Abstract
Natural products have played a prominent role in the history of organic chemistry, and they continue to be important as drugs, biological probes, and targets of study for synthetic and analytical chemists. In this perspective, we explore how connecting Nature’s small molecules to the genes that encode them has sparked a renaissance in natural product research, focusing primarily on the biosynthesis of polyketides and nonribosomal peptides. We survey monomer biogenesis, coupling chemistries from templated and non-templated pathways, and the broad set of tailoring reactions and hybrid pathways that give rise to the diverse scaffolds and functionalization patterns of natural products. We conclude by considering two questions: What would it take to find all natural product scaffolds? What kind of scientists will be studying natural products in the future?
Introduction: Why do natural products still matter?
Nature’s small molecules have played a prominent role in the history of organic chemistry. Over the past century, the complexity and diversity of natural product scaffolds and functional groups have been an inspiration to chemists for developing spectroscopic technology and devising principles and strategies for total synthesis, both biomimetic and abiotic. For much of the past half century, natural products and their semisynthetic derivatives have also been an important source of drugs for the pharmaceutical industry.1 In spite of their historical importance, natural product discovery has been in decline over the last two decades, calling into question why natural products still matter in an age where advances in synthetic methodology have made almost any molecule, natural or unnatural, a reasonable synthetic target.
Natural products still matter for at least four reasons. First, they continue to inspire synthetic and analytical chemists.2,3 Current challenges in chemical synthesis include constructing libraries with the architectural and functional group complexity of natural products,4,5 designing catalysts to carry out site-selective oxidations commonly found in natural product pathways,6,7 and screening catalysts based on natural molecules (e.g., peptides) to carry out chemical transformations.8 In the coming years, regio- and stereoselective synthetic catalysts are likely to complement enzymes in ‘hybrid’ pathways for natural product derivatives and natural product-like molecules.
Second, they remain a major source of human medicines.9 34% of all small molecule new chemical entities (NCEs) between 1981 and 2006 were natural products or their semisynthetic derivatives; these molecules comprise 68% (74/109) of antibacterial NCEs and 54% (45/83) of anticancer NCEs.1 Despite the decline in discovery efforts, the contribution of natural products and their semisynthetic derivatives to NCEs has remained steady over the last two decades.1
Third, they have led to important biological insights. Eons of evolution have optimized natural products’ structures, often leading to sub-nanomolar potency and profound specificity. Their ability to perturb a single node in the cellular network makes them useful as biological probes. For example, rapamycin led to the discovery of the serine/threonine protein kinase mTOR (mammalian target of rapamycin) and helped to establish its role in signaling pathways leading to protein synthesis and cell proliferation.10 Many natural products have thousands of NCBI database references from their use as biological probes; examples include phorbol esters (46266), cycloheximide (25907), colchicine (16703), cytochalasin (11358), and okadaic acid (4603). Natural products have also demonstrated important capabilities of small molecules. Staurosporine11 proved that certain pharmacophores – in spite of their small surface area – could selectively inhibit kinases over other ATP-binding enzymes, a notion that has been realized with synthetic molecules like imatinib.12 In the coming years, new technologies for identifying the cellular targets of natural products will make them increasingly useful as drug candidates and biological probes.13
Fourth, there are many more natural products to discover.14–16 Efforts to mine new ecological niches17–20 and microbial taxa21,22 have uncovered a wealth of novel molecules. Bacterial genome sequences have shown that a single strain has the capacity to produce 25–30 different molecules, which has spawned new efforts to discover the ~90% of natural products that remain ‘cryptic’.23–31 Plant natural product biosynthesis remains rich in growth opportunities; recent advances hold promise for manipulating pathways in plants and reconstituting plant pathways in microbial hosts.32–34 New DNA sequencing,35 MS36,37 and NMR38,39 technologies will accelerate discovery by making it easier to use a combination of chemistry and bioinformatics to go from a complex cell extract to each of its pure, structurally characterized components.
This perspective focuses primarily on the biosynthesis of templated natural products: polyketides40 and nonribosomal peptides.41 While important advances have been made in understanding how terpenoids, oligosaccharides, and other classes of natural products are constructed, we only cover them briefly here; interested readers are encouraged to consult several recent reviews on the biosynthesis of these nontemplated natural products.42–46 Given the scope of this review, we have not been able to include all of the relevant studies and references; we apologize to those authors whose work was inadvertently omitted.
