Abstract
Antimalarial drugs impose strong pressure on Plasmodium falciparum parasites and leave signatures of selection in the parasite genome 1,2. Search for signals of selection may lead to genes encoding drug or immune targets 3. The lack of high-throughput genotyping methods, inadequate knowledge of parasite population history, and time-consuming adaptations of parasites to in vitro culture have hampered genome-wide association studies (GWAS) of parasite traits. Here we report genotyping of DNA from 189 culture-adapted P. falciparum parasites using a custom-built array with thousands of single nucleotide polymorphisms (SNPs). Population structure, variation in recombination rate, and loci under recent positive selection were detected. Parasite half maximum inhibitory concentrations (IC50) to seven antimalarial drugs were obtained and used in GWAS to identify genes associated with drug responses. The SNP array and genome-wide parameters provide valuable tools and information for new advances in P. falciparum genetics.
Keywords: malaria, single nucleotide polymorphism (SNP), genome-wide association study, recombination, drug resistance, population structure
Drug resistance in P. falciparum parasites has evolved and spread rapidly, leading to the loss of chloroquine (CQ) and sulfadoxine-pyrimethamine (SP) as first-line treatments in most endemic areas. Resistance to all antimalarial drug classes has been reported, including recently the artemisinin (ART) derivatives 4–7. Mutations in the P. falciparum CQ resistance transporter gene (pfcrt) and the genes encoding dihydrofolate reductase (pfdhfr) and dihydropteroate synthase (pfdhps) have been shown to confer resistance to CQ and SP, respectively. Additionally, copy number and/or point mutations at the gene encoding a homolog of human P-glycoprotein (pfmdr1) on chromosome 5 have been associated with parasite response to mefloquine (MQ), quinine (QN), ART, and other antimalarial drugs, although other unknown genes may have roles in the responses 8. P. falciparum resistance to antimalarial drugs has occurred only since widespread deployment of the drugs (i.e. within the past 60 years), and there may have not been enough time for recombination to break down completely linkages between causal alleles and nearby genetic markers. Indeed, by scanning for regions of high LD, the chromosome segment carrying the pfcrt locus was correctly identified using 342 genome-wide microsatellite (MS) markers and 92 parasite isolates collected from different parts of the world 1. Here we report the first genome-wide P. falciparum maps of population recombination events, signatures of recent positive selection, and GWAS of multiple drug resistant phenotypes and SNP genotypes obtained using a custom-built SNP typing microarray.
RESULTS
We collected and adapted 189 independent P. falciparum isolates into in vitro culture, including 146 from the Asia (Thailand and Cambodia), 26 from Africa, 14 from America, and 3 from Papua New Guinea (Supplementary Table 1). We developed a custom 3K oligo probe array based on the molecular inversion probe (MIP) technology (Affymetrix Inc, Santa Clara, CA) 9 to interrogate 3354 SNPs we identified previously 3. The MIP array provides a simple and reliable method to genotype the 23 megabase (mb) P. falciparum genome with a coverage averaging ~one SNP per 7 kilobase (kb). Among the 3257 (97.1%) SNPs called, 2763 (82.4%) had call rate >90%, and only seven were different from those in the 3D7 genome sequence (0.2%). One thousand, eight hundreds and eighty-nine (58.3%) SNPs had a minor allele frequency (MAF) greater then 2% among all the parasites; 1216 (37.3%) SNPs had MAF >2% in the Asian population; 1637 (50.3%) SNPs had MAF >2% in the African population; and 813 (24.9%) had MAF >2% in the American populations.
