Abstract
The vitamin D receptor (VDR) and its ligand 1,25D, play an important role in regulating cell growth and cell fate. We examined the effect of 1,25D on cell morphology, cell proliferation, cell cycle progression and apoptosis on mesenchymal multipotent cells. Multipotent cells were treated with and without 1,25D in a time and dose dependent manner. Changes in cell morphology were evaluated by a green fluorescence fluorocrome. Cell proliferation was determined by the Formazan assay and PCNA antigen expression. The expression of genes related to the cell cycle was analyzed by DNA microarrays, RT2PCR arrays and western blots. Apoptosis was evaluated by TUNEL assay, and the expression of pro and anti-apoptotic related genes by RT2PCR arrays and western blots. 1,25D inhibited cell proliferation, induced cell cycle arrest, and promoted accumulation of cells in G0/G1 phase without inducing apoptosis. An increase in cell size was associated with a decrease in the GTPase Rho and the atypical Rho family GTPase Rhou/Wrch-1 expression without inducing Wnt-1 expression. Survivin expression was also increased and may represent a novel 1,25D mediated pathway regulating tissue injury and fibrosis. The data provide a mechanistic explanation for the anti-proliferative and anti-apoptototic properties of 1,25D in mesenchymal multipotent cells.
Keywords: VDR, PCNA, Rho, Wrch-1, Bcl-2, survivin
1. Introduction
Vitamin D, a fat-soluble pro-hormone, is obtained from dietary sources or from de novo synthesis in the skin as a result of ultraviolet light-induced photolytic conversion of 7-dehydrocholesterol to previtamin D3, followed by thermal isomerization to vitamin D3 [1] [2]. The first step in the metabolic activation of vitamin D is hydroxylation of carbon 25 (25OHD), which occurs primarily in the liver. The second and more regulated step in vitamin D bioactivation is the formation of 1α, 25-dihydroxyvitamin D3 (1,25D), also known as calcitriol, which occurs mainly in the kidney [3] via the 25OHD-1α-hydroxylase enzyme, although numerous cells and tissues express 1α- hydroxylase [4]. 1,25D binds to the vitamin D receptor (VDR), a nuclear receptor and a member of the steroid/thyroid hormone superfamily receptors, which acts as a ligand-activated transcription factor [5]. VDR functions as a heterodimer with retinoid X receptor; upon ligand binding, VDR undergoes a conformational change that promotes retinoid X receptor-VDR heterodimerization [6]. The binded heterodimer translocates to the nucleus, where VDR binds to the vitamin D response elements, induces chromatin-modifying enzymatic activities, and ultimately modulates gene transcription [7]. The VDR appears to be ubiquitous, supporting the vitamin D endocrine system's involvement in a wide range of physiological functions [8] [9] [10] [11].
It has been extensively reported that 1,25D can, in general, inhibit the growth of normal and malignant cell types. Previous studies on the mechanism behind this growth-inhibitory action have revealed that 1,25D-induced inhibition of cell growth is probably mediated via different combinations of pathways and mechanisms in varying cell types [12]. In view of its potent antiproliferative and prodifferentiating action, and the presence of the VDR in a large variety of cells, 1,25D or select analogs could be considered a promising intervention to prevent and treat hyperproliferative disorders [11] [12]. Elucidation of the molecular mechanism underlying cell cycle related antiproliferative action of 1,25D could help identify new biomarkers for increased disease -specific risk in vitamin D- deficient persons as well as targeting treatments with novel vitamin D analogs.
Studies of the effects of calcitriol on apoptosis are contradictory and depend mostly on the cell model applied. It has been reported that 1,25D induced apoptosis in peripheral blood mononuclear cell (PBMC) obtained from healthy subjects and in inflammatory bowel disease (IBD) patients [13]. By contrast, other studies have reported an inhibitory effect of calcitriol on hepatocyte apoptosis in rat allograft by upregulating antiapoptosis-associated genes [14]. Concerning the effects of 1,25D on cell morphology/phenotype, it has been reported that 1,25D profoundly affects the phenotype of breast cancer cells, suggesting that it reverts the myoepithelial features associated with more aggressive forms and poor prognosis in human breast cancer, increasing cell and nuclear size, and inducing a change from a rounded to a flattened morphology [15]. C3H 10T1/2 is a well-known and widely used mesenchymal multipotent cell line in vitro model that has been shown by us and others to have the potential to differentiate into a variety of specialized cells such as osteocytes, chondrocytes, adipocytes, endothelial and smooth muscle cells (SMCs) [16] [17] [18], [19], [20], [21], [22], [23] and they have been used as a cell model to study the effects of myostatin [24] and 1,25D on fibrosis [20] and cytokinesis [23]. C3H 10T1/2 cells are also considered an important model to systematically identify and analyze specific gene products that play an early role in cell proliferation and cell fate [20], [26]. Moreover, there are no reports in the literature assessing the effects of 1,25D on apoptosis, cell morphology/phenotype in C3H 10T1/2 mesenchymal multipotent cells.
In this study all C3H 10T1/2 mesenchymal multipotent cells were primed with 5′-azacytidine [17], [19], [20], [24], [26], [28] allowing us to evaluate the independent effects of 1,25D in comparison to no 1,25D at an early stage when cells still have the ability to undergo differentiation [19], [20]. C3H 10T1/2 cells have the ability to differentiate into multiple cell lineages according to the cell culture conditions such as incubation medium, cell density, and/or the addition of factors such as androgens [17], [28] or TGFβ factors [19] [24]. This is in contrast to progenitor cells that are already committed to a single cell lineage or terminally differentiated cells [29] [30] [31]. This approach allowed us to explore potential novel 1,25D mediated pathways that could provide the foundation for future adjuvant therapeutic approaches to address select hypovitaminosis D related chronic disease states.
Given the rapid advances in regenerative medicine such as the promise of induced pluripotent stem cells, a better understanding of 1,25D in mesenchymal multipotent cells is poised to accelerate the translation of novel basic science findings into human clinical trials. Thus, the aim of the present study was to provide evidence of the molecular mechanism (s) by which 1,25D affects cell morphology, cell proliferation, cell cycle progression and apoptosis using a well known and widely used mesenchymal multipotent cell line.
2. Material and Methods
2.1. Cell Culture
Mouse C3H 10T1/2 multipotent mesenchymal cells (MMCs) (ATCC, Manassas, VA, USA) grown in DMEM supplemented with 10% dialyzed fetal bovine serum at 37°C and 5% CO2 were treated with 20 μM 5′-azacytidine (AZCT) for 2 days to induce differentiation by epigenetic changes [16], [17] [19], [20]. Cells were split in a 1:2 ratio, allowed to recover for 2 days, and replated at 60–70% confluence in T75 flasks, eight-well chamber slides or six-well plates. The next day, cells were incubated with and without 100 nM of 1,25D (Sigma–Aldrich, St. Louis, MO, USA) dissolved in less than 0.1% ethanol as vehicle in DMEM 10% dialyzed fetal bovine serum for 4 and 7 days. For proliferation studies, mesenchymal multipotent cells primed with AZCT were incubated with 1,25D (10 nM to 500 nM) for 4 days, in the same way control groups were incubated with 0.1% ethanol in DMEM 10% dialyzed fetal bovine serum (Invitrogen, Carlsbad, CA, USA). Because of the short half-life of 1,25D, the cell culture media with and without 1,25D (100 nM-500nM) was replaced every 24h.
2.2. Cell proliferation assay
Cell proliferation was determined in 96-well plates by the Formazan dye assay (Promega Corp., Madison, WI, USA). Cells were grown at an initial density of 4,000 cells/well; and then treated for 4 days with 1,25D in a concentration range from 10 to 500 nM. At the end of the incubation time, 100 μl of Formazan substrate buffer was added to the cultures for 3 h at 37 °C in 5% CO2, and the absorbance at 490 nm was read. For cell counting, cells were removed by trypsinization and the number of viable cells was counted in a hemocytometer with the use of Trypan blue staining [32].
Cell proliferation was also evaluated / confirmed by changes in gene expression of a related cell proliferation antigen after 4 days incubation time with or without 1,25D. The cell proliferation antigen applied was the proliferating cell nuclear antigen (PCNA) a protein essential for DNA replication, repair and cell proliferation [33].
2.3. Cell Morphology / Phenotype
Changes in cell morphology / phenotype were determined in living cells in culture by the PKH2 Green Fluorescent Cell Linker assay (Sigma-Aldrich, Saint Louis, MO, USA) [34]. After an initial and stable incorporation of the PKH2-GL (2× 10-6 M) into the lipid regions of the cell membrane, cells were counterstained with DAPI 0.02% (Invitrogen, Carlsbad, CA, USA). Cells were grown at an initial density of 8,000 cells/well in 8-wells chamber slides and were treated for 4 and 7 days respectively with or without 1,25D (100 nM). Serial pictures were taken every 24h with a fluorescent microscope equipped with green and blue filters. The serial pictures at 200× and 400× magnification were taken with a Leica DMLB fluorescence microscope coupled to Spot RT digital camera.
The area and the number of cells per field were analyzed using ImagePro-Plus 5.1 software (Media Cybernetics, Silver Spring, MD, USA). At least 10 pictures were taken per treatment group per wells done in duplicate. The experiments were repeated at least three times.
