Abstract
Tissue-specific transcriptional regulation is central to human disease1. To identify regulatory DNA active in human pancreatic islets, we profiled chromatin by FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements)2–4 coupled with high-throughput sequencing. We identified ~80,000 open chromatin sites. Comparison of islet FAIRE-seq to five non-islet cell lines revealed ~3,300 physically linked clusters of islet-selective open chromatin sites, which typically encompassed single genes exhibiting islet-specific expression. We mapped sequence variants to open chromatin sites and found that rs7903146, a TCF7L2 intronic variant strongly associated with type 2 diabetes (T2D)5, is located in islet-selective open chromatin. We show that rs7903146 heterozygotes exhibit allelic imbalance in islet FAIRE signal, and that the variant alters enhancer activity, indicating that genetic variation at this locus acts in cis with local chromatin and regulatory changes. These findings illuminate the tissue-specific organization of cis-regulatory elements, and show that FAIRE-seq can guide identification of regulatory variants important for disease.
Pancreatic islets are composed of endocrine cells that secrete insulin, glucagon, and other polypeptide hormones. Islet cells are essential for glucose homeostasis, and thus elucidating the transcriptional control of islet-cell function and growth has implications for understanding diabetes pathogenesis and treatment6,7. Gene regulatory elements often function by recruiting DNA-associated proteins to specific loci, a process that typically results in local nucleosome eviction8. Nucleosome loss, or formation of “open chromatin”, is an evolutionarily conserved indicator of regulatory activity and can be used as a molecular tag to isolate regions of the genome bound by regulatory factors9.
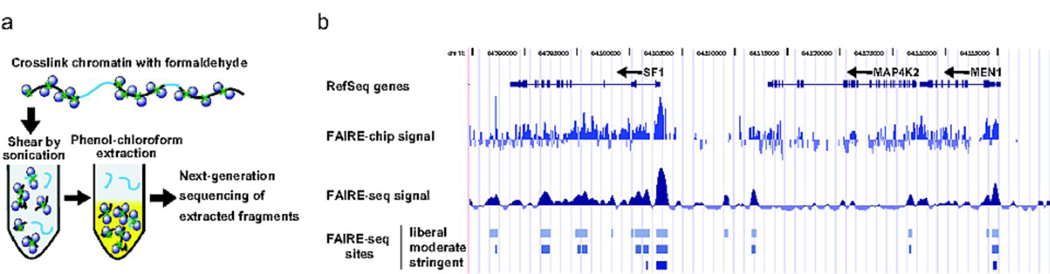
For three samples of purified human pancreatic islets, we used FAIRE2–4 to identify sites of open chromatin (Fig. 1a, Table 1, Supplementary Table 1 online). We technically validated FAIRE-seq by its high concordance to patterns determined by hybridization of the same FAIRE samples to tiling DNA microarrays (Fig. 1b, Supplementary Fig. 1a online). Furthermore, we found that despite differences in age, cause of death, genotype, islet isolation procedures, and level of exocrine cell contaminants, the majority of regions identified by FAIRE in any one sample were also detected in the others (Supplementary Fig. 1b online). Thus, FAIRE-seq is a robust method for characterizing chromatin in islets.
Figure 1. FAIRE-seq in human pancreatic islets.
(a) Chromatin is cross-linked using formaldehyde, sonicated, and subjected to phenol-chloroform extraction. DNA fragments recovered in the aqueous phase are then sequenced. (b) Reads obtained from sequencing were highly concordant with FAIRE signal obtained from tiling microarrays covering the ENCODE pilot project regions. Arrows indicate the direction of gene transcription.
Table 1.
FAIRE-Seq sequence depth and enrichment sites in 3 human islet samples
| FAIRE-seq sites |
||||
|---|---|---|---|---|
| Sequence reads | Liberal | Moderate | Stringent | |
| Sample 1 | 39,359,429 | 205,922 | 99,361 | 18,189 |
| Sample 2 | 25,176,624 | 213,972 | 91,455 | 9,601 |
| Sample 3 | 60,515,180 | 202,783 | 81,546 | 33,305 |
FAIRE was performed on three human pancreatic islet samples (Methods). Sample 3 had the highest purity (Supplementary Table S1 online), and was thus sequenced at greater depth and used for subsequent analysis. Aligned reads were used to call FAIRE sites at three thresholds using F-Seq.