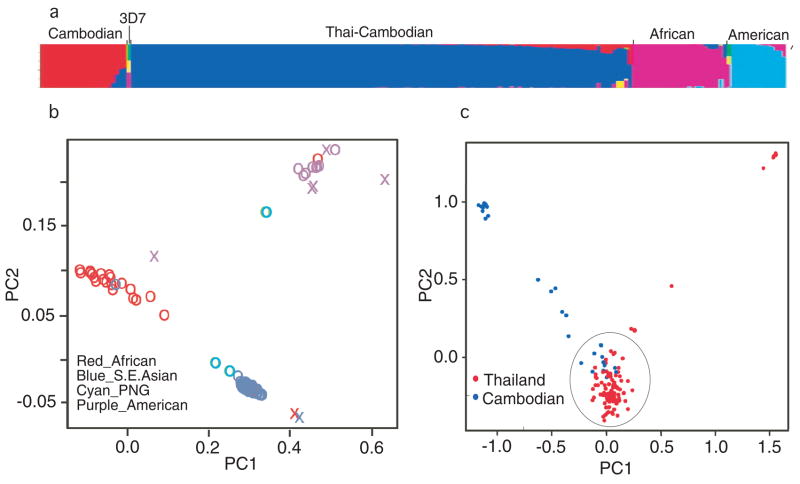
We tested for genetic heterogeneity that may be associated with geography. STRUCTURE analyses 10 showed that the parasites could be clustered into continental populations, with a group of Cambodian parasites separated from the majority of the those of Thailand and Cambodia (Fig. 1a). Similarly, principal component analysis (PCA) using EIGENSOFT11 identified significant axes of variations partitioning the parasites into clusters of Asia, Africa, America (Fig. 1b) as well as distinct groups of parasites from Thai-Cambodian regions (Fig. 1c). The clustering of Cambodian parasites, which were collected from sites within a radius of ~50 km, into different groups suggests either a recent population admixture or possibly the presence of SNPs that could distinguish parasites with different phenotypes. These population clusterings were corroborated with Wright’s Fst values (Africa vs Asia 0.054; Africa vs America 0.136; Asia vs America 0.028, and between the two Cambodian populations 0.254). The large Fst value for the Cambodian populations was due to fixation of ~75% SNPs (765/1024) in the outlier Cambodian population.
Figure 1.
Population structure and principal component analysis (PCA) of Plasmodium falciparum parasite populations. (a) Population partitions using STRUCTURE (v2.2) 10. The Cambodian group (red) consists of parasites CP195, CP201, CP216, CP285, CP286, CP291, CP313, CP268, CP325, CP305, CP307, CP256, CP238, and CP211. (b) PCA plot of all the parasites. Parasite continental origins are as color-coded and ‘X’ indicates outliers. PNG, Papua New Guinea. (c) PCA plot of the Thai-Cambodian parasites showing outliers from the region.
Using genome-wide SNPs, we generated population recombination maps for all 14 chromosomes. Interestingly, the five largest chromosomes (9–14) had relatively fewer recombination events than the smaller chromosomes (Supplementary Fig. 1). Similar to those observed on chromosome 3 12, many recombination hot- or cold-spots appeared to be ‘conserved’ among populations. There were several loci with extremely high levels of recombination activity, including a locus at one end of chromosome 1 and a segment on chromosome 7 containing pfcrt (from 400 to 800 kb) that had a mosaic recombination pattern. The chromosome 7 recombination hotspots flanked a central 100 kb segment (containing pfcrt) with a reduced recombination activity, suggesting a recent selective sweep. In contrast, balancing selection on the nearby var and other genes may favor higher rates of allelic exchange.
We mapped chromosomal loci potentially under selection using relative extended haplotype homozygosity (REHH) 13,14, integrated haplotype score (iHS) 15, and cross population extended haplotype homozygosity (XP-EHH) 14,16. We generated genome-wide maps of selection for parasite populations from Asia, Africa, and America, separately, and detected many loci that were under significant positive selection (Fig. 2). Examples of recent positive selection from REHH included the locus on chromosome 7 containing pfcrt, a locus on chromosome 11 containing pfama-1, and a locus on chromosome 13 containing an ABC transporter (PF13_0271) (Fig. 2a and Supplementary Table 2). The pfcrt gene is under CQ selection 1; pfama-1 is a target of host immune response 17; and the gene encoding the ABC transporter on the chromosome 13 was predicted to transport iron into mitochondrion (PlasmoDB). Other signals evident in Fig. 2a were likely to represent regions containing genes for either antigens and/or putative transporters that may be under immune or drug selection pressures (Supplementary Table 2).
Figure 2.
