]
- Subramaniam, S., Slater, S., Karberg, K., Chen, R., Valentin, H.E., and Wong, Y.-H. (2001). Nucleic acid sequences to proteins involved in tocopherol synthesis. International patent application WO 01/79472.
- Tan, B. (1989). Palm carotenoids, tocopherols and tocotrienols. J. Am. Oil Chem. Soc. 66, 770–776. [Google Scholar]
- Tangney, C.C. (1997). Vitamin E and cardiovascular disease. Nutr. Today 32, 13–22. [Google Scholar]
- Traber, M.G., and Sies, H. (1996). Vitamin E and humans: Demand and delivery. Annu. Rev. Nutr. 16, 321–347. [DOI] [PubMed] [Google Scholar]
- USDA Foreign Agriculture Service (2003). Oil Seeds: World Market and Trade. (Washington, DC: U.S. Department of Agriculture), pp. 1–26.
- Van Eenennaam, A., Valentin, H.E., Karunanandaa, B., Hao, M., Aasen, E., and Levering, C. (2003). Methyltransferase genes and uses thereof. International patent application WO 03/016482.
- Wang, Q., and Dubois, P. (2003). Seed specific 7Sα promoter for expressing genes in plants. International patent application WO 03/020016.