2.4. Apoptosis
The apoptotic index was determined by the TdT-mediated dUTP nick end labeling (TUNEL) method, based on the ability of terminal TdT to catalyze addition of digoxenin-dUTP and dATP to 3′-OH ends of cleaved DNA. Cells were seeded on 8-well chamber slides and were incubated with or without 1,25D (100 nM) for 4 days and then fixed in 2% p-formaldehyde in 1× PBS. Cells were treated with 2% H2O2 for quenching endogenous peroxidase activity, digoxigenin-conjugated nucleotides and TdT, and finally anti-digoxigenin-peroxidase. Sections were stained with 0.5% diaminobenzidine/0.01% H2O2, and counterstained with haematoxylin. As a negative control, PBS (1×) buffer was substituted for the TdT enzyme. Quantification of the apoptotic index was done at 400 ×. In all cases, 20 fields were randomly selected and the apoptotic index of each field was calculated as the percent of TUNEL-positive cells [32].
2.5. DNA Microarray Analysis of Cell Cycle Target Genes
Total cellular RNA was isolated with Trizol-Reagent (Invitrogen, Carlsbad, CA, USA) from C3H 10T1/2 cells undergoing differentiation with 5′-azacytidine (AZCT), and treated with or without 1,25D (100nM) for 4 days. Isolated RNA was subjected to cDNA gene arrays (Oligo GEArray, Cell Cycle gene array (OMM-020) SABiosciences Corp., Frederick, MD, USA). The Mouse Cell Cycle Microarray profiles the expression of 112 genes key to cell cycle regulation such as CDKs and CDK-modifying proteins, including cyclins, CDK inhibitors, CDK phosphatases, and CDK kinases, as well as genes in the APC ubiquitin-conjugation complexes. Biotin-labeled cDNA probes were synthesized from total RNA, denatured, and hybridized overnight at 60°C in GE Hybridization solution to membranes spotted with Cell cycle related genes. Membranes were washed, and chemiluminescent analysis was performed per the manufacturer's instructions. Raw data were analyzed using GE Array Expression Analysis Suite (SABiosciences Corp., Frederick, MD). Fold changes in relative gene expression were presented after background correction and normalization with a housekeeping gene [24], [35].
2.6. RT2 Profiler PCR Array Analysis of Cell Cycle and Apoptosis Target Genes
RT2 profiler PCR SABiosciences analysis of cell cycle and apoptosis target genes were applied in triplicates in order to confirm the GE Array data (cell cycle) and to complement the TUNEL and western blot results (apoptosis). Aliquots of total cellular RNA isolated with Trizol-Reagent from C3H 10T1/2 cells undergoing differentiation with 5′-azacytidine (AZCT) were treated with or without 1,25D (100nM) for 4 days. They were then subjected to reverse transcription, and the resulting cDNA was analyzed by: RT2 profiler PCR Mouse Cell Cycle Array (PAMM-020) and by RT2 profiler PCR mouse apoptosis (PAMM-012) (SABiosciences Corp., Frederick, MD, USA). The Mouse Cell Cycle RT2 Profiler™ PCR Array contains genes that both positively and negatively regulate the cell cycle, the transitions between the each of the phases, DNA replication, checkpoints and arrest. The Mouse Apoptosis RT2 Profiler™ PCR Array profiles the expression of genes involved in apoptosis, or programmed cell death. Each array contains a panel of 84 primer sets related to the cell cycle and to apoptosis plus 5 housekeeping genes and 2 negative controls. Real time PCRs were performed as follows: melting for 10 min at 95°C, 40 cycles of two-step PCR including melting for 15 sec at 95°C, annealing for 1 min at 60°C. The raw data were analyzed using the ΔΔCt method following the manufacturer's instructions (SABiosciences Corp., Frederick, MD, USA) [24], [35].
2.7. Real-Time Quantitative PCR
Total RNA was extracted using Trizol-Reagent (Invitrogen, Carlsbad, CA) and equal amounts (1μg) of RNA were reverse transcribed using a RNA PCR kit (Applied Biosystems, Foster City, CA, USA). Mouse gene PCR primer sets (RT2) for PCNA, Casp3, Wrch-1 and Wnt1 were obtained from SuperArray Bioscience (SABiosciences Corp., Frederick, MD, USA). The QIAGEN Sybr Green PCR kit with HotStar Taq DNA polymerase (QIAGEN, Valencia, CA, USA) was used with i-Cycler PCR thermocycler and fluorescent detector lid (Bio-Rad, Hercules, CA, USA) [35].
The protocol included melting for 15 min at 95°C, 40 cycles of three-step PCR including melting for 15 sec at 95°C, annealing for 30 sec at 58°C, elongation for 30 sec at 72°C with an additional detection step of 15 sec at 81°C, followed by a melting curve from 55–95°C at the rate of 0.5°C per 10 sec. Samples of 25 ng cDNA were analyzed in quadruplicate in parallel with GAPDH controls; standard curves (threshold cycle vs. log pg cDNA) were generated by log dilutions of from 0.1 pg to 100 ng standard cDNA (reverse-transcribed mRNA from C3H 10T1/2 cells in AM). Experimental mRNA starting quantities were then calculated from the standard curves and averaged using i-Cycler, iQ software as described previously [20]. The ratios of marker experimental gene (e.g. PCNA, Casp3 and Wrch-1 mRNA) to GAPDH mRNA were computed and normalized to control (untreated) samples as 100%.
2.8. Immunocytochemical analyses of PCNA antigen
After incubating the MMCs for 4 days with and without 1,25D, cells were washed five times with PBS (1×) and fixed by immersion in 2% p-formaldehyde, quenched with H2O2, blocked with normal horse serum and incubated with anti-proliferating Cell Nuclear Antigen (PCNA) monoclonal antibody (1:400) (Millipore, Temecula, CA, USA). Detection was based on a secondary biotinylated antibody (1:200), followed by the addition of the streptavidin-horseradish peroxidase ABC complex (1:100), Vectastain (Elite ABC System, Vector Laboratories, Burlingame, CA, USA) and 3,3′-diaminobenzidine and H2O2 mixture (Sigma, St. Louis, MO). The MMCs were counterstained with Mayer's hematoxylin solution (Sigma-Aldrich, St. Louis, MO, USA). In negative controls, we either omitted the first antibody or used a rabbit non-specific IgG.
2.9. Western Blot and Densitometry Analysis
Cell lysates (25-40μg of protein) were subjected to western blot analyses by 4-15% Tris-HCl polyacrylamide gel electrophoresis (PAGE) (Bio-Rad, Hercules, CA, USA) in running buffer (Tris / Glycine / SDS). Proteins were transferred overnight at 4°C, to nitrocellulose membranes in transfer buffer (Tris / Glycine / Methanol). Next day the non-specific binding was blocked by immersing the membranes into 5% non-fat dried milk, 0.1% (v/v) Tween 20 in PBS for 2 hours at room temperature. After several washes with washing buffer (PBS Tween 0.1%), membranes were incubated with the primary antibodies for 3 hours at room temperature or overnight at 4°C, monoclonal antibodies were as follow: a) PCNA (1/500) (Millipore, Temecula, CA, USA) b) glyceraldehide-3-phosphate dehydrogenase (GAPDH) (1/10,000) (Chemicon International, Temecula, CA, USA), c) Bcl-2 (1/500) (BD Biosciences, San Jose, CA, USA), d) Cyclin D1 (1/500), e) Cyclin D3 (1/500), f) CDk4 (1/500) (Cell Signaling Technology, Inc., Danvers, MA, USA) and Rho (1/500) (Abcam Inc., Cambridge, MA, USA). Polyclonal antibodies were used for a) CDk2 (1/500), b) p21 (1/500) and c) p27 (1/500) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). The washed membranes were incubated for 1 hour at room temperature with 1/3,000 dilution (anti-mouse) or 1:2,000 dilution (anti-rabbit) of secondary antibody linked to horseradish peroxidase, respectively (Cell Signaling Technology, Inc., Danvers, MA, USA). After several washes, the immunoreactive bands were visualized using the SuperSignal western blotting chemiluminescence detection system (Thermo Fisher Scientific, Inc., Rockford, IL, USA). The densitometry analysis of the bands was done with the Scion Image software beta 4.02 (Scion Corp., Frederick, MD, USA).
2.10. Qualitative and Quantitative Immunocytochemical Analyses
The immunoreactivity was quantified by Image Analysis using ImagePro-Plus 5.1 software (Media Cybernetics, Silver Spring, MD, USA). Fields are count blindly by two independent observers and each experiment was repeated three times. For PCNA and TUNEL determinations, the number of positive cells at 400× was counted in a computerized grid and results were expressed as a % of positive cells/total cells per field. In all cases, 8 fields at 400× were analyzed per wells.
Cell size was determined by outlining and measuring the area of each cell per field. Approximately 100 cells were measured for each experiment and the results were converted in cell size histograms plotting the number of cells per area expressed in μm2. To test the precision of the QIA, we evaluated both inter- and intra-assay variability. Standard deviation was 0.39 for assays in triplicate, CV=1.9% for intra-assay determinations and CV 4.0% for inter-assay.
The results expressed as Mean ± S.E.M. represent the average of three independent experiments [19], [20] and [24].