Loci subject to positive selection in Plasmodium falciparum populations from Africa, Asia and America. (a) plots of −Log P values showing loci significantly under positive selection. Arrowheads point to loci containing the genes encoding chloroquine resistance transporter (pfcrt) on chromosome 7, the apical membrane antigen (pfama-1) on chromosome 11, and an ABC transporter on chromosome 13, respectively; Dots above the dash lines indicate significant. (b) plots of integrated haplotype scores (iHS) showing loci under selection. Arrowheads indicate the pfcrt and pfama-1 loci on chromosome 7 and 11, respectively. SNPs with |iHS| values≥2.3 were those above the horizontal line in each graph. Each dot represents an |iHS| value from a window of 21 SNPs (a core SNP plus 10 SNPs on each side). (c) plots of −logP values from cross population extended haplotype homozygosity (XP-EHH) analyses. AF/AM, comparison of African and American populations; AF/AS, comparison of African and Asian populations; AS/AM, comparison of Asian and American populations. The horizontal lines indicate significant P-values (<0.05), and the arrowhead points to the pfcrt locus on chromosome 7 and PFE1445c locus on chromosome 5, respectively.
Similarly, iHS detected strong selection signals at the pfcrt and pfama-1 loci (Fig. 2b), consistent with REHH results. Additional interesting iHS signals included PFA0655w (encoding a member of SURFIN) 18 on chromosome 1 and a putative metabolite/drug transporter (PF14-0260) on chromosome 14. If we used an iHS score of 2.3 as a significant cutoff value (approximately top 1% of theoretical iHS distribution), we identified many SNPs that were also identified using REHH (Supplementary Table 2 and Supplementary Table 3). We also performed XP-EHH to detect selective sweeps that drive some alleles to fixation in one population but remains polymorphic in others. Indeed, many extended haplotypes were detected between populations (Fig. 2c and Supplementary Table 4). Again, the pfcrt locus had highly significant P-values, particularly in comparison of African v.s. American (AF/AM) and African v.s Asian (AF/AS) populations. Another gene with very significant XP-EHH P-value was PFE1445c on chromosome 5 that encoded a Plasmodium conserved protein (Fig. 2 and Supplementary Table 4). There were also several large extended haplotypes (519126-922368 bp on chromosome 7, 831749-925515 bp on chromosome 8, 319075-495408 bp on chromosome 9) between African and American populations. A total of 11 genes under significant selection were detected by all the three methods (Supplementary Table 5), although the signatures at the chromosome 7 locus may be due to selective sweeps and hitchhiking 1. Other genes such as PFC0940c and PFE1445c were highly polymorphic with predicted transmembrane domains and were likely conserved antigen genes in Plasmodium.
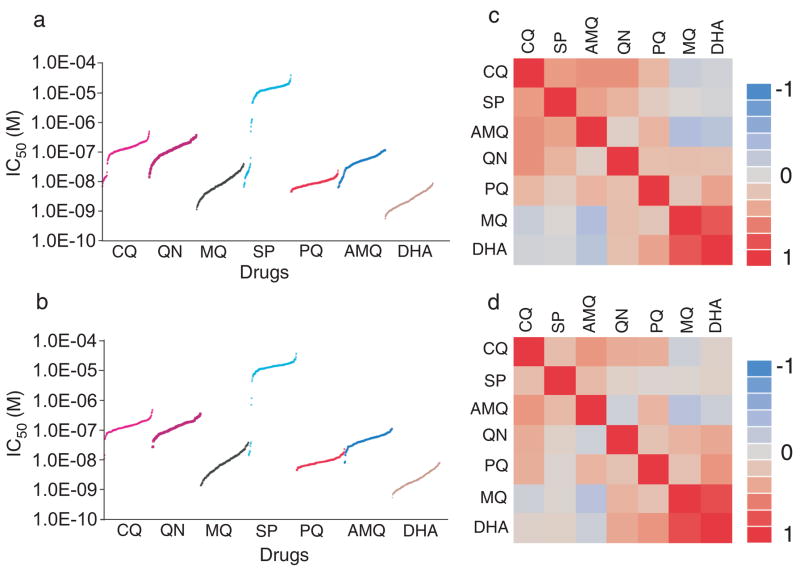
To detect genes associated with drug responses, we measured IC50 of CQ, QN, MQ, SP, dihydroartemisinin (DHA), amodiaquine (AMQ), and piperaquine (PQ) from 185 culture-adapted parasites using a SYBR green method 19 (Fig. 3a and Supplementary Table 1) and conducted GWAS. Except for CQ and SP that had bimodal distributions of IC50 values, the distributions of IC50 values for the other five drugs were more unimodal (Fig. 3a). All the parasites were sensitive to PQ and DHA. The range of IC50 values for PQ was small (5 folds) while the IC50 range for SP was large (~56,000 folds). The IC50 ranges for the other drugs were 10-fold or higher (16 fold for DHA; 17 fold for AMQ; 26 fold for QN; 34 fold for MQ; 70 fold for CQ). Parasites from the Thai-Cambodian population had similar distributions of IC50 values to those of the worldwide population, except that there were only 2 and 6 (out of 143) parasites that were sensitive to CQ and SP, respectively (Fig. 3b). We also compared the IC50 values of the parasites from the two genetically distinct Cambodian populations and found that the average IC50 for the all drugs were not significantly different (unpaired t-test; data not shown).