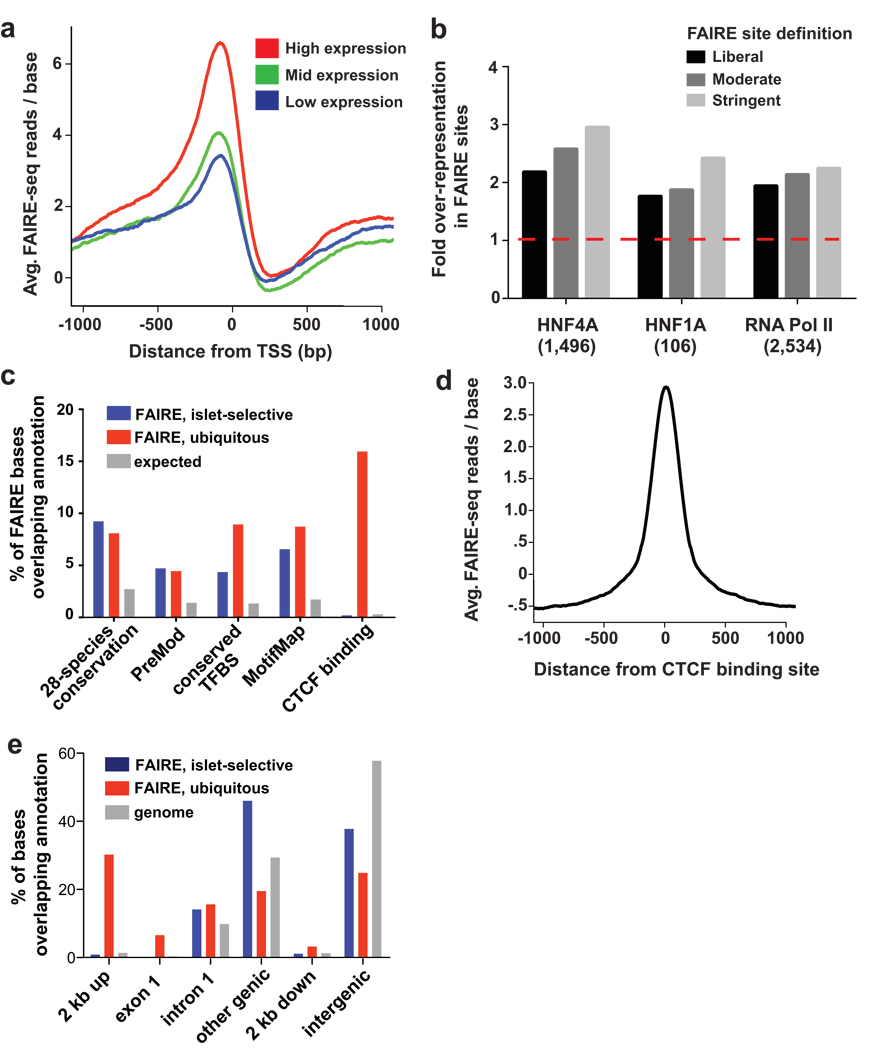
Several lines of evidence indicate that FAIRE reliably identifies active regulatory elements in islets. Consistent with FAIRE in fibroblasts2, the most enriched FAIRE regions were found near known Transcription Start Sites (TSS) (Fig. 2a). Overall, there was a positive relationship between FAIRE signal near TSS and transcript levels in human islets10 (Fig. 2a). Furthermore, promoters previously shown to bind RNA Polymerase II and the transcription factors HNF4A and HNF1A in islets11 were enriched by FAIRE more frequently than other promoter regions (Fig. 2b).
Figure 2. Both proximal and distant FAIRE sites harbor functional regulatory elements.
(a) Genes with high expression in islets (top 20%; red) have more FAIRE enrichment at promoters than genes with moderate (middle 20%; green) or low (bottom 20%; blue) expression. (b) Promoters (−750/+250 bp) bound by RNA Pol II, HNF4A or HNF1A in human islets11 are significantly over-represented among islet FAIRE sites (red dash indicates expected value; all bars: P<0.001). (c) Intergenic islet-selective and ubiquitous FAIRE sites that are located >2 kb from a TSS are enriched for evolutionary conserved sequences (P<0.001), predicted regulatory modules (PreMod, P<0.001), and transcription factor binding sites (conserved TFBS and MotifMap, both P<0.001). CTCF binding, however, is enriched in ubiquitous FAIRE sites only. Over half of intergenic open chromatin sites are coincident with an experimentally or computationally determined functional annotation (expected value for random sites: 27%). (d) Open chromatin is most enriched directly at sites of experimentally determined CTCF binding. (e) In contrast to ubiquitous FAIRE sites, islet-selective FAIRE sites are rarely located within 2 kb upstream of a TSS or in exon 1, and are instead located predominantly in more distal regions. Shown is the percentage of bases covered by each annotation category in islet-selective FAIRE sites (blue), ubiquitous FAIRE sites (red), and the mappable genome (gray).
To extend this observation, we identified regulatory regions that were utilized selectively in islets relative to other cell types. We compared FAIRE-seq data from islets to data from five non-islet cell lines (HeLa-S3, HUVEC, GM12878, HepG2 and K562; Methods) and found that 45% of islet open chromatin sites were unique to islets among this group of cell types. We refer to these sites as islet-selective open chromatin. We identified 340 RefSeq genes with islet-selective open chromatin in the TSS or gene body (Supplementary Table 2 online). This relatively short list included 24 well characterized genes that are selectively expressed in islets (Supplementary Table 3 online), including genes involved in human diabetes (PDX1, ABCC8, SLC30A8, G6PC2, GAD2) and islet developmental regulators (NEUROD1, NKX6-1, PAX6, ISL1)6,7,12–14. Therefore, islet-selective open chromatin detected by FAIRE identifies loci integral to islet-cell biology and disease.
Many sites of open chromatin detected by FAIRE are located in intergenic regions, far (>2 kb) from a known TSS. For these distal sites, evidence also points strongly toward a regulatory function. First, distal intergenic open chromatin sites were enriched in evolutionary conserved sequences, predicted transcription factor binding sites and regulatory modules, regardless of whether the open chromatin was islet-selective or ubiquitous (shared by all six cell types) (Fig. 2c, Supplementary Fig. 1d and Supplementary Table 4 online, Methods). Second, ubiquitous intergenic open chromatin often coincided with binding sites of CTCF15–17 (observed 16%, expected 0.39%, P<0.001) (Fig. 2c), a transcriptional regulator and insulator protein that binds to a large number of genomic sites, many of which are shared in different cell types16. Open chromatin at CTCF sites was centered at the location of the CTCF binding15, suggesting that FAIRE signal is indicative of interactions between regulatory factors and DNA (Fig. 2d). Third, intergenic islet-selective (but not ubiquitous) open chromatin preferentially harbors DNA-binding motifs of pancreatic islet transcription factors, including RFX, TCF1/HNF1, HNF3B, FOXD, and MAF (P<0.01, Supplementary Table 4 online). Notably, whereas ~36% of ubiquitous open chromatin was located within 2 kb upstream of a TSS or in the first exon, only 1% of islet-selective open chromatin was located in these regions (Fig. 2e, Supplementary Fig. 1c online). Collectively, these findings indicate that distal FAIRE sites harbor regulatory elements, and consistent with recent studies of histone modification patterns in enhancer regions18 suggest that most cell-type specific open chromatin is located far from known TSS.