2.11. Statistical Analysis
All data are presented as Mean +/- S.E.M, and between-group differences were analyzed using ANOVA. If the overall ANOVA revealed significant differences, then pair-wise comparisons between groups were performed by Tukey multiple comparison test. All comparisons were two-tailed, and P values less than 0.05 were considered statistically significant. The in vitro experiments were repeated thrice, and data from representative experiments are shown. Specifically, the DNA microarrays tests were done twice and the results confirmed by RT2 Profiler PCR arrays in triplicates and in some cases by qRT-PCR in triplicates.
3. Results
3.1. 1,25 D inhibits cell proliferation of C3H 10T1/2 mesenchymal multipotent cells
Even though we seeded the same number of cells, incubation with 1,25D (100nM) for 4 days shows a noticeable decrease in cell number per field compared to control cells (no 1,25D) (Fig 1 A). To assess whether 1,25D inhibits C3H 10T1/2 multipotent cell replication, an initial number of 4,000 cells/well (low confluence) were seeded in 96 well plates and incubated in triplicates with increasing concentrations of 1,25D, from 10 to 500 nM, for 4 days, and at the end of the incubation time, cell proliferation was evaluated by the formazan assay (Fig 1B). Starting at 25 nM, 1,25D induced a statistically significant reduction in cell number reaching a plateau at 100nM. For that reason, 100nM was the dose applied throughout the study to evaluate the effects of 1,25D on mesenchymal multipotent cells.
Figure 1. 1,25D inhibits cell proliferation in Multipotent mesenchymal C3H 10T1/2 in a dose dependent manner.
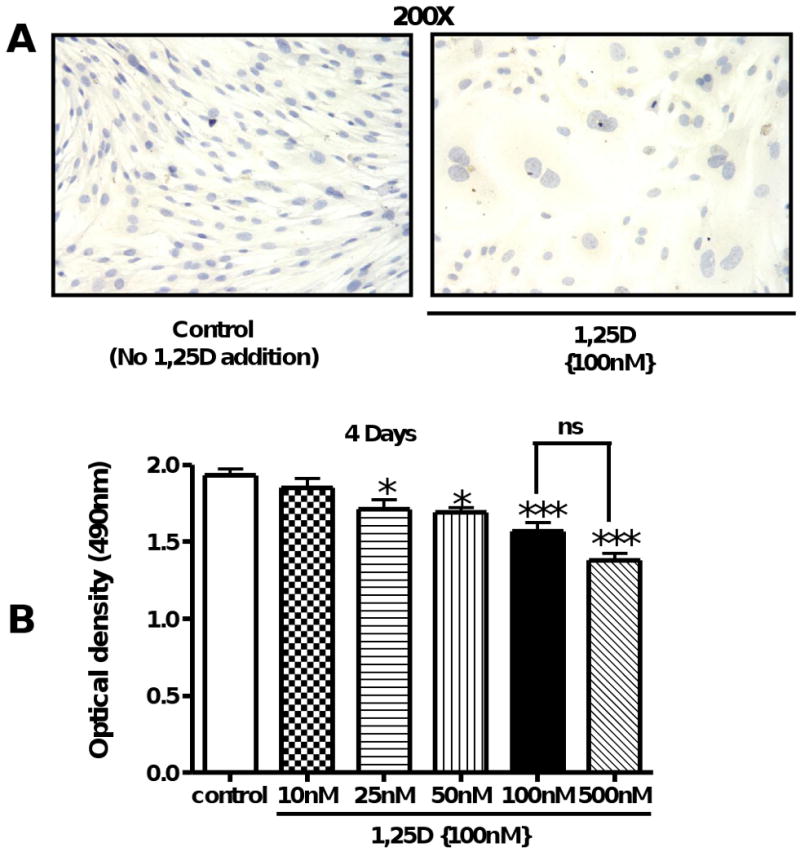
(A) Cells treated with or without 1,25D (100 nM) for 4 days were fixed with 2% p-formaldehyde and counterstained with hematoxyline (200×). (B) C3H 10T1/2 cells primed with 5′-azacytidine (20 μM) at an initial number of 4,000 cells/well were incubated for 4 days, as follows: control: no addition of 1,25D and 1,25D: increasing concentrations of 1,25D from 10 – 500 nM. Results are expressed as Mean±SEM. *P<0.05; ***P<0.001
In order to confirm the anti-proliferative effect of 1,25D on mesenchymal multipotent cells, PCNA expression was evaluated. Incubation of cells with 1,25D decreased the expression of proliferating cell nuclear antigen (PCNA) as assessed by several approaches; Fig 2A, left panel shows representative pictures at 200× and 400× magnification of the decreased expression of PCNA upon incubation with 1,25D (100nM) for 4 days by immunocytochemistry (ICC). The visual inspection was confirmed by quantitative image analysis (Fig 2 A, right panel) where PCNA expression was significantly decreased by 3-fold with respect to the control (p<0,001). The decreased expression of PCNA was further confirmed by real time PCR (PCNA/GAPDH ratio: -2.05) (Fig 2B) and by western blot (Fig 2C, left) with the corresponding densitometry analysis (Fig 2C, right).
Figure 2. 1,25D downregulates the expression of proliferating cell nuclear antigen (PCNA).
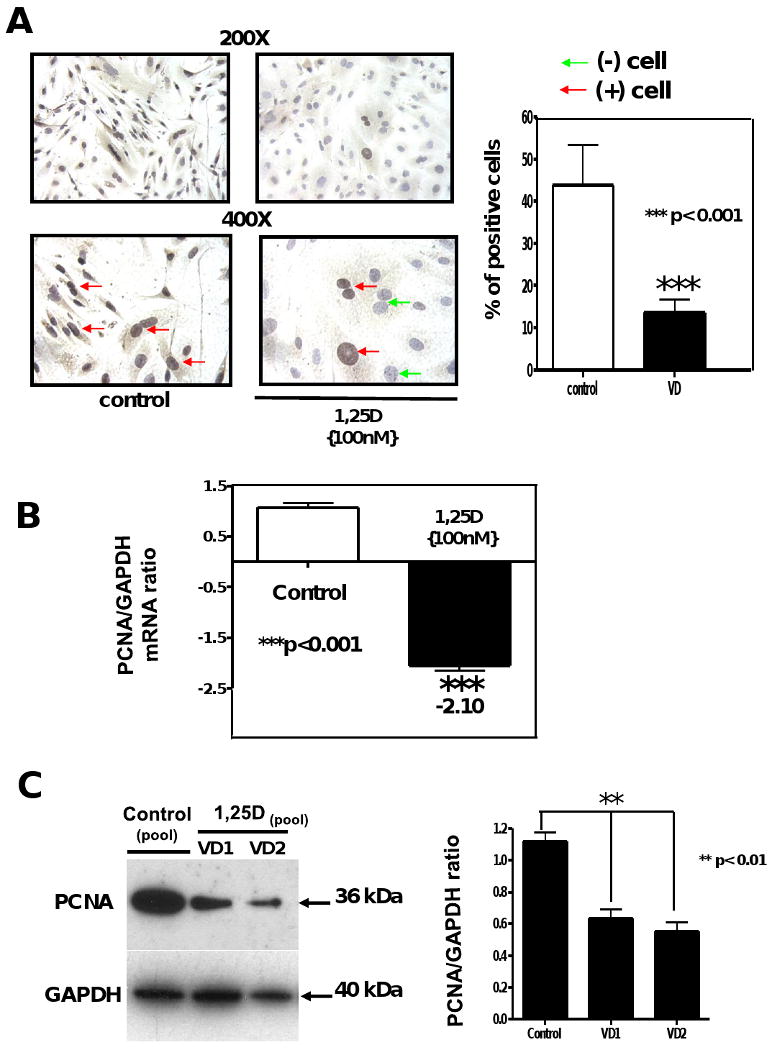
Cells treated as in Figure 1 were incubated with or without 100nM of 1,25D for 4 days. Immunocytochemistry for PCNA were performed at the end of the incubation time (A). Mean±SEM corresponds to experiments done in triplicates of the percentage of positive cells (brown nuclear staining) by quantitative image analysis. ***p<0.001. (B) Total RNA isolation followed by real time PCR was applied in other aliquots for the 4 days incubations normalized by GAPDH housekeeping gene. Mean±SEM corresponds to experiments done in triplicates ***p<0.001. (C) Western blots analysis was performed for extracts from the 4 days incubation time (left) and the corresponding densitometry analysis (right). 200× and 400×.
3.2. 1,25 D modifies cell morphology increasing cell size of C3H 10T1/2 mesenchymal multipotent cells
In order to verify our previous visual observations regarding changes in cell number and size upon incubation of multipotent cells with 1,25D, cells were pre-labeled with a cell membrane marker, the PKH2 Green Fluorescent Cell Linker and DAPI to visualize cell boundaries and cell nuclei respectively and later incubated with or without 1,25D for 4 and 7 days. 4 days incubation with 1,25D significantly increased the morphology/phenotype/size and decreased cell number of mesenchymal multipotent cells. (Fig 3 A, upper panel). The visual inspection was further confirmed by QIA (quantitative image analysis) shown in Fig 3 A, lower panel where the histogram shows a shift from smaller cells to larger cells in the population indicating that 1,25 D significantly increased cell size and reduce cell number per field with respect to the control. Similar results were obtained in a parallel experiment where cells were incubated for 7 days. Representative pictures can be observed in Fig 3 B, upper panel and the corresponding QIA (quantitative image analysis) Fig 3 B, lower panel. Incubation of mesenchymal multipotent cells with 1,25D (100nM) for 7 days increased cell size, modified cell morphology and reduced cell number per field.