Figure 3.
In vitro parasite responses (IC50) to seven antimalarial drugs. (a) IC50 values to seven different antimalarial drugs from 185 parasites were sorted from the lowest to the highest values. Note gaps in IC50 values in parasite responses to chloroquine (CQ) and sulfadoxine-pyrimethamine (SP), but continuous distributions for the other drugs. IC50 curves for each drug as marked in the figure; (b) similar plots as in (a) for parasites from Thai-Cambodian population; (c), (d) multivariate analyses showing correlations between responses to seven different drugs for all the parasites (c) and Thai-Cambodian parasites (d). Dihydroartemisinin (DHA) and mefloquine (MQ) had strong positive correlation; CQ, amodiaquine (AMQ), piperaquine (PQ), quinine (QN), and SP also had positive correlation to some degree; whereas AMQ/DHA, AMQ/MQ had negative correlation.
Delayed parasite clearance following artesunate treatment or artemisinin combination therapy (ACT) has been reported from patients at the Thai-Cambodian border 6,7. The Cambodian parasites did have a significantly higher mean IC50 value to DHA (5.2±1.5 nM) compared to the parasites from Thailand (2.0±1.0 nM) and America (2.5±1.1 nM) (t- tests, P<0.001), but not from Africa (3.1±2.4 nM) (P=0.09). As reported previously 20,21, multivariate analyses showed a strong positive correlation (R2= 0.78) between IC50 values of MQ and DHA, some positive correlations between CQ IC50 values and those of SP (R2= 0.47), AMQ (R2= 0.52), and QN (R2= 0.52), and slight negative relationships between DHA/AMQ, MQ/AMQ, CQ/MQ, and CQ/DHA among all the parasites (Fig. 3c) and those from the Thai-Cambodian population (Fig. 3d). The strong positive correlation between the responses to DHA and MQ suggests either co-selection by the drugs and/or a common resistance mechanism. This association may also be partly explained by pfmdr1 amplification.
We performed GWAS on individual populations using PLINK 22 and EIGENSOFT (Figure 4 and Supplementary Table 6). Quantile-Quantile plots suggested effective correction of potential population structure (Supplementary Fig. 2). Although several genes were associated with responses to CQ, QN, DHA and MQ, only MAL7P1.27-9 (pfcrt), PFA0665w-18 (pfsurfin) and PFE1150w-4 (pfmdr1) had a minor allele frequency higher than 15%. All of the three genes were also under positive selection. The association of pfcrt with CQ response is well established 23. Likewise, the association of pfmdr1 with QN is consistent with the linkage of QN response to polymorphisms in the gene 24 and with altered QN IC50 values in parasites engineered to have wild type pfdmr1 allele replaced with a mutant allele 25. Association of PFA0665w (SURFIN) with responses to antimalarial drugs has not been reported previously. SURFIN was reported to be co-transported with PfEMP1 and RIFIN to the infected erythrocyte surface 18 and could be part of a protein complex involved in binding or transport chemical compounds. There were also two Plasmodium conserved genes (PF11_0079 and PFC0460w) with significant P-values from both EIGENSOFT and PLINK. However, the associations of some of these candidate genes could be due to linkage to genes nearby that might be the real actors. The functions of these candidate genes in the associated loci and their contributions to antimalarial drug resistance require further studies
Figure 4.