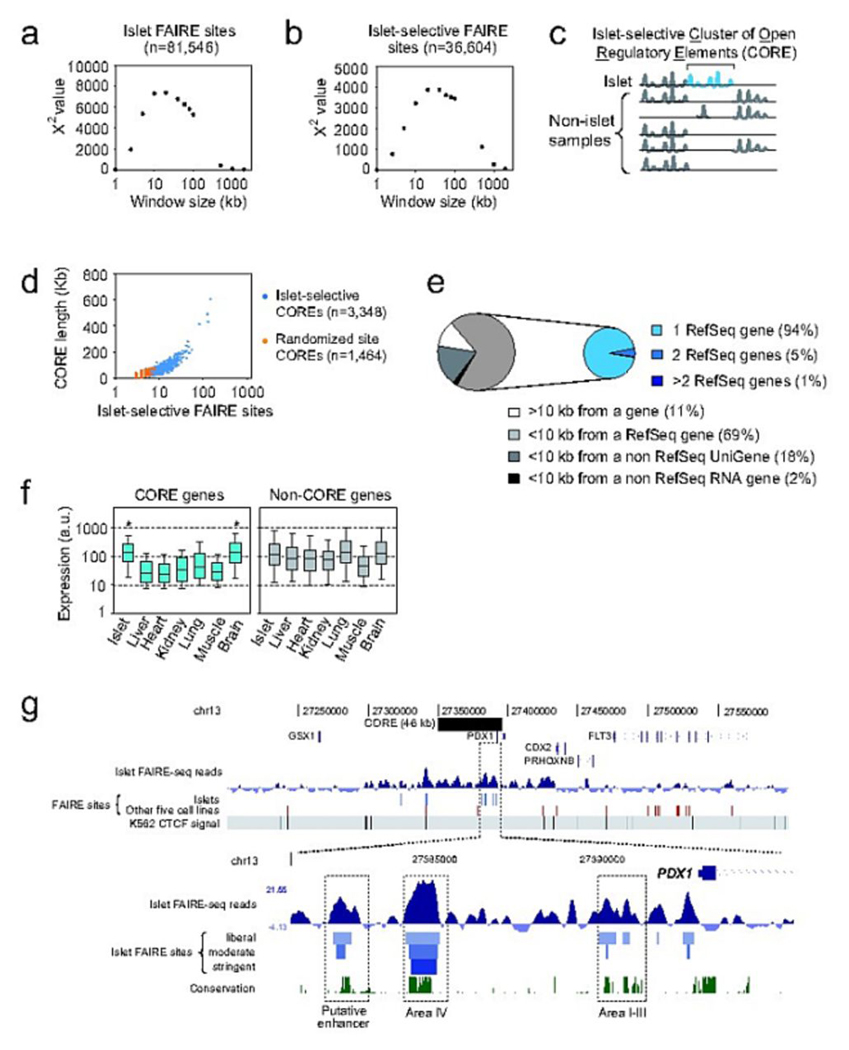
We next sought to link these distal islet-selective elements to specific genes. We examined whether islet-selective sites exhibit a higher-order organization that could point to the existence of functional domains. We found that open chromatin sites were not evenly distributed throughout the genome, but instead were located in physically linked clusters (Fig. 3a). Clustering was also observed with islet-selective open chromatin (Fig. 3b). We identified 3,348 domains containing at least three islet-selective open chromatin sites separated by less than 20 kb, which we call islet-selective COREs (Clusters of Open Regulatory Elements) (Fig. 3c, Supplementary Table 5 online, Methods). Islet-selective COREs had a median size of 25 kb, with the largest containing 148 FAIRE sites spanning 602 kb (Fig. 3d). Consistent with CTCF binding to insulator elements separating chromatin domains19, the frequency of CTCF binding sites was two-fold higher outside than within COREs (P=1.3×10−48). This suggested that islet-selective COREs were functional chromatin domains and provided an avenue to assigning open chromatin sites to genes.
Figure 3. Islet-selective FAIRE sites form Clusters of Open Regulatory Elements (COREs).
(a) FAIRE sites are highly clustered. We divided the genome in windows of varying size (x axis), and calculated a χ2 statistic to determine if the number of windows with 0, 1 or >1 FAIRE sites differed from randomly distributed sites. The highest significance was observed in ~20 kb windows. (b) Same as (a) but for islet-selective sites. (c) We defined islet-selective clusters of open chromatin regulatory elements (COREs) as three or more islet-selective FAIRE sites separated from each other by <20 kb. (d) We identified 3,348 islet-selective COREs (blue points). Fewer COREs were generated using randomized FAIRE sites (orange points), and they were smaller than in vivo COREs (e) Most islet-selective COREs were associated with a single gene. (f) RefSeq genes associated with islet-selective COREs were on average inactive in non-islet human tissues, except for brain. Asterisks indicate P<1×10−5 (one-way ANOVA). (g) Chromatin landscape of the PDX1 locus showing an extended cluster of islet-selective FAIRE sites, contrasting with a closed conformation of the adjacent gut-specific homeodomain gene CDX2. The top panel depicts the density of FAIRE-Seq reads centered on the genomic average density value, the location of moderate stringency FAIRE sites in islets (blue) or in any of the 5 non-islet cells (red), and the binding sites of the CTCF insulator protein in K562 cells. CTCF sites demarcate regions that show broadly consistent FAIRE-Seq enrichment patterns. The bottom panel shows a closer view of a portion of the PDX1 islet-selective CORE, with islet-selective open chromatin sites at previously characterized regulatory elements (Area I–III, Area IV) and in an evolutionarily conserved putative enhancer.
Islet-selective COREs were located within 10 kb of at least one RefSeq gene in 69% of cases (randomized COREs=54%; P=1.5×10−35, Fig. 3e). Of these, 94% were associated with only one gene, and most were contained within 2 kb of gene boundaries (Supplementary Fig. 2 online) suggesting single-gene regulatory function (expected=84%; P=6.2×10−23, Fig. 3e, Methods). Compared to six other primary tissues, genes overlapping islet COREs had higher expression in islets and brain (one-way ANOVA, both tissues P<1×10−5, Fig. 3f), consistent with the neuroendocrine nature of islet-cells20. Islet-selective COREs were also enriched in genes linked to islet-specific functions, including transcription factors, ion channels, and secretory apparatus components (Table 2, Supplementary Table 6 online). Thus, islet-selective COREs are typically linked to single genes that are expressed in an islet-selective manner.