Figure 3. 1,25D decreases cell number and increases cell size of multipotent mesenchymal cells.
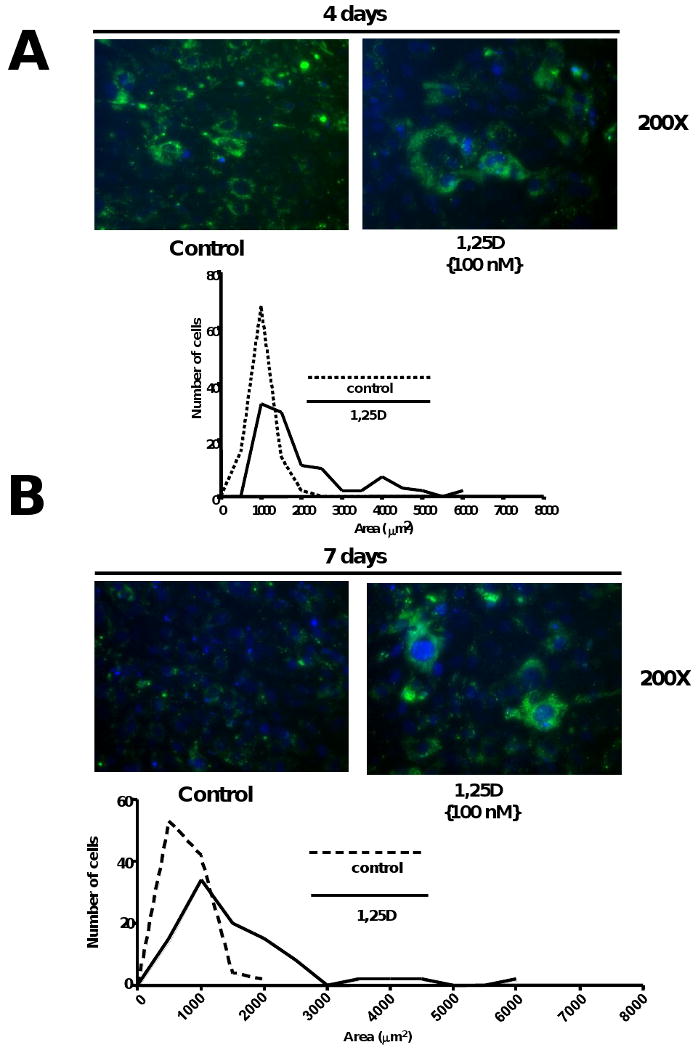
Cells treated as in Figure 1 were preincubated with Green Fluorescent Cell Linker (PKH2) and counterstaing with DAPI. Next day, cells were incubating with or without 1,25D for 4 and 7 days respectively. Representative pictures are presented for control (No 1,25D addition) and 1,25D (100nM) at 4 days (A upper panel) and at 7 days (B, upper panel) with the corresponding histograms showing the cell size distribution and expressing the number of cells per area (μm2). (A and B, lower panels)
3.3. Addition of 1,25 D downregulate the expression of Rho a member of the Ras super family of low molecular weight GTPases and the atypical GTPase Rhou/Wrch1 without increasing Wnt-1 expression in C3H 10T1/2 mesenchymal multipotent cells
In order to clarify the mechanism by which 1,25 D changes cell morphology, the expression of a novel gene Wrch-1 involved in regulating cell morphology, cytoskeletal organization, and cell proliferation was evaluated. We found that upon incubation with 1,25D (100nM) for 4 days the mRNA level of Rhou/Wrch-1 were decreased by 4.5-fold as determined by real-time PCR (qRT-PCR) compared to control (no 1,25D addition) (Fig 4 A, upper panel). In addition, Fig 4, panel B left shows decreased expression upon 1,25D incubation for 4 days of Rho, a member of the GTpase family which has been linked to biological pathways related to cytoskeletal dynamics, polarity, cell survival / apoptosis and cell proliferation, by western blot with the corresponding densitometric analysis Fig 4, panel B right.
Figure 4. 1,25D regulates cell size through downregulation of the GTPase Rho and the atypical rho family GTPase Rhou/Wrcth-1 without intervention of Wnt1.
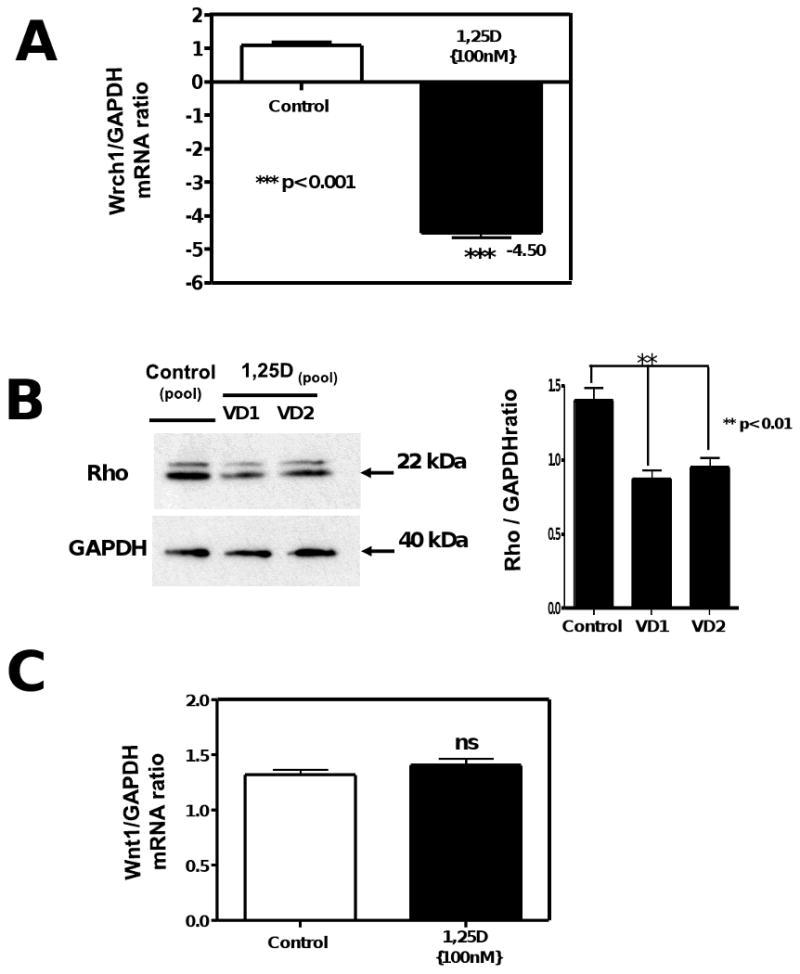
Total RNA isolation from cells treated as in Figure 2, was subjected to real time PCR for Wrcth1 normalized by GAPDH housekeeping gene (A). Mean±SEM corresponds to experiments done in triplicates ***p<0.001. (B) Western blots analysis was performed for extracts from the 4 days incubation time for Rho normalized with GAPDH with the correspondent densitometric analysis. In another set of aliquots the RNA samples were utilized for real time PCR for Wnt1 normalized by GAPDH housekeeping gene (C).
Fig 4, panel C, shows no changes in mRNA expression of Wnt-1 by real-time PCR (qRT-PCR) after addition of 1,25D (100nM) for 4 days, demonstrating no activation of Wnt-1 signaling by 1,25D.
3.4. 1,25 D exerts an anti-apoptotic effect in C3H 10T1/2 mesenchymal multipotent cells
To investigate whether 1,25D might control cell number through the regulation of programmed cell death, we incubated the cells with 1,25D (100nM) for 4 days and assessed selected apoptotic markers by different approaches. Fig 5 shows there was not significant effect of 1,25 D on the number of apoptotic cells determined by TUNEL assay (Fig 5 A, left) followed by quantitative image analysis (Fig 5 A, right). In contrast, Fig 5 B shows that incubation of multipotent cells with 1,25D (100nM) for 4 days exerts an anti-apoptotic effect decreasing the expression of pro-apoptotic genes such as caspase 3 by 2.12-fold (p<0.001) as assessed by real time PCR. Moreover, the addition of 1,25D (100nM) to the cell culture promotes the upregulation of anti-apoptotic markers such as Bcl-2 evaluated by western blot (Fig 5 C, left) with corresponding densitometry analysis (Fig 5 C, right) and confirmed by RT2 PCR arrays, Table 1, Bcl-2 (+2.9).
Figure 5. 1,25D exerts an antiapoptotic effect in C3H 10T1/2 multipotent mesenchymal cells.