Genome-wide scan for SNPs associated with responses to antimalarial drugs in the Asian population. Values of −Log P for four drugs were plotted against chromosomal positions. The arrowheads indicate SNPs with Bonferroni corrected P<0.05. (a) plots from EIGENSOFT; (b) plots from PLINK. CQ, chloroquine; QN, quinine; MQ, mefloquine; DHA, dihydroartemisinin.
Our in vitro assays suggest that P. falciparum strains from different continents remain sensitive to DHA and PQ, although parasites from Cambodia are generally more resistant to the drugs. Many genes under recent positive selection were identified, some of which could be drug or immune targets. The candidate genes associated with responses to the antimalarial drugs require further verification due to small parasite sample size and low minor allele frequencies. Gene copy number variation has been reported to contribute to parasite drug response 26,27 and need to be investigated too. The high throughput MIP array, estimates of genome-wide recombination events and recent positive selection maps provided important tools and information for GWAS to identify genes controlling various malaria traits.
ONLINE METHODS
Parasite collection
All the parasites used in this study were culture-adapted clonal lines collected from 23 different countries. Some of the Asian parasites and all the parasites from Africa, America, and PNG were described previously 28,29. Thirty-four parasites from Cambodia were collected in a clinical study approved by the IRBs of the National Institute Allergy and Infectious Diseases, USA; the Ministry of Health of the Kingdom of Cambodia; and the Guangzhou University of Chinese Medicine, Guangzhou, the People’s Republic of China, with informed consent obtained from all subjects. The identity and clonality of the parasites were verified using multiple microsatellites before drug assays.
DNA extraction and SNP genotyping using MIP array
Parasite culture and genomic DNA extraction were as described 30. Genomic DNA isolated from Plasmodium falciparum grown in culture was genotyped using the custom designed 3K Malaria Panel (Affymetrix Inc, Santa Clara, CA). Samples were prepared with the Malaria 3K Panel following the GeneChip® Scanner 3000 Targeted Genotyping System Protocol and hybridized to Universal 3K Tag arrays (Affymetrix Inc, Santa Clara, CA). The only modification in the assay protocol was to normalize samples to a starting concentration of 65ng/μL that equates to a total gDNA input of 871ng. Following hybridization and scanning, genotypes were assigned using the GeneChip® Targeted Genotyping Analysis Software (Affymetrix Inc.) with the following changes to the default clustering parameters: MinHetToHalfRatio=0.5, and MinAssayCallRate=90. Genotypes were scored and stored in Excel sheets for further analyses.
Drug assays and IC50 calculation
Drug assays were performed as described previously30,31. To ensure high quality of phenotypic data, we repeated all drug assays at least 3 times independently using the same drug stock solutions. CQ, QN, MQ and DHA were purchased from Sigma-Aldrich (St. Louis, USA); SP was obtained from Roche (Indianapolis, USA); AMQ was bought from LGC Promochem (UK), and PQ was obtained from Guangzhou University of Traditional Chinese Medicine, China. The same stock solution for each drug (10mM in ethanol, except SP in dimethylsulfoxide) was used in all drug assays. The 3D7 parasite was included in all drug assays as a control for plate-to-plate variation.
Structure, Fst and principal component analysis
We applied PCA, a Bayesian clustering approach, as implemented in the program EIGENSOFT 11 and STRUCTURE (v2.2) 10, respectively, and Fst to investigate potential population structure. We used Wright’s population differentiation estimator Fst to ensure ploidy independence. To run the STRUCTURE program, we applied the same conditions described previously 12. Briefly, ten runs of 50,000 burn-ins and 100,000 iterations were performed for K=1 to 10 using the admixture model. For PCA, we used the LD correction and calculated the top 10 eigenvectors or principal components (PCs) from the genotypes of the African, Asian, and American populations. We identified and removed isolates that were greater than 6 standard deviations from the PC mean along any of the top 5 PCs and repeated the PCA calculation and outlier detection for 10 iterations.
Estimate of recombination events
Nonparametric estimates of the number of recombination event (Rh) were calculated using the Myers and Griffiths method as described previously 12. The 14 chromosomes were analyzed individually for African, Asian, American and the Cambodian populations.