Table 2.
Biological processes enriched among genes associated with islet-selective COREs
| PANTHER Biological Process | P-Value | GO Biological Process | P-Value |
|---|---|---|---|
| mRNA transcription regulation | 2.25E-24 | Small GTPase signal transduction | 5.72E-08 |
| Proteolysis | 1.45E-22 | Vesicle-mediated transport | 5.85E-07 |
| mRNA transcription | 4.95E-21 | Ubiquitin cycle | 9.13E-07 |
| Protein modification | 4.32E-20 | Regulation of signal transduction | 3.44E-06 |
| G-protein mediated signaling | 7.57E-16 | Small GTPase mediated signal transduction | 7.57E-06 |
| Cation transport | 8.83E-14 | Transcription from RNA II promoter | 9.15E-06 |
| Protein phosphorylation | 1.49E-12 | Secretion | 1.05E-05 |
The seven most enriched GO and PANTHER biological process terms are listed. A more comprehensive list is shown in Supplementary Table 6 online.
A subset of islet-selective COREs spanned remarkably broad distances at loci encoding critical regulators of pancreas development and function (Fig. 3g, Supplementary Table 7 and Supplementary Figs. 2 and 3 online). For example, an islet-selective CORE spanned a 46-kb domain containing PDX1, a master regulator of pancreas development and β-cell function7 (Fig. 3g). At this locus, FAIRE sites coincided with previously characterized evolutionarily conserved enhancers named “Area I–IV”21,22 and with uncharacterized putative enhancer sites (Fig. 3h). Other islet-selective COREs included a 94-kb domain 3′ of NKX6-1, an essential regulator of β-cell differentiation23, one located in a cluster of brain-enriched snoRNA and miRNAs24, and another in conserved sequences >400 kb from any annotated gene (Supplementary Fig. 3 and Supplementary Table 7 online for additional examples). Such domains contrasted with loci devoid of open chromatin and known to be inactive in islets (Supplementary Fig. 3q–s). This dataset thus provides a rich resource to dissect cis regulation in pancreatic islets.
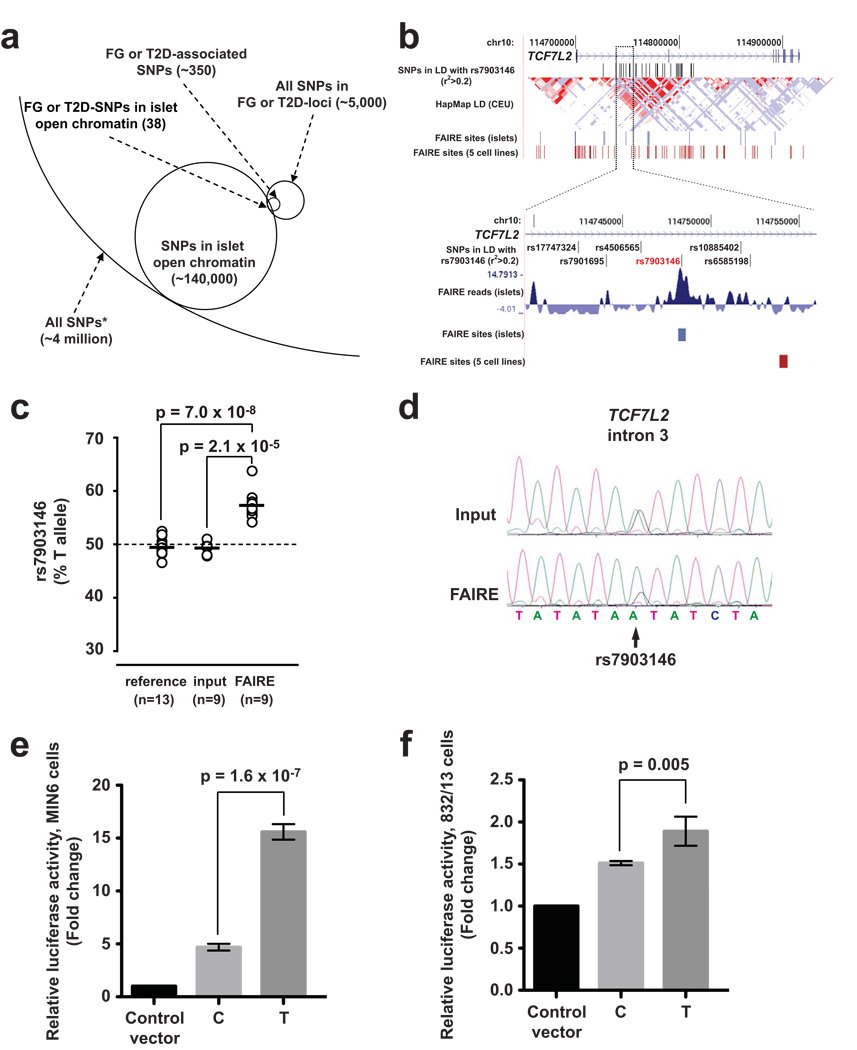
Recent genome-wide association studies for T2D susceptibility have implicated sequence variants at multiple loci, many of which may impair islet-cell function13,14,25–27. Many T2D susceptibility loci do not contain strongly associated variants in protein-coding regions, suggesting that the underlying functional variants regulate gene activity. Furthermore, at each locus, most associated SNPs are not expected to directly affect disease risk and are instead in linkage disequilibrium with one or more functional variant(s). We sought to use our open chromatin map to guide identification of functional regulatory SNPs. We identified known SNPs mapping to islet FAIRE sites and focused on 20 loci harboring variants associated with T2D or fasting glycemia (FG)5,13,14,25,28 (Fig. 4a, and Supplementary Table 8 online). Of 350 SNPs in strong linkage disequilibrium with a reported SNP associated with T2D or FG (Methods), 38 SNPs at 10 loci overlapped islet FAIRE regions (Fig. 4a, and Supplementary Table 8 online). Notably, rs7903146 in TCF7L2, which shows consistent T2D association in samples across diverse ethnic groups29, is located in an islet-selective open chromatin site (Fig. 4b, and Supplementary Fig. 4a online).