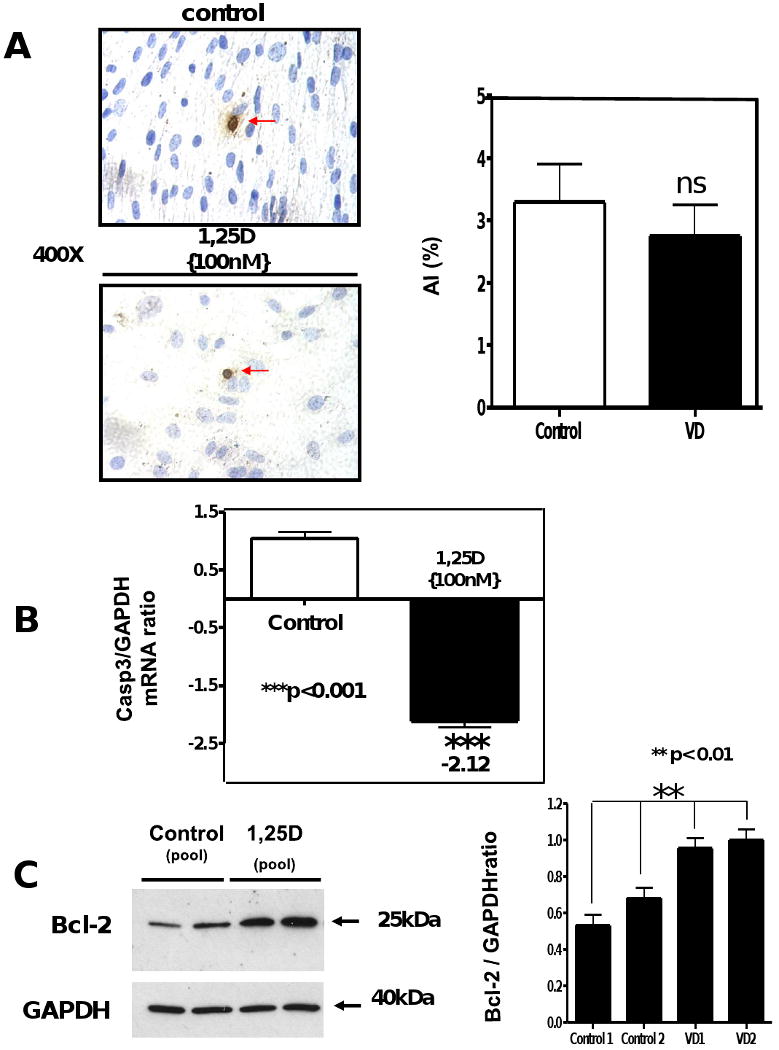
Cells treated as in Figure 2, were seeded in 8-well chamber slides for TUNEL assay or in 6-well plates for RNA and protein isolation. At the end of the incubation period, samples were subjected to TUNEL assay where the apoptotic index (AI) was obtained for control and VD incubated samples (A). In another set of samples, total RNA isolation was followed by real time PCR for Caspase 3 normalized by GAPDH housekeeping gene. Mean±SEM corresponds to experiments done in triplicates ***p<0.001. (B) Western blots analysis was performed for extracts from the 4 days incubation time for Bcl-2 normalized with GAPDH with the corresponding densitometric analysis (C).
Table 1. 1,25D modulates the transcriptional expression of apoptotic related genes in C3H 10T1/2 multipotent mesenchymal cells.
Total RNA from cells treated as in Figure 2 was subjected to RT-real time PCR by the Mouse Apoptosis RT2 Profiler™ PCR Array and the ratios between the 1,25D treated and 1,25D-untreated cells corrected by GAPDH were calculated for the assays performed in triplicates.
| Symbol | Description | Real-time PCR array ratios (4 days) |
|---|---|---|
| Api 5/Aac11 | Apoptosis Inhibitor 5 | + 2.16 |
| Atf5 | Activated transcription factor 5 | -2.81 |
| Bcl-2 | B-cell leukemia/lymphoma 2 | +2.9 |
| Birc5 | Baculoviral IAP repeat-containing 5 (survivin) | +4.53 |
| Casp 3 | Caspase 3 | -2.30 |
| BAX | Bcl2-associated X protein | 1.18 |
| BAD | Bcl-associated death promoter | 1.22 |
| BAK | BCL2-antagonist/killer | 1.36 |
| BOK | Bcl-2-related ovarian killer protein | 1.29 |
The anti-apoptotic effects of 1,25D on C3H 10T1/2 were further analyzed at the transcriptional level by applying the RT2 profiler real time apoptosis PCR array (PAMM-012). Table 1 shows the ratios of determinations done in triplicate after continuous incubation with and without 1,25D for 4 days. Table 1 shows that C3H 10T1/2 cells incubated with 1,25D (100nM) for 4 days increased the total mRNA expression of Apoptosis Inhibitor-5/antiapoptosis clone 11 (Api5/Aac11) (+2.16), which is a critical determinant of dE2F1-induced apoptosis in vivo and in vitro. Table 1 shows also an increased expression of Bcl-2 (+2.9) in agreement with the western blot shown in Fig 5; and Baculoviral IAP repeat containing 5 (Birc5) (+4.53) also known as “survivin”, a protein that inhibits apoptosis and regulates cell division.
In addition, incubation of multipotent mesenchymal cells with 1,25D (100nM) decreases the expression of select pro-apoptotic genes such as Activated transcription factor 5 (Atf5) (-2.81), which plays an essential role in hematopoietic and glioma cell survival and neuronal cell differentiation. The decreased gene expression of caspase 3 (-2.30) (an executioner / effector caspase) upon incubation with 1,25D was further confirmed by RT2 PCR arrays. Table 1 also shows that several members of the Bcl-2 family of genes, specifically the pro-apoptotic members such as Bax (+1.18); BAD (+1.22); Bak (+1.36) and Bok (+1.29), remain unchanged upon 1.25D incubation.
3.5. 1,25 D promotes cell cycle arrest in C3H 10T1/2 mesenchymal multipotent cells
The effect of 1,25D (100nM) on cell cycle components was evaluated at the transcriptional level applying the mouse cell cycle DNA gene array (OMM-020). Table 2 shows the differential cDNA gene expression between the 1,25D-treated and -untreated cells, where the computer-generated ratios of spot intensities, normalized by housekeeping genes are tabulated. DNA microarray analysis shows a general decrease in the gene expression of cyclins such as Cyclin A2 (-2.76), B1 (-2.15), B2 (-2.9), D1 (-2.32), E1 (-4.39), and F (-3.24), and Cyclin dependent kinases such as Cdk2 (-2.05) and genes related to check points such as Chek1 (checkpoint kinase 1) (-4.7). We also observed a decreased expression of PCNA (proliferating cell nuclear antigen) (-2.05), which is in agreement with the previous data provided by immunocytochemistry (ICC), real-time PCR and western blot; as well as a decreased expression of Caspase 3 (-2.05) and Skp2 (S-phase kinase-associated protein 2) (-2.19), a gene that specifically recognizes phosphorylated cyclin-dependent kinase inhibitor 1B (p27) predominantly in S phase. In order to confirm and to expand the DNA microarray data with a more sensitive procedure, we performed real-time PCR arrays using the RT2 profiler PCR SuperArray set of primers and procedures (PAMM-020). The ratios for triplicate determinations are shown in Table 3 for 4 days after continuous incubation with or without 1,25D (100nM). The agreement between the ratios obtained by DNAmicroarrays and RT2PCR arrays is in general adequate and provides a reasonable assessment of up- and down- gene regulation. The results of the RT2PCR arrays provided confirmatory information related to the cell cycle inhibitory effects of 1,25D on multipotent cells. A substantial decrease in the mRNA expression of numerous cyclins such as Cyclin A2 (-4.39), B1 (-4.87) and B2 (-3.44), and D1 (-2.30) which is a positive regulator of G0/G1 cell cycle progression and a well-known promoter of cell proliferation); Cyclin E1 (-3.21); F (-5.40); cyclin dependent kinases such as Cdk2 (-2.05) and Cdk4 (-2.25) and genes related to cell cycle check points such as Chek1 (checkpoint kinase 1) (-3.0). Incubation of multipotent cells with 1,25D inhibits G1 to S phase progression by down-regulating the expression of the F-box protein Skp2 (p45) (-2.19) that induces cell growth inhibition via G1 arrest.
Table 2. 1,25D regulates the transcriptional expression of cell cycle related genes in C3H 10T1/2 multipotent cells.
Total RNA from cells treated as in Figure 2 was subjected to cDNA microarrays by the Oligo GEArray Cell Cycle gene array, and the ratios between the 1,25D treated, and 1,25D-untreated cells corrected by GAPDH were calculated for the assays performed in duplicates.
| Symbol | Description | DNA microarray Ratios (4 days) |
|---|---|---|
| Ccna2 | Cyclin A2 | -2.76 |
| Ccnb1 | Cyclin B1 | -2.15 |
| Ccnb2 | Cyclin B2 | -2.9 |
| Ccnd1 | Cyclin D1 | -2.32 |
| Ccne1 | Cyclin E1 | -4.39 |
| Ccnf | Cyclin F | -3.24 |
| Cdk2 | Cyclin-dependent kinase 2 | -2.05 |
| Chek1 | Checkpoint kinase 1 homolog (S. pombe) | -4.7 |
| Casp3 | Caspase 3 | -2.05 |
| Skp2 | S-phase kinase-associated protein 2 (p45) | -2.19 |
| PCNA | Proliferating cell nuclear antigen | -2.05 |
Table 3. Confirmation of the effects of 1,25D on the transcriptional expression of cell cycle related genes in C3H 10T1/2 multipotent cells by real-time PCR arrays.