Detection of recent positive selection
We used long-range haplotype (LRH) and integrated haplotype score (iHS) to detect loci under recent natural selection in parasite genome 13,15. For LRH analysis, we compare the REHH extending 100kb in both directions from a core SNP. For iHS, extended haplotype homozygosity (EHH) was calculated with a window size of 10 SNPs in each direction from the core SNP, and then EHH was integrated using physical distance resulting in the integrated EHH (iHH) for each allele at the core SNP. The log ratio of the major allele iHH to minor allele iHH was taken and, conditioning on minor allele frequency, standardized to have mean = 0 and variance = 1, resulting in the iHS score for the core SNP. Theoretical cutoffs for the 1% of signals genome-wide was considered as strong signals to indicate candidate selection regions. Isolates from three different geographic locations were tested separately. For XP-EHH analysis, we calculated EHH and the log ratio iHH for the pair-wise tests of the African, Asian and American populations as described 16. The log ratios were standardized to have mean 0, variance 1, and assigned P values assuming a normal distribution. SNPs with P-values less than 0.05 where considered strong signals.
Genome-wide association analysis
The individual populations were analyzed for association to the seven antimalarial drugs using EigenstratQTL in the EIGENSOFT program, utilizing PCA to control for population structure within the populations. Population structure was corrected using three, one, and zero significant PCs in the PCA for the Asian, African, and American populations, respectively. The correction is a function of sample position and the regression of genotypes at PC position for that sample, which adjusted genotypes and phenotypes and effectively eliminated population structure within each individual population. The correction for the genotype of sample i at SNP j is: Where aj is the ancestry/position of individual j in the PC.
Test statistic is (N − K)* correlation (corrected genotypes, corrected phenotypes)^2, where N = number of isolates (N = 133), and K = number of PCs used for correction (K = 3). The correlation between corrected genotypes and corrected phenotypes were obtained with the top 3 PC’s as fixed effects. Nominal P-values were determined using the Chi-sq distribution, df =1. Bonferroni P-values were determined as 1-(1-nominal P-value)^number of successful tests.
Association analysis was also performed using software PLINK 22. Because PLINK does not have PCA correction within its test, population outliers from PCA analysis (those outside the circle in Fig 1c) were removed before association analysis. A linear regression was fitted to test for each SNP for its association with in vitro IC50 values of the seven antimalarial drugs. Significant SNPs (P<0.05) were determined after Bonferroni correction. Quantile-Quantile plots for both methods were obtained by contrasting uncorrected and corrected (if applicable) experimental P value distributions to the expected uniform 0 to 1 distribution.
Supplementary Material
Acknowledgments
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, and funds from the Canadian Institute of Health Research #11284, National Academies Keck Genome Initiative, and the Human Frontiers in Science Program #RGP54/2006 for P.A. KC and NW are supported by the Wellcome Trust. S.L. was supported by the 973 National Basic Research Program of China, #2007CB513103. We also thank Dr. Jetsumon Sattabongkot for help in parasite shipping; Ms. Josephine Dunn and Mr. Louie Zhang for assistance in parasite culture; and NIAID intramural editor Brenda Rae Marshall for assistance.
Footnotes
Accession codes. The SNPs have been submitted to NCBI databases with accession codes rs45247208 to rs45245700 and also at PlasmodDB.
URLs. PlasmoDB, http://plasmodb.org/plasmo/; PLINK, http://pngu.mgh.harvard.edu/~purcell/plink/; EIGENSOFT, http://helix.nih.gov/Applications/eigensoft.html; REHH, http://www.broadinstitute.org/mpg/sweep/; XP-EHH: http://hgdp.uchicago.edu/Software/
COMPETING INTEREST STATEMENT
The authors declare that they do not have any competing financial interests.
AUTHOR CONTRIBUTION STATEMENTS
J.M. parasite culture and DNA extraction, drug assay, data analysis, writing; R.A.M statistical design, data analysis and writing; H.J. parasite collection, culture, and drug assay; S.L. parasite culture and drug assay; S.R., D.E.S. and S.P. array development and genotyping; M.W. drug assay software; K.C., P.W., S.K., N.J.W., R.U., L.C., M.H., F.O., H.L., J.P., X.W., G.L, S. Seila, S. Sokunthea, D. S., field studies and parasite collection; R.M.F. and T.E.W., field work and writing; P.A. statistical analysis and design, and writing; X-z. S. project design, data analysis, and writing.