Figure 4. Allele-specific open chromatin and enhancer activity at the TCF7L2 locus.
(a) Schematic representation of how FAIRE-seq enables the identification of human sequence variants located in islet open chromatin. From ~4 million SNPs present in dbSNP with average heterozygosity >1%, 38 SNPs associated with T2D or fasting glycemia mapped to islet open chromatin sites. The analysis was carried out with all SNPs in strong linkage disequilibrium (r2>0.8) with an FG- or T2D-associated variant, which are labeled as FG or T2D SNPs, and FAIRE-seq sites identified with a liberal threshold. (b) Among TCF7L2 variants in linkage disequilibrium with rs7903146 (r2>0.2, top panel), only rs7903146 maps to an islet-selective FAIRE site. (c) In all 9 human islet samples that were heterozygous for rs7903146, the risk allele T was more abundant than the non-risk C allele in the open chromatin fraction, in contrast to input DNA or gDNA from unrelated heterozygous individuals. (d) Allelic imbalance for open chromatin at rs7903146 was verified in independent assays using quantitative Sanger sequencing (see also Supplementary Fig. 4b). (e) The risk allele T of rs7903146 exhibits greater enhancer activity than the non-risk allele C in MIN6 cells and (f) 832/13 cells. Standard deviations represent four independent clones for each allele. Results for inserts in the reverse direction are provided in Supplementary Fig. 4. P-values were calculated by two-sided t-test.
The presence of rs7903146 in a FAIRE-enriched site allowed us to test directly whether sequence variation at this locus correlates with chromatin state in islet cells. We tested 31 human islet samples and identified nine individuals heterozygous at rs7903146. Using two independent assays, FAIRE-isolated DNA from heterozygous individuals exhibited a T:C allelic ratio that was significantly greater than observed from input genomic DNA or from genomic DNA from unrelated heterozygote individuals (Real-time PCR: input: 49.3±1.0% T allele, FAIRE: 57.3±2.8% T allele, P=2.1×10−5, Fig. 4c; Quantitative sequencing: input: 57.5 ± 2.7% T allele, FAIRE: 66.2 ± 4.6% T allele, P=0.004, Fig. 4d and Supplementary Fig. 4b online). Thus, in human islet cells, the chromatin state at rs7903146 is more open in chromosomes carrying the T allele, which is associated with increased T2D risk5.
Next, we created allele-specific luciferase reporter constructs and measured enhancer activity in two islet β-cell lines, MIN6 and 832/13. Allelic differences in enhancer activity were observed in both cell lines. The T allele showed significantly greater enhancer activity compared to the C allele in both orientations (Forward: MIN6 P=1.6×10−7, 832/13 P=0.005; Reverse: MIN6 P=3.1×10−7, 832/13 P=3.1×10−4, Fig. 4e,f, Supplementary Fig. 4c,d online). However, allele-specific differences were not observed in the human embryonic kidney cell line 293T (Supplementary Fig 4e online). These data suggest that sequence variation at TCF7L2 affects T2D susceptibility by altering cis regulation and local chromatin structure in islet cells. The results are consistent with a previous report of association between the T allele and increased TCF7L2 transcripts in islets29, although the allele-specific changes described here can potentially impact different genomic regulatory functions, including transcriptional rates, promoter usage, or splicing.
To our knowledge, this study represents the first high-resolution atlas of regulatory elements in pancreatic islets. The unbiased maps generated by FAIRE-seq reveal new insights regarding the organization of tissue-specific cis-regulatory elements. Many earlier studies have shown that the genome is functionally organized in chromosomal territories1,30–32. Our observations extend previous findings by uncovering the existence of a large number of cell-selective regulatory domains associated with single genes, and provide a foundation for mechanistic understanding of transcriptional regulation of genes important for pancreatic islet cells. Identification of regulatory sites in a disease-relevant primary tissue also serves to dramatically reduce the genomic space when searching for functional non-coding sequence variants that influence T2D susceptibility. More generally, the current study provides a framework to move forward from the identification of large sets of disease-associated variants toward understanding the subset of functional variants that underlie disease risk.
Data accession numbers
Human islet FAIRE-seq raw data, alignments in the form of mean-centered base counts, and enriched sites at the three thresholds for all three samples can be obtained from Gene Expression Omnibus (GSE17616).
Methods
Islet sample preparation
All experiments were performed according to protocols approved by the Institutional ethical committees of the Hospital Clinic de Barcelona, Geneva University Hospitals, Istituto Scientifico Ospedale San Raffaele, and Hospital Universitari de Bellvitge. All samples were isolated from multiorgan donors without a history of glucose intolerance after informed consent from family members. Information on samples used for FAIRE-Seq are provided in Supplementary Table 1 online. Pancreatic islets were isolated according to established procedures33. After isolation, islets were cultured in CMRL 1066 containing 10% FCS and shipped at room temperature in the same medium. Samples 1 and 2 were processed upon arrival, while sample 3 and subsequent samples used in locus-specific assays were recultured in RPMI 1640 containing 10% FCS for three days before performing FAIRE. Islets were rinsed with PBS three times, and crosslinked for 10 min in 1% formaldehyde at room temperature with constant shaking. After adding glycine (final concentration 125 mM), islets were rinsed with PBS containing protease inhibitor cocktail (Roche) at 4°C, snap frozen, and stored at −80°C. Islet purity was assessed by dithizone staining34 immediately prior to fixation. The accuracy of dithizone staining was verified by immunofluorescence analysis using DAPI, anti-insulin, and anti-glucagon antibodies35.