Section 1: Natural Products 2.0 is based on connecting molecules to the genes that encode them
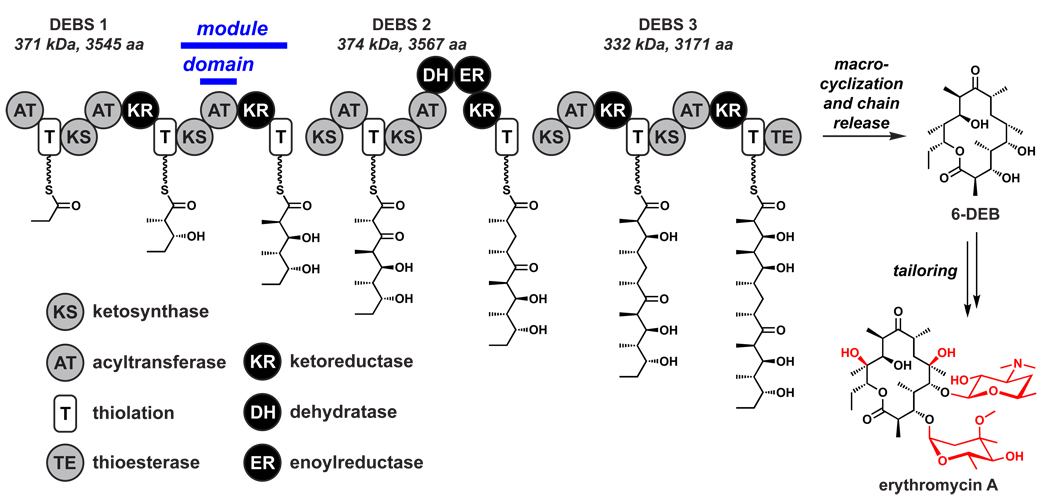
By the end of the 1980s, natural product biosynthetic pathways were being deciphered by elegant studies based on sophisticated feeding experiments with selectively labeled precursors. However, a pair of landmark papers in 1990 and 1991 dramatically changed the paradigm.47,48 Katz and colleagues at Abbott Laboratories in Chicago, and Leadlay and coworkers at the University of Cambridge, independently reported the cloning of three giant genes that encoded three subunits of 6-deoxyerythronolide B synthase, the enzyme that synthesizes the 14-membered macrolactone scaffold of the erythromycin antibiotics. The sequences of these genes offered the unanticipated revelation that the 200 kDa erythromycin synthase consists of seven ‘modules’, each consisting of 3–6 independently folded protein domains, for a total of 28 domains. These domains were distributed across three enormous proteins to form a multi-domain enzyme that resembles an assembly line (Figure 1). A parallel set of discoveries that nonribosomal peptides such as penicillins/cephalosporins, vancomycin, cyclosporine, and daptomycin are built by similar assembly line enzymes49 altered the paradigm for understanding how Nature synthesizes peptidic natural products. As we will describe in Section 5, the subsequent discovery of the genes that encode natural products such as rapamycin,50,51 FK506,52,53 epothilone,54 and bleomycin55 revealed their synthetases to be hybrids between polyketide synthase (PKS) and nonribosomal peptide synthetase (NRPS) enzymes, demonstrating the logic of these assembly line biosynthetic systems to be compatible.56
Figure 1.
The erythromycin synthase.47,48 The three proteins of the erythromycin synthase harbor 28 domains organized into seven modules; each module is responsible for inserting a building block into the growing chain. Following macrocyclization and concomitant release from DEBS 3, 6dEB undergoes two hydroxylations and two glycosylations (highlighted in red) to yield erythromycin A.
The breathtaking discovery that a single elegant model – assembly line enzymology57 – could explain the synthesis of thousands of diverse natural product scaffolds was enabled by the tendency of genes encoding a natural product to be physically clustered in the genomes of their microbial producers (Figure 2). (The genes for plant natural product pathways are not physically clustered, so efforts to decipher plant natural product pathways have lagged behind those to investigate bacterial and fungal pathways.) Thus, genes have become as important as chemistry in categorizing known natural products and identifying likely unknown variants still to be discovered. There are now ~1000 bacterial genomes sequenced and >2000 in progress (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi, accessed October 2009), and many thousands of PKS and NRPS gene clusters in the genomic databases. Connecting polyketides and nonribosomal peptides to the genes that encode them marked the dawn of the current era of natural products research.