References
- 1.Wootton JC, et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- 2.Roper C, et al. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124. doi: 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- 3.Mu J, et al. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat Genet. 2007;39:126–130. doi: 10.1038/ng1924. [DOI] [PubMed] [Google Scholar]
- 4.Zalis MG, Pang L, Silveira MS, Milhous WK, Wirth DF. Characterization of Plasmodium falciparum isolated from the Amazon region of Brazil: evidence for quinine resistance. Am J Trop Med Hyg. 1998;58:630–637. doi: 10.4269/ajtmh.1998.58.630. [DOI] [PubMed] [Google Scholar]
- 5.Baird JK. Effectiveness of antimalarial drugs. N Engl J Med. 2005;352:1565–1577. doi: 10.1056/NEJMra043207. [DOI] [PubMed] [Google Scholar]
- 6.Wongsrichanalai C, Meshnick SR. Declining artesunate-mefloquine efficacy against falciparum malaria on the Cambodia-Thailand border. Emerg Infect Dis. 2008;14:716–719. doi: 10.3201/eid1405.071601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noedl H, et al. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 8.Hayton K, Su X-z. Drug resistance and genetic mapping in Plasmodium falciparum. Curr Genet. 2008 doi: 10.1007/s00294-008-0214-x. [DOI] [PubMed] [Google Scholar]
- 9.Hardenbol P, et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol. 2003;21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 10.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 12.Mu J, et al. Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biol. 2005;3:e335. doi: 10.1371/journal.pbio.0030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 14.Sabeti PC, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pickrell JK, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escalante AA, Lal AA, Ayala FJ. Genetic polymorphism and natural selection in the malaria parasite Plasmodium falciparum. Genetics. 1998;149:189–202. doi: 10.1093/genetics/149.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winter G, et al. SURFIN is a polymorphic antigen expressed on Plasmodium falciparum merozoites and infected erythrocytes. J Exp Med. 2005;201:1853–1863. doi: 10.1084/jem.20041392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu S, Mu J, Jiang H, Su X-z. Effects of Plasmodium falciparum mixed infections on in vitro antimalarial drug tests and genotyping. Am J Trop Med Hyg. 2008;79:178–184. [PMC free article] [PubMed] [Google Scholar]
- 20.Brockman A, et al. Plasmodium falciparum antimalarial drug susceptibility on the north-western border of Thailand during five years of extensive use of artesunate-mefloquine. Trans R Soc Trop Med Hyg. 2000;94:537–544. doi: 10.1016/s0035-9203(00)90080-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basco LK, Le Bras J. In vitro activity of artemisinin derivatives against African isolates and clones of Plasmodium falciparum. Am J Trop Med Hyg. 1993;49:301–307. doi: 10.4269/ajtmh.1993.49.301. [DOI] [PubMed] [Google Scholar]
- 22.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fidock DA, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferdig MT, et al. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol Microbiol. 2004;52:985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- 25.Sidhu AB, Valderramos SG, Fidock DA. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005;57:913–926. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- 26.Cowman AF, Galatis D, Thompson JK. Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the pfmdr1 gene and cross-resistance to halofantrine and quinine. Proc Natl Acad Sci U S A. 1994;91:1143–1147. doi: 10.1073/pnas.91.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dharia NV, et al. Use of high-density tiling microarrays to identify mutations globally and elucidate mechanisms of drug resistance in Plasmodium falciparum. Genome Biol. 2009;10:R21. doi: 10.1186/gb-2009-10-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su X-z, Kirkman LA, Fujioka H, Wellems TE. Complex polymorphisms in an approximately 330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell. 1997;91:593–603. doi: 10.1016/s0092-8674(00)80447-x. [DOI] [PubMed] [Google Scholar]
- 29.Mu J, et al. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol Microbiol. 2003;49:977–989. doi: 10.1046/j.1365-2958.2003.03627.x. [DOI] [PubMed] [Google Scholar]
- 30.Jiang H, et al. Genome-wide compensatory changes accompany drug- selected mutations in the Plasmodium falciparum crt gene. PLoS ONE. 2008;3:e2484. doi: 10.1371/journal.pone.0002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raj DK, et al. Disruption of a plasmodium falciparum multidrug resistance-associated protein (PFMRP) alters its fitness and transport of antimalarial drugs and glutathione. J Biol Chem. 2008 doi: 10.1074/jbc.M806944200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.