Total RNA from cells treated as in Table 2 was subjected to RT-real time PCR by the RT2 profiler PCR mouse Cell cycle array and the ratios between the 1,25D treated and 1,25D-untreated cells corrected by GAPDH were calculated for the assays performed in triplicates.
| Symbol | Description | Real time PCR Array Ratios (4 days) |
|---|---|---|
| Ccna2 | Cyclin A2 | -4.39 |
| Ccnb1 | Cyclin B1 | -4.87 |
| Ccnb2 | Cyclin B2 | -3.44 |
| Ccnd1 | Cyclin D1 | -2.85 |
| Ccne1 | Cyclin E1 | -3.21 |
| Ccnf | Cyclin F | -4.40 |
| Cdk2 | Cyclin-dependent kinase 2 | -2.05 |
| Cdk4 | Cyclin-dependent kinase 4 | -2.25 |
| Chek1 | Checkpoint kinase 1 homolog (S. pombe) | -3.0 |
| Skp2 | S-phase kinase-associated protein 2 (p45) | -2.60 |
In order to verify the effect of 1,25D not only at the transcriptional but also at the translational level, several of the most important regulators of cell cycle progression were assessed to further confirm our previous results by western blot. Figure 6, A shows the confirmatory result of the decreased expression of Cyclin D1 and Cyclin D3 upon incubation with 1.25D (100nM). Figure 6, B a decreased expression of Cdk2 (cyclin dependent kinase 2) and Cdk4 (cyclin dependent kinase 4) upon incubation with 1.25D (100nM). Figure 6, C shows that the expression of the cell cycle inhibitor p21 was negligible in the untreated C3H 10T1/2 cells, and 1,25D exposure led to much higher levels of p21 but not p27, making p21 the main primary mediator of the 1,25D antiproliferative effect in this multipotent cell model.
Figure 6. 1,25D promotes cell cycle arrest in C3H 10T1/2 mesenchymal multipotent cells by decreasing the expression of cyclins, cyclin dependent kinases (Cdk) and Cdki (cyclin dependent kinases inhibitors).
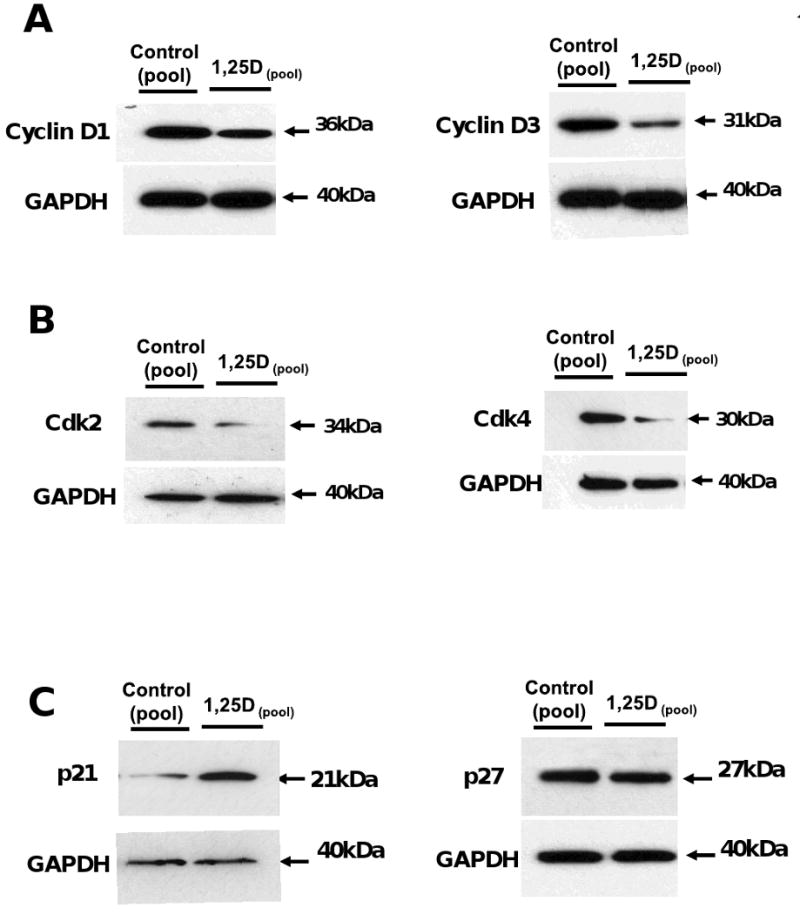
Cells treated as in Figure 2, were seeded on 6-well plates for protein isolation Western blots analysis was performed for extracts from the 4 days incubation time for Cyclin D1 and D3 (A), for Cdk2 and Cdk4 (B) and p21 and p27 (C) all of them normalized for housekeeping genes with GAPDH.
4. Discussion
Our data demonstrate that the addition of 1,25D to mesenchymal multipotent cells in a time and dose dependent manner exerts a potent antiproliferative effect, inducing cell cycle arrest, promoting the accumulation of cells in G0/G1 phase without inducing apoptosis. This antiproliferative effect is characterized by a reduction in the number of cells and an increase in cell size compared to the untreated group. Specifically, we demonstrate for the first time, that supplementation with 1,25D decreases cell proliferation and increases cell size through the activation of Rho and Rhou/Wrcth-1 pathways without inducing apoptosis. In addition, we demonstrate that 1,25D regulates genes involved in cell cycle progression and key pro- and anti-apoptotic pathways as well increases survivin expression a potentially novel 1,25D mediated pathway for regulating tissue injury and fibrosis. These advances in our understanding of key mechanistic pathways regulating antiproliferative and anti-apoptotic effects of 1,25D in a multipotent stem cell model provides an important foundation for future therapeutic interventions such as 1,25D analogs to selected activation of induced pluripotent stem cells in the prevention or treatment of presently considered irreversible hypovitaminosis D related chronic medical conditions such as neoplasias and hyperproliferative disorders including premature organ fibrosis. Although the molecular mechanisms underlying the 1,25D antiproliferative effect have been widely studied, the molecular mechanisms in many relevant models remain poorly characterized. In this manuscript we first focused on unfolding the inhibitory effect of supplementing mesenchymal multipotent cells with 1,25D by different approaches. These multipotent mesenchymal cells upon incubation with AZCT are able to differentiate into multiple phenotypes, such as contractile striated muscle cells, differentiated adipocytes and chondrocytes [16] [17] [19] [20]. The number of muscle and fat cells that appear in treated cultures depends on the concentration of 5-azacytidine (20μM) and the incubation time. Differentiated cells are not observed until 10-15 days after AZCT treatment, when the cells have passed through several division stages [17] [19]. In our case, we performed our experiments after 4 days when the cells were still in a pre-differentiated stage, which allowed us to study the effects of 1,25D on cell proliferation when cells were still in a division stage.
We showed that incubation of multipotent cells in a time and dose manner decreases cell proliferation as demonstrated by the decreased expression of PCNA, a protein found in the nucleus that acts as a cofactor of DNA polymerase delta and increases the processivity of leading strand synthesis during DNA replication. The antibody against PCNA recognizes a 36-kd protein that is present during the S phase [36]; however, in response to DNA damage it is ubiquitinated and is associated in the RAD6-dependent DNA repair pathway, so its detection will mark cell proliferation and DNA repair processes [37]. The data presented in this manuscript also demonstrate that 1,25D decreases cell number and increases cell size, a phenomenon that can be partially attributed to a reduced expression of Rho and Rhou/Wrch1, but not Wnt-1.
The Rho family of proteins plays a central role in regulating cell shape, polarity and locomotion through their effects on actin polymerization, actomyosin contractility, cell adhesion, and microtubule dynamics [38]. Rho proteins also promote reorganization of the actin cytoskeleton and regulate cell shape, attachment, and motility. It has been shown that overexpression of this gene is associated with tumor cell proliferation and metastasis. Rhou, also known as Wrch-1 (Wnt-regulated Cdc42 homolog) is an atypical member of the Cdc42 subgroup of Rho GTPases. Rhou mRNA level has been shown by Tao et al. to increase in response to Wnt-1 signaling in Wnt-1 transformed mouse mammary epithelial cells [39]. Previous work with fibroblasts and epithelial cells in culture has shown that Cdc42 contributes to G1 cell cycle progression by promoting entry into G1 and progression to S phase when expressed in quiescent fibroblasts, whereas inhibition blocks serum-induced G1 progression [40]. Moreover, a short hairpin RNA-mediated knockdown of Wrch-1 was shown to perturb cystogenesis in 3D culture, suggesting that tight regulation of Wrch-1 activity is necessary for normal epithelial morphogenesis [41].
Exploring putative mechanisms posited by Eelen and collaborators [12], we found the addition of 1,25D to mesenchymal multipotent cells blocks the transition from G1 to S1-phase of the cell cycle accumulating cells in G1 by decreasing the expression of cyclins such as Cyclin A2, B1, B2, D1, D3, E1 and F; as well as cyclin dependent kinases (Cdk) such as Cdk2 and Cdk4. Of particular interest is that in this multipotent cell model, 1,25D G1/S-blocking effect was accompanied by an increased expression of p21, a Cdk-inhibitor. However, 1,25D does not increase the expression of p27, making p21 a primary candidate for the antiproliferative effect in C3H 10T1/2 multipotent cells. It is interesting that the stable expression of p27 upon incubation of cells with 1,25D was accompanied by a decreased expression of the F-box protein Skp2 (S-phase kinase-associated protein 2) which, when expressed, positively regulates the G1-S transition by controlling the stability of several G1 regulators, such as the cell cycle inhibitor p27, down-regulating Skp2 and as a consequence causing sustained stabilization of p27, which is a key elements in the antiproliferative and anticancer actions of vitamin D.