FAIRE
For all but 2 samples (see below), FAIRE was performed as described2 with modifications. Frozen pellets with ~3000 crosslinked islets were thawed on ice in 1 mL lysis buffer (2% Triton X-100, 1% SDS, 100 mM NaCl, 10 mM Tris-Cl at pH 8.0, 1 mM EDTA) and disrupted with five 1-minute cycles using 0.5 mm glass beads (BioSpec). Samples were sonicated for 10–20 rounds of 30 pulses (1 second on/0.5 second off) using a Branson Sonifier 450D at 15% amplitude. After 10 rounds the efficiency of sonication was assessed, and further rounds were performed when needed to ensure that the majority of chromatin fragments were in the 200–1000 bp range. Debris was cleared by centrifugation at 15,000 g for 5 minutes at 4°C. Nucleosome-depleted DNA was extracted with phenol-chloroform followed by ethanol precipitation and RNase A (100 µg/mL) treatment2.
For samples 1 and 2 we employed a modified procedure that yielded less consistent chromatin fragmentation. Cells were incubated with 50 mM HEPES (pH 8.0), 140 mM NaCl, 1 mM EDTA, 0.1% SDS, 0.1% sodium deoxycholate, then centrifuged at 9,000 g 10 min. The pellet was resuspended in 50 mM HEPES (pH 8.0), 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, and then processed as described above.
Sequence analysis
Libraries were generated from gel-purified ~200 bp DNA fragments. After adaptor ligation and PCR-based amplification, samples were sequenced on the Illumina Genome Analyzer II platform using standard procedures. Sequence reads were aligned to the human reference genome (hg18) using Mapping and Assembly with Qualities (MAQ) with default mapping parameters 36. Post-alignment processing removed all reads that had an overall MAQ mapping quality <30 and artificially extended each read to a final length of 200 bp. We counted filtered reads mapping to each base in the genome to obtain a read density for each base. To facilitate display, read densities were centered on the mean read density of each chromosome.
Sites of FAIRE-seq enrichment were assessed with F-Seq37, which uses a kernel density estimate to calculate genomic regions where the continuous probability is greater than a user-defined standard deviation threshold over the mean across a local background. We used a feature length of 1,000 and three standard deviation (s.d.) thresholds resulting in three sets of enriched regions for each sample. The most liberal threshold was set for each sample using an empirical estimation of the upper bounds on the number of nucleosome-depleted regions genome-wide (roughly 200,000). For sample 3, the thresholds used were s.d.=6 (liberal), s.d.=8 (moderate) and s.d.=10 (stringent). For samples 1 and 2, which were sequenced to a lower depth, the thresholds used were s.d.=4 (liberal), s.d.=5 (moderate) and s.d.=8 (stringent).
We estimated the mappable proportion of the reference genome in two ways, using 120 million randomly generated reads (2.7×109 mappable bases, ~85% of hg18) and using PeakSeq (2.8×109 mappable bases, ~89% of hg18)38. We independently calculated genome coverage using 125 million reads obtained from islet FAIRE and found it to be 97.9% and 97.7% concordant, respectively.
Sequence reads obtained from five major ENCODE cell lines (GM12878, HeLa-S3, HUVEC, K562, and HepG2) were aligned to the reference genome (hg18) using MAQ36 and filtered as described above. Sites of enrichment were determined using F-Seq37, using the same parameters as islet samples 1 and 2.
Microarray analysis
Islet FAIRE preparations from samples 1 and 2 were fluorescently labeled and hybridized to a tiling DNA microarray covering 1% of the genome selected for the ENCODE pilot project1. Sites of enrichment were called using ChIPOTle39. For Receiver Operating Characteristic (ROC) curve analysis, sites of enrichment were called using a p-value threshold of 1×10−12.
Regulatory feature analysis
We recorded the percentage of bases underlying FAIRE-seq sites that overlapped 28-species conserved elements40, predicted regulatory modules (PreMod)41 and transcription factor binding sites (TRANSFAC42 and MotifMap43). For each set of peaks we permuted positions across the mappable genome 1,000 times and re-calculated the overlap. P values were calculated from permutations that had a higher degree of overlap than the observed set of peaks. We used Clover to test for over-represented transcription factor binding motifs in sequences underlying intergenic FAIRE-seq enrichment44. Sequences were separated by chromosome and analyzed for motifs from JASPAR45 and TRANSFAC42, as well as the CTCF motif15. Significance was calculated by comparing to the mappable intergenic portion of the tested chromosome, and motifs reaching a p-value threshold of .01 were reported.
FAIRE-Seq and expression level analysis
We used RMA-normalized signals from a previously reported experiment using HG-U133A and HG-U133B GeneChips with five non-diabetic islet samples10, and obtained an average value for each probe. The five samples were selected by hierarchically clustering expression data from 7 non-diabetic individuals. We excluded two samples (Sydp2 and SydPI) that had poor concordance with the others and showed low expression of known islet genes. We counted the number of FAIRE-Seq reads mapping to each base in a 1 kb window surrounding each RefSeq TSS, grouped RefSeq genes by their average islet expression level, and, for each group, calculated the average mean-centered FAIRE read density at every base in the window.
Islet-selective and ubiquitous site definitions
An islet FAIRE-seq site was considered islet-selective if the site did not overlap a site from any of the five additional tested cell types. Note that such sites are not expected to be necessarily unique to islets. An islet site was considered ubiquitous if the site overlapped a FAIRE-seq site in all five additional cell types. Moderate stringency FAIRE-seq site thresholds were used for all datasets.
Selection of genes with islet-selective open chromatin
For each RefSeq transcript we assessed the region 2 kb upstream through 2 kb downstream and calculated the percentage of bases that overlapped a moderate islet FAIRE enrichment site. We calculated the same value in the combined data from five non-islet cell lines, and selected genes that were more or equally enriched in islets compared to combined non-islet data.
Clusters of Open Regulatory Elements (COREs)
We identified 3,348 regions with at least three islet-selective sites (as defined above) located <20 kb from each other using Galaxy46. These criteria were based on the typical size of islet-selective clusters (Fig. 3b). For comparisons we created a set of randomized COREs of identical size and mappability. To assess CTCF binding enrichment, we obtained CTCF binding sites from multiple cell types17, calculated the frequency within COREs (0.007 sites/kb), in randomized COREs (0.013 sites/kb), and in the mappable genome (0.013 sites/kb), and tested for significance using a two-sided χ2 test.