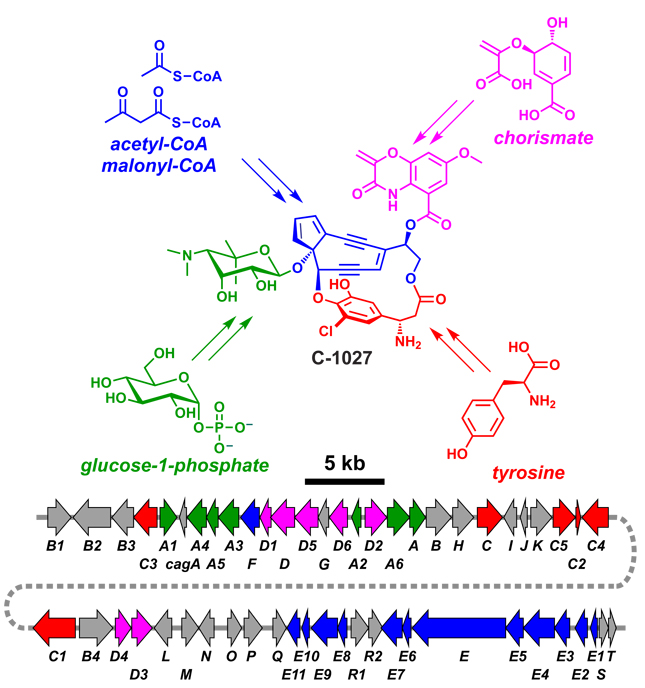
Figure 2.
Biosynthetic genes are physically clustered in bacterial genomes. The gene cluster for the enediyne C-1027 is shown.190 Genes are color coded according to the portion of the molecule their protein products contribute to synthesizing, with unassigned ORFs in gray.
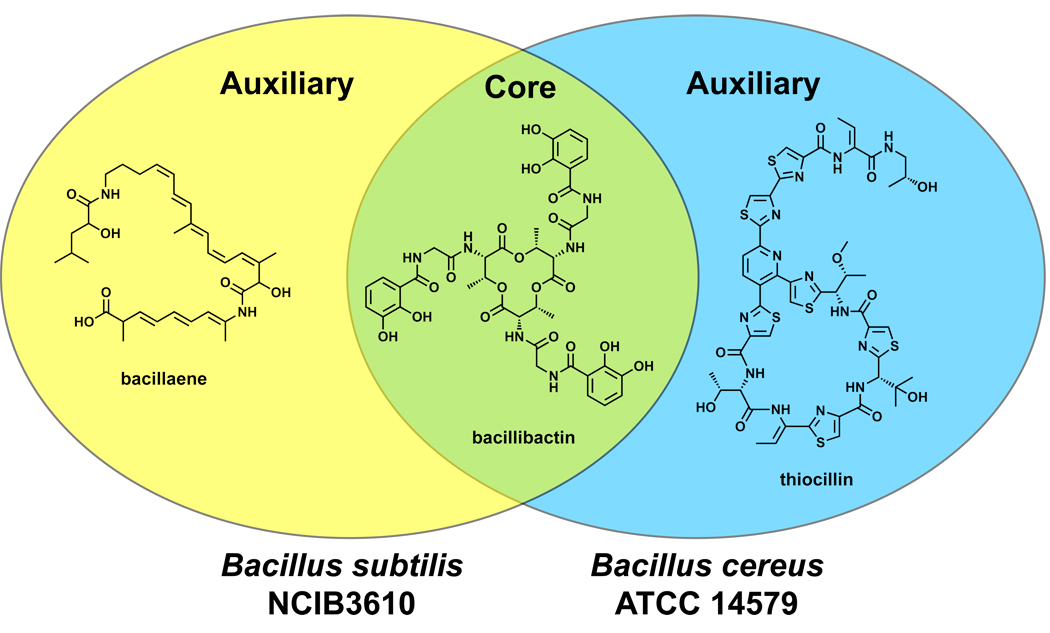
The traditional chemical categorizations of classes of natural products – polyketides, peptides, oligosaccharides, terpenoids, and alkaloids – will still have purchase for structural and functional properties. But genetic insights are beginning to provide an equally valid organizational scheme in which natural products can be grouped by the enzymes that couple their constituent monomers or tailor their nascent scaffolds. While natural products are classically defined as secondary metabolites diverted from primary metabolic pathways, the emerging principle that any group of genomes has shared (core) and unique (auxiliary) genes58 may lead to a new classification system in which genetically encoded small molecules are similarly classified as core or auxiliary with respect to a group of genomes (Figure 3).