In this present work, we found the decrease in cell proliferation was not accompanied by an increase in cell apoptosis. On the contrary, addition of 1,25D to the cell cultures exerts an anti-apoptotic effect, characterized by increased expression of the anti-apoptotic protein Bcl-2, a gene that has been thought to promote cell survival in cancer patients [42], [43]. Moreover, 1,25D induced a decreased expression of the executioner pro-apoptotic gene caspase 3. These results are in agreement with Zhang et al [44] who found that 1,25D inhibits hepatocyte apoptosis in rat allografts by regulating apoptotic-associated genes.
Reaffirming the antiapoptotic effect of 1,25D in our cell model is the fact that the gene expression of Apoptosis inhibitor 5 (Api 5/Aac11) increases when the cells have been incubated with 1,25D. Api56/Aac11 is a critical determinant of dE2F1-induced apoptosis in vivo and in vitro. It has been described that this functional interaction occurs in multiple tissues, and it is conserved from flies to humans. Interestingly, Api5/Aac11 acts downstream of E2F and suppresses E2F-dependent apoptosis without generally blocking E2F-dependent transcription. [45].
It is interesting to note that the gene expression of the proapoptotic members of the Bcl-2 family such as Bax, Bad, Bak and Bok remains unchanged when the cells have been incubated with 1,25D; reinforcing the antiapoptotic effect of 1,25D in this multipotent cell model.
We also found an increased expression of Survivin, the Baculoviral IAP repeat-containing 5 (Birc5). Survivin contains a baculovirus inhibitor of apoptosis repeat (BIR) a protein domain that is a member of the inhibitor of apoptosis protein (IAP) family. Survivin inhibits apoptosis via its BIR domain, by directly or indirectly interfering with the function of caspases. Survivin is also a chromosomal passenger protein that is required for cell division and is expressed in embryonic tissues as well as in the majority of human cancers, but is not expressed in most normal adult tissues [46]. Recently, emerging evidence suggest that survivin is involved in tissue injury and in wound healing [47] [48], which is in agreement with our previous publication regarding the antifibrotic effects of 1,25D in the same multipotent cell system [20].
A previous report by Wei and colleagues has identified the pro-apoptotic role of activated transcription factor 5 (ATF5) and identified Cyclin D3 as an ATF5-targeted apoptosis-related gene [49]. The same article showed that the ectopic expression of ATF5 in HeLa cells markedly increased cisplatin-induced apoptosis and the cleavage of Caspase-3, and induced Cyclin D3 mRNA expression via cooperation with E2F1 transcription factor.
In summary, the addition of 1,25D, the active form of vitamin D, to multipotent mesenchymal cells induced a decrease in cell proliferation, characterized by an increase in cell size, a phenomenon possibly linked to decrease in Rho and Rhou/Wrch-1 expression; it also induced cell cycle arrest, promoting accumulation of cells in G0/G1 phase without inducing apoptosis. The data presented in this manuscript provide a mechanistic explanation for the anti-proliferative and anti-apoptototic properties of 1,25D observed in mesenchymal multipotent cells and provides the mechanistic foundation for not only traditional therapeutic interventions such as 1,25D analogs but select activation of induced pluripotent stem cells in targeted clinical studies.
Acknowledgments
This work was supported by National Institutes of Health (NIH). National Center for Research Resources (NCRR) / Research Centers in Minority Institutions (RCMI) U54RR022762, RR019234 and RR003026, and National Center on Minority Health & Disparities (NCMHD) UCLA/Drew Project EXPORT MD000545 and 2P20MD000182.
Abbreviations
- 1,25D
1α, 25-dihydroxyvitamin D3
- PCNA
proliferating cell nuclear antigen
- VDR
vitamin D receptor
- Rho
Ras homolog
- Wrch-1
Wnt-regulated Cdc42 homolog
- Bcl-2
B-cell leukemia/lymphoma 2
- GAPDH
glyceraldehyde-3-phosphate-dehydrogenase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Feldman D, Pike JW, Glorieux FH. Vitamin D. 2nd. Elsevier Academic Press; 2005. Chapter 11. [Google Scholar]
- 2.Dusso AS, Brown AJ. Mechanism of vitamin D action and its regulation. Am J Kidney Dis. 1998;32:S13–S24. doi: 10.1053/ajkd.1998.v32.pm9808140. [DOI] [PubMed] [Google Scholar]
- 3.Dusso AS, Brown AJ, Slatopolsky E. Vitamin D. Am J Physiol Renal Physiol. 2005;289:F8–F28. doi: 10.1152/ajprenal.00336.2004. [DOI] [PubMed] [Google Scholar]
- 4.Zehnder D, Bland R, Williams MC, McNinch RW, Howie AJ, Stewart PM, Hewison M. Extrarenal expression of 25-hydroxyvitamin d(3)-1 alpha-hydroxylase. J Clin Endocrinol Metab. 2001;86(2):888–94. doi: 10.1210/jcem.86.2.7220. [DOI] [PubMed] [Google Scholar]
- 5.Carlberg C, Quack M, Herdick M, Bury Y, Polly P, Toell A. Central role of VDR conformations for understanding selective actions of vitamin D(3) analogues. Steroids. 2001;66:213–221. doi: 10.1016/s0039-128x(00)00150-1. [DOI] [PubMed] [Google Scholar]
- 6.Lemon BD, Freedman LP. Selective effects of ligands on vitamin D3 receptor- and retinoid X receptor-mediated gene activation in vivo. Mol Cell Biol. 1996;16:1006–1016. doi: 10.1128/mcb.16.3.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rachez C, Freedman LP. Mechanisms of gene regulation by vitamin D3 receptor: a network of coactivator interactions. Gene. 2000;246:9–21. doi: 10.1016/s0378-1119(00)00052-4. [DOI] [PubMed] [Google Scholar]
- 8.Mathieu C, Bouillon R, Rutgeerts O, Vandeputte M, Waer M. Potentia l role of 1,25(OH)2 vitamin D3 as a dose-reducing agent for cyclosporine and FK 506. Transplant Proc. 1994;26(6):30–31. [PubMed] [Google Scholar]
- 9.Casteels K, Waer M, Bouillon R, Depovere J, Valckx D, Laureys J, Mathieu C. 1,25-Dihydroxyvitamin D3 restores sensitivity to cyclophosphamide-induced apoptosis in non-obese diabetic (NOD) mice and protects against diabetes. Clin Exp Immunol. 1998;112(2):181–7. doi: 10.1046/j.1365-2249.1998.00568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malluche HH, Mawad H, Koszewski NJ. Update on vitamin D and its newer analogues: actions and rationale for treatment in chronic renal failure. Kidney Int. 2002;62:367–374. doi: 10.1046/j.1523-1755.2002.00450.x. [DOI] [PubMed] [Google Scholar]
- 11.Dusso AS, Thadhani R, Slatopolsky E. Vitamin D receptor and analogs. Semin Nephrol. 2004;24(1):10–6. doi: 10.1053/j.semnephrol.2003.08.018. [DOI] [PubMed] [Google Scholar]
- 12.Eelen G, Gysemans C, Verlinden L, Vanoirbeek E, De Clercq P, Van Haver D, Mathieu C, Bouillon R, Verstuyf A. Mechanism and potential of the growth-inhibitory actions of vitamin D and analogs. Curr Med Chem. 2007;14(17):1893–910. doi: 10.2174/092986707781058823. [DOI] [PubMed] [Google Scholar]
- 13.Martinesi M, Treves C, d'Albasio G, Bagnoli S, Bonanomi AG, Stio M. Vitamin D derivatives induce apoptosis and downregulate ICAM-1 levels in peripheral blood mononuclear cells of inflammatory bowel disease patients. Inflamm Bowel Dis. 2008;14(5):597–604. doi: 10.1002/ibd.20354. [DOI] [PubMed] [Google Scholar]
- 14.Zhang A, Wang Y, Xie H, Zheng S. Calcitriol inhibits hepatocyte apoptosis in rat allograft by regulating apoptosis-associated genes. Int Immunopharmacol. 2007;7(8):1122–8. doi: 10.1016/j.intimp.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Pendás-Franco N, González-Sancho JM, Suárez Y, Aguilera O, Steinmeyer A, Gamallo C, Berciano MT, Lafarga M, Muñoz A. Vitamin D regulates the phenotype of human breast cancer cells. Differentiation. 2007;75(3):193–207. doi: 10.1111/j.1432-0436.2006.00131.x. [DOI] [PubMed] [Google Scholar]
- 16.Rainier S, Feinberg AP. Capture and characterization of 5-aza-2′-deoxycytidine-treated C3H/10T1/2 cells prior to transformation. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6384–8. doi: 10.1073/pnas.85.17.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh R, Artaza JN, Taylor WE, Gonzalez-Cadavid NF, Bhasin S. Androgens stimulate myogenic differentiation and inhibit adipogenesis in C3H 10T1/2 pluripotent cells through an androgen receptor-mediated pathway. Endocrinology. 2003;144(11):5081–8. doi: 10.1210/en.2003-0741. [DOI] [PubMed] [Google Scholar]