RefSeq, ncRNA, or Non-RefSeq Unigene transcripts were assigned to a CORE when the transcriptional start or end site was within 10 kb of either end of the CORE. When comparing CORE overlaps with one or more genes we required a minimum overlap of one bp. We used DAVID47 to test enrichment of biological processes in CORE genes, and used all RefSeq genes as a background. We employed all biological processes from GO and PANTHER48, removing GO terms associated with >1,000 genes from the analysis.
For gene expression comparisons, we obtained gcRMA-normalized signals from Human U133A and GNF1H microarrays from seven tissues (pancreatic islet, liver, heart, kidney, lung, skeletal muscle and whole brain)49. We required that probes reported a signal of >100 in at least one tissue. We used one-way ANOVA to assess expression differences between tissues for CORE genes and for an identical number of randomly selected genes that did not overlap a CORE.
Definition of T2D susceptibility variants
We identified 20 loci where SNPs have been reported to show genome-wide association (P<5×10−8) with T2D or fasting glycemia14,25,26,50–54. For each locus we identified variants in HapMap CEU release 22 in strong linkage disequilibrium (r2>0.8) with the reported reference SNP; 350 variants satisfied these criteria and were termed ‘T2D-associated SNPs’. We then defined the region of association for each locus by manually identifying recombination hotspots from HapMap release 22 data flanking the associated SNPs. We identified SNPs in dbSNP v129 with an average heterozygosity >1% in these regions. SNPs that overlapped FAIRE sites in sample 3 were recorded.
Detection of allelic imbalance in open chromatin and PCR analysis of FAIRE
We genotyped 31 human islet genomic DNA (gDNA) samples using TaqMan SNP Genotyping Assays (Applied Biosystems). Nine samples were heterozygous for rs7903146. For these samples we used TaqMan SNP Genotyping Assays to determine the allelic ratio of DNA fragments containing rs7903146 in FAIRE and input DNA. All reactions were performed in triplicate in a volume of 25 µl using 5 ng DNA quantified with a Nanodrop 1000 (Thermo Scientific). A standard curve was generated by mixing gDNA from samples with known genotype to generate seven allelic ratios - 10:90, 20:80, 40:60, 50:50, 60:40, 80:20, and 90:10. The relative abundance of T and C alleles in each experimental sample was estimated from the standard curve, and compared to input DNA from the same samples and gDNA from unrelated heterozygous individuals. Allelic ratio was also assessed in seven samples heterozygous for rs7903146 by quantitative Sanger sequencing in FAIRE and input DNA (oligonucleotides shown in Supplementary Table 9 online). ImageJ was used to quantify the area under the curve of peaks in the chromatogram. Data are expressed as mean ± s.d. and were assessed with two-sided unpaired t-tests for gDNA vs. FAIRE, or paired t-tests for input vs. FAIRE.
To confirm islet-selective FAIRE enrichments, we employed real-time PCR with SYBR Green detection as described55,56. We performed triplicate measurements from 5 ng FAIRE DNA, and used a serial dilution of input DNA as the standard curve. We expressed FAIRE enrichment values relative to the enrichment values in the same sample at a local negative control region.
Luciferase reporter assays
A 240 bp fragment surrounding rs7903146 was PCR-amplified from DNA of individuals homozygous for either the T or C allele of rs7903146 (oligonucleotides shown in Supplementary Table S9 online). PCR fragments were cloned in both orientations in the multiple cloning site of the minimal promoter-containing firefly luciferase reporter vector pGL4.23 (Promega). Four independent clones for each allele for each orientation were verified by sequencing and transfected in duplicate into MIN657 and 832/13 (Chris Newgard, Duke University) β-cell lines, and into HEK293T cells. Cells were co-transfected with a phRL-TK Renilla-luciferase vector to control for transfection efficiency. Transfections were performed with lipofectamine 2000 (for MIN6 and HEK 293T; Invitrogen) or FUGENE-6 (for 832/13; Roche Diagnostics). Cells were assayed 48 hours after transfection using the Dual Luciferase Assay (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity and then divided by values for a pGL4.23 minimal promoter empty vector control. A two-sided t-test was used to compare luciferase activity between alleles. Experiments in MIN6 and 832/13 cells were carried out on a second independent day and yielded comparable allele-specific results.
Supplementary Material
Acknowledgements
We thank Xavi Garcia for experimental support in rs7903146 functional studies, Ignasi Moran for insights and help in the analysis of COREs, Dr Pierre Maechler (University of Geneva Medical Center) for generously providing insulin release data, Drs. Lorenzo Piemonti and Rita Nano (San Raffaele Scientific Institute), and Dr. Montserrat Nacher (IDIBELL) for human islets. This work was supported by the European Union VI Framework Programme project Eurodia to JF, Ministerio de Ciencia e Innovación (SAF2008-03116) to JF, Juvenile Diabetes Research Foundation (26-2008-633 to JF, 31-2008-416 to TB, 6-2005-1178 and 31-2008-416 to AS), the US NHGRI ENCODE project (U54 HG004563 subcontract to JDL), and R01 DK072193 to KLM. KLM is a Pew Scholar in the Biomedical Sciences.