Figure 3.
Core vs. auxiliary metabolites. Bacillus subtilis NCIB 3610 and Bacillus cereus ATCC 14579 both harbor the bacillibactin gene cluster and produce this iron-binding molecule (siderophore);307 however, only B. subtilis NCIB 3610 encodes and produces bacillaene,105 while only B. cereus ATCC 14579 encodes and produces thiocillin.241,242 Thus, with respect to these two bacterial strains, bacillibactin is a core metabolite while bacillaene and thiocillin are auxiliary metabolites.
The next five sections of this review are organized like a typical biosynthetic pathway for a natural product: Section 2 surveys natural product monomers and their biosynthetic origins, Sections 3 and 4 discuss how these monomers are coupled into scaffolds by templated and non-templated systems, Section 5 looks at the hybrid natural products that arise from collaborations between pathways with compatible chemistries, and Section 6 examines how nascent scaffolds are tailored by group transfer and oxidative enzymes. Section 7 covers a special category of tailoring reactions: posttranslational modifications that convert ribosomally synthesized peptides into natural products. Section 8 looks at Nature’s remarkable ability to control the oxidation state of intermediates and to use their intrinsic reactivity to set up cascade reaction sequences. Sections 9 and 10 conclude by considering two questions: What would it take to find all of Nature’s natural product scaffolds? What kind of scientists will be studying natural products in the future?
Section 2: Investment in building blocks for natural product assembly lines: monomer diversity
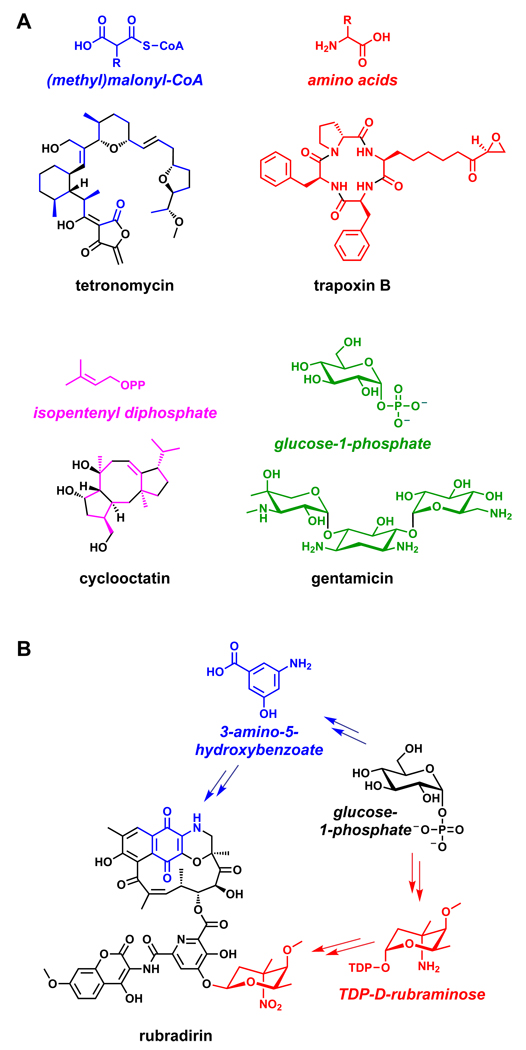
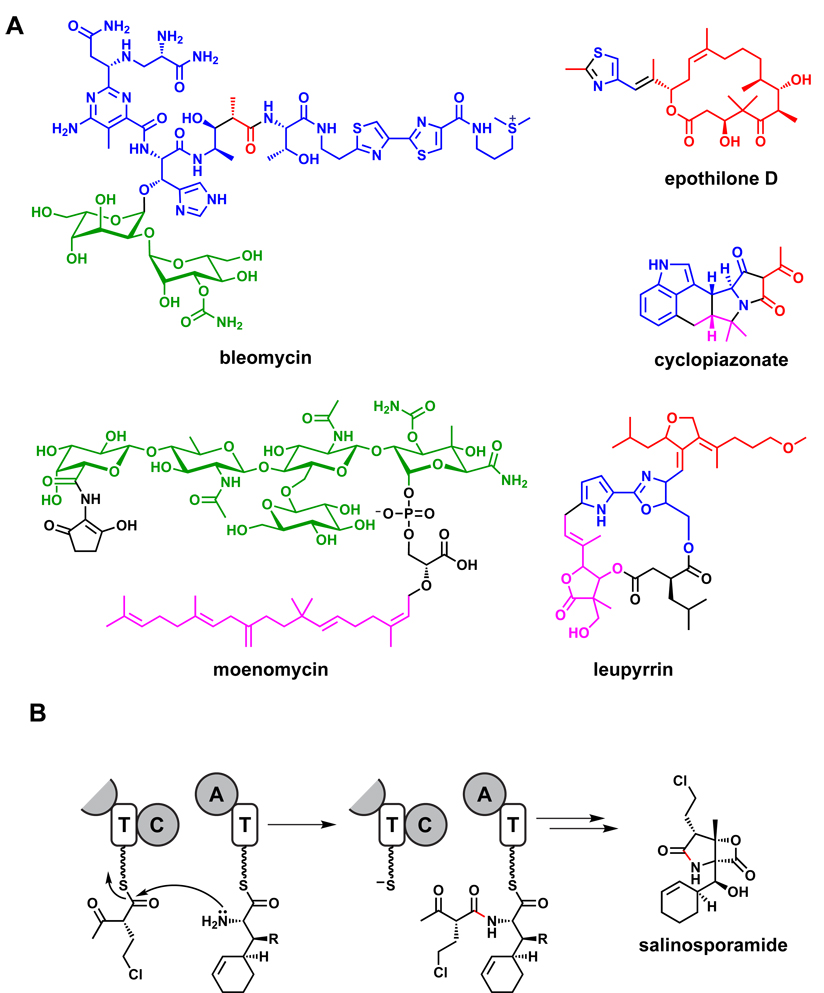