- 18.Pittenger MF, Martin BJ. Mesenchymal stem cells and their potential as cardiac therapeutics. Circ Res. 2004;95(1):9–20. doi: 10.1161/01.RES.0000135902.99383.6f. Review. [DOI] [PubMed] [Google Scholar]
- 19.Artaza JN, Bhasin S, Magee TR, Reisz-Porszasz S, Shen R, Groome NP, Meerasahib MF, Gonzalez-Cadavid NF. Myostatin inhibits myogenesis and promotes adipogenesis in C3H 10T1/2 mesenchymal multipotent cells. Endocrinology. 2005;146(8):3547–57. doi: 10.1210/en.2005-0362. [DOI] [PubMed] [Google Scholar]
- 20.Artaza JN, Norris KC. Vitamin D reduces the expression of collagen and key profibrotic factors by inducing an antifibrotic phenotype in mesenchymal multipotent cells. J Endocrinol. 2009;200(2):207–21. doi: 10.1677/JOE-08-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher I, Boland G, Tuan RS. Wnt-3A enhances bone morphogenetic protein-2-mediated chondrogenesis of murine C3H 10T1/2 mesenchymal cells. J Biol Chem. 2002;277:30870–30878. doi: 10.1074/jbc.M109330200. [DOI] [PubMed] [Google Scholar]
- 22.Wilson EM, Rotwein P. Control of MyoD function during initiation of muscle differentiation by an autocrine signaling pathway activated by insulin-like growth factor-II. J Biol Chem. 2006;281(40):29962–71. doi: 10.1074/jbc.M605445200. [DOI] [PubMed] [Google Scholar]
- 23.Brackman D, Lillehaug JR, Aksnes L, Aarskog D. Modulation of cell proliferation and cell cycle, and inhibition of cytokinesis by 1,25-dihydroxyvitamin D3 in C3H/10T1/2 fibroblasts. J Steroid Biochem Mol Biol. 1993;46(2):155–62. doi: 10.1016/0960-0760(93)90290-d. [DOI] [PubMed] [Google Scholar]
- 24.Artaza JN, Singh R, Ferrini MG, Braga M, Tsao J, Gonzalez-Cadavid NF. Myostatin promotes a fibrotic phenotypic switch in multipotent C3H 10T1/2 cells without affecting their differentiation into myofibroblasts. J Endocrinol. 2008;196(2):235–49. doi: 10.1677/JOE-07-0408. [DOI] [PubMed] [Google Scholar]
- 25.Schmittwolf C, Kirchhof N, Jauch A, Durr M, Harder F, Zenke M, Muller AM. In vivo haematopoietic activity is induced in neurosphere cells by chromatin-modifying agents. EMBO Journal. 2005;24:554–566. doi: 10.1038/sj.emboj.7600546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor SM, Jones PA. Multiple new phenotypes induced in 10T1/2 and 2T3 cells treated with azacytidine. Cell. 1979;17:769–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- 27.Asakura A, Komaki M, Rudnicki M. Muscle satellite cells are multipotential stem cells that exhibit myogenic, osteogenic, and adipogenic differentiation. Differentiation. 2001;68:245–253. doi: 10.1046/j.1432-0436.2001.680412.x. [DOI] [PubMed] [Google Scholar]
- 28.Jasuja R, Ramaraj P, Mac RP, Singh AB, Storer TW, Artaza J, Miller A, Singh R, Taylor WE, Lee ML, Davidson T, Sinha-Hikim I, Gonzalez-Cadavid N, Bhasin S. Delta-4-androstene-3,17-dione binds androgen receptor, promotes myogenesis in vitro, and increases serum testosterone levels, fat-free mass, and muscle strength in hypogonadal men. J Clin Endocrinol Metab. 2005;90(2):855–63. doi: 10.1210/jc.2004-1577. [DOI] [PubMed] [Google Scholar]
- 29.Seale P, Rudnicki MA. A new look at the origin, function, and stem-cell status of muscle satellite cells. Dev Biol. 2000;218:115–124. doi: 10.1006/dbio.1999.9565. [DOI] [PubMed] [Google Scholar]
- 30.Asakura A. Stem cells in adult skeletal muscle. Trends Cardiovasc Med. 2003;13:123–128. doi: 10.1016/s1050-1738(03)00024-0. [DOI] [PubMed] [Google Scholar]
- 31.Atkinson BL, Fantle KS, Benedict JJ, Huffer WE, Gutierrez-Hartmann A. Combination of osteoinductive bone proteins differentiates mesenchymal C3H/10T1/2 cells specifically to the cartilage lineage. J Cell Biochem. 1997;65(3):325–39. [PubMed] [Google Scholar]
- 32.Artaza JN, Reisz-Porszasz S, Dow JS, Kloner RA, Tsao J, Bhasin S, Gonzalez-Cadavid N. Alterations in myostatin expression are associated with changes in cardiac left ventricular mass but not ejection fraction in the mouse. J Endocrinol. 2007;194(1):63–76. doi: 10.1677/JOE-07-0072. [DOI] [PubMed] [Google Scholar]
- 33.Essers J, Theil AF, Baldeyron C, van Cappellen WA, Houtsmuller AB, Kanaar R, Vermeulen W. Nuclear dynamics of PCNA in DNA replication and repair. Mol Cell Biol. 2005;25(21):9350–9. doi: 10.1128/MCB.25.21.9350-9359.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horan PK, Slezak SE. Stable cell membrane labeling Nature. 1989;340(6229):167–8. doi: 10.1038/340167a0. [DOI] [PubMed] [Google Scholar]
- 35.Singh R, Artaza JN, Taylor WE, Braga M, Yuan X, Gonzalez-Cadavid NF, Bhasin S. Testosterone inhibits adipogenic differentiation in 3T3-L1 cells: nuclear translocation of androgen receptor complex with beta-catenin and T-cell factor 4 may bypass canonical Wnt signaling to down-regulate adipogenic transcription factors. Endocrinology. 2006;147(1):141–54. doi: 10.1210/en.2004-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peled A, Shezen E, Schwartz D, Shav-Tal Y, Kushtai G, Lee BC, Gothelf Y, Krupsky M, Zipori D. Nuclear antigen expressed by proliferating cells. Hybridoma. 1997;16(4):325–34. doi: 10.1089/hyb.1997.16.325. [DOI] [PubMed] [Google Scholar]
- 37.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419(6903):135–41. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 38.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 39.Tao W, Pennica D, Xu L, Kalejata RF, Levine AJ. Wrch-1, a novel member of the Rho gene family that is regulated by Wnt-1. Genes Dev. 2001;15:1796–1807. doi: 10.1101/gad.894301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olson MF, Ashworth A, Hall A. An essential role for Rho, Rac and CDC42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 41.Brady DC, Alan JK, Madigan JP, Fanning AS, Cox AD. The Transforming Rho Family GTPase Wrch-1 Disrupts Epithelial Cell Tight Junctions and Epithelial Morphogenesis. Molecular and Cellular Biology. 2009;29(4):1035–1049. doi: 10.1128/MCB.00336-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ilievska Poposka B, Smickova S, Jovanovska Crvenkovska S, Zafirovska Ivanovska B, Stefanovski T, Petrusevska G. Bcl-2 as a prognostic factor for survival in small-cell lung cancer. Prilozi. 2009;29(2):281–94. [PubMed] [Google Scholar]
- 43.Tsamandas AC, Kardamakis D, Tsiamalos P, Liava A, Tzelepi V, Vassiliou V, Petsas T, Vagenas K, Zolota V, Scopa CD. The potential role of Bcl-2 expression, apoptosis and cell proliferation (Ki-67 expression) in cases of gastric carcinoma and correlation with classic prognostic factors and patient outcome. Anticancer Res. 2009;29(2):703–9. [PubMed] [Google Scholar]
- 44.Zhang A, Wang Y, Xie H, Zheng S. Calcitriol inhibits hepatocyte apoptosis in rat allograft by regulating apoptosis-associated genes. Int Immunopharmacol. 2007;7(8):1122–8. doi: 10.1016/j.intimp.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 45.Morris EJ, Michaud WA, Ji JY, Moon NS, Rocco JW, Dyson NJ. Functional identification of Api5 as a suppressor of E2F-dependent apoptosis in vivo. PLoS Genet 17. 2006;2(11):196. doi: 10.1371/journal.pgen.0020196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiou SK, Jones MK, Tarnawski AS. Survivin - an anti-apoptosis protein: its biological roles and implications for cancer and beyond. AS Med Sci Monit. 2003;9(4):125–9. [PubMed] [Google Scholar]
- 47.Zhang M, Lin L, Lee SJ, Mo L, Cao J, Ifedigbo E, Jin Y. Deletion of caveolin-1 protects hyperoxia-induced apoptosis via survivin-mediated pathways. Am J Physiol Lung Cell Mol Physiol. 2009;297(5):L945–53. doi: 10.1152/ajplung.00081.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li JY, Gu X, Yin HZ, Zhou Y, Zhang WH, Qin YM. Protective effect of ischemic preconditioning on hepatic ischemia-reperfusion injury by advancing the expressive phase of survivin in rats. Hepatobiliary Pancreat Dis Int. 2008;7(6):615–20. [PubMed] [Google Scholar]
- 49.Wei Y, Jiang J, Sun M, Chen X, Wang H, Gu J. ATF5 increases cisplatin-induced apoptosis through up-regulation of cyclin D3 transcription in HeLa cells. Biochem Biophys Res Commun. 2006;339(2):591–6. doi: 10.1016/j.bbrc.2005.11.054. [DOI] [PubMed] [Google Scholar]