Footnotes
Author contributions
JF and JDL conceived the study. KJG, TN, LP, JMS, KLM, JDL, and JF designed the experiments, interpreted results, and wrote the manuscript. TN conducted FAIRE experiments, developed and performed allelic imbalance assays. PGG optimized the FAIRE protocol and performed microarray studies. KJG, JMS, and PGG performed sequence analysis and KJG, LP, TN and JMS performed data analysis. LP conducted the analysis of CORES. MPF and TMP conducted reporter assays. PM conducted high-throughput sequencing. AS, DB, TB, and EM provided purified human islet samples.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hogan GJ, Lee CK, Lieb JD. Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet. 2006;2:e158. doi: 10.1371/journal.pgen.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagy PL, Cleary ML, Brown PO, Lieb JD. Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc Natl Acad Sci U S A. 2003;100:6364–6369. doi: 10.1073/pnas.1131966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant SF, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 6.Bell GI, Polonsky KS. Diabetes mellitus and genetically programmed defects in beta-cell function. Nature. 2001;414:788–791. doi: 10.1038/414788a. [DOI] [PubMed] [Google Scholar]
- 7.Oliver-Krasinski JM, Stoffers DA. On the origin of the beta cell. Genes Dev. 2008;22:1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nature Reviews Genetics. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 9.Wallrath LL, Lu Q, Granok H, Elgin SC. Architectural variations of inducible eukaryotic promoters: preset and remodeling chromatin structures. Bioessays. 1994;16:165–170. doi: 10.1002/bies.950160306. [DOI] [PubMed] [Google Scholar]
- 10.Gunton JE, et al. Loss of ARNT/HIF1beta mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell. 2005;122:337–349. doi: 10.1016/j.cell.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 11.Odom DT, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Lorenzo TP, Peakman M, Roep BO. Translational mini-review series on type 1 diabetes: Systematic analysis of T cell epitopes in autoimmune diabetes. Clin Exp Immunol. 2007;148:1–16. doi: 10.1111/j.1365-2249.2006.03244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarthy MI, Zeggini E. Genome-wide association scans for Type 2 diabetes: new insights into biology and therapy. Trends Pharmacol Sci. 2007;28:598–601. doi: 10.1016/j.tips.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 15.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xi H, et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bao L, Zhou M, Cui Y. CTCFBSDB: a CTCF-binding site database for characterization of vertebrate genomic insulators. Nucleic Acids Res. 2008;36:D83–D87. doi: 10.1093/nar/gkm875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuddapah S, et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atouf F, Czernichow P, Scharfmann R. Expression of neuronal traits in pancreatic beta cells. Implication of neuron-restrictive silencing factor/repressor element silencing transcription factor, a neuron-restrictive silencer. J Biol Chem. 1997;272:1929–1934. doi: 10.1074/jbc.272.3.1929. [DOI] [PubMed] [Google Scholar]
- 21.Fujitani Y, et al. Targeted deletion of a cis-regulatory region reveals differential gene dosage requirements for Pdx1 in foregut organ differentiation and pancreas formation. Genes Dev. 2006;20:253–266. doi: 10.1101/gad.1360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerrish K, Van Velkinburgh JC, Stein R. Conserved transcriptional regulatory domains of the pdx-1 gene. Mol Endocrinol. 2004;18:533–548. doi: 10.1210/me.2003-0371. [DOI] [PubMed] [Google Scholar]
- 23.Sander M, et al. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- 24.Edwards CA, et al. The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol. 2008;6:e135. doi: 10.1371/journal.pbio.0060135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohlke KL, Boehnke M, Abecasis GR. Metabolic and cardiovascular traits: an abundance of recently identified common genetic variants. Hum Mol Genet. 2008;17:R102–R108. doi: 10.1093/hmg/ddn275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyssenko V, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359:2220–2232. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 28.Bouatia-Naji N, et al. A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science. 2008;320:1085–1088. doi: 10.1126/science.1156849. [DOI] [PubMed] [Google Scholar]
- 29.Helgason A, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet. 2007;39:218–225. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 30.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 31.Dillon N. Gene regulation and large-scale chromatin organization in the nucleus. Chromosome Res. 2006;14:117–126. doi: 10.1007/s10577-006-1027-8. [DOI] [PubMed] [Google Scholar]
- 32.Gilbert N, et al. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 33.Bucher P, et al. Assessment of a novel two-component enzyme preparation for human islet isolation and transplantation. Transplantation. 2005;79:91–97. doi: 10.1097/01.tp.0000147344.73915.c8. [DOI] [PubMed] [Google Scholar]
- 34.Latif ZA, Noel J, Alejandro R. A simple method of staining fresh and cultured islets. Transplantation. 1988;45:827–830. [PubMed] [Google Scholar]
- 35.Boj SF, Parrizas M, Maestro MA, Ferrer J. A transcription factor regulatory circuit in differentiated pancreatic cells. Proc Natl Acad Sci U S A. 2001;98:14481–14486. doi: 10.1073/pnas.241349398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boyle AP, Guinney J, Crawford GE, Furey TS. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008;24:2537–2538. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rozowsky J, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buck MJ, Nobel AB, Lieb JD. ChIPOTle: a user-friendly tool for the analysis of ChIP-chip data. Genome Biol. 2005;6:R97. doi: 10.1186/gb-2005-6-11-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller W, et al. 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res. 2007;17:1797–1808. doi: 10.1101/gr.6761107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blanchette M, et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wingender E, et al. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie X, Rigor P, Baldi P. MotifMap: a human genome-wide map of candidate regulatory motif sites. Bioinformatics. 2009;25:167–174. doi: 10.1093/bioinformatics/btn605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frith MC, et al. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giardine B, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 48.Thomas PD, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gudmundsson J, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 51.Prokopenko I, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41:77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandhu MS, et al. Common variants in WFS1 confer risk of type 2 diabetes. Nat Genet. 2007;39:951–953. doi: 10.1038/ng2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Unoki H, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008 doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 54.Yasuda K, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40:1092–1097. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 55.Luco RF, et al. A conditional model reveals that induction of hepatocyte nuclear factor-1alpha in Hnf1alpha-null mutant beta-cells can activate silenced genes postnatally, whereas overexpression is deleterious. Diabetes. 2006;55:2202–2211. doi: 10.2337/db05-1534. [DOI] [PubMed] [Google Scholar]
- 56.Luco RF, Maestro MA, Sadoni N, Zink D, Ferrer J. Targeted deficiency of the transcriptional activator Hnf1alpha alters subnuclear positioning of its genomic targets. PLoS Genet. 2008;4:e1000079. doi: 10.1371/journal.pgen.1000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ishihara H, et al. Pancreatic beta cell line MIN6 exhibits characteristics of glucose metabolism and glucose-stimulated insulin secretion similar to those of normal islets. Diabetologia. 1993;36:1139–1145. doi: 10.1007/BF00401058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.