The enormous sweep of polyketide functional group diversity comes from a meagerly diverse set of acyl-coenzyme A (acyl-CoA) building blocks (Figure 4).59 Malonyl-CoA, diverted from fatty acid biosynthesis, and methylmalonyl-CoA (the key monomer for erythromycin and many other polyketides) lead to the introduction of C2 or alpha-branched C3 units during each PKS chain elongation step. As we will discuss in Section 3, most of the diverse polyketide functional groups come from an array of embedded tailoring enzymes that determine whether the β-ketoacyl thioester that results from coupling a new monomer to the growing chain is carried forward as a β-ketone, a β-hydroxyl, an α,β-olefin or an unreactive β-methylene. In some cases, specialized acyl-CoA building blocks are synthesized and used as monomers.60 These specialized monomers are generally produced from primary metabolic building blocks by a sub-pathway of enzymes encoded in the gene cluster. These include dihydroxycyclohexenyl-CoA (FK506, rapamycin),61 chloroethylmalonyl-CoA (salinosporamide),62 and methoxymalonyl-CoA (many polyketides, including FK506, ansamitocin, soraphen, geldanamycin, oxazolomycin, and tautomycin).63 Even with these additional acyl-CoAs as starter units, the diversity input for polyketides is small, at first glance surprisingly so.
Figure 4.
Natural product building blocks. (A) Highlighted are the two- and three-carbon building blocks of the polyketide tetronomycin, the amino acid building blocks of the nonribosomal peptide trapoxin B, the five-carbon building blocks of the polyketide cyclooctatin, and the hexose building blocks of the oligosaccharide gentamicin. (B). Glucose-1-phosphate is the precursor for two of the building blocks of rubradirin, 3-amino-5-hydroxybenzoate and TDP-D-rubraminose.308
In contrast to the paucity of building blocks for PKSs, NRPSs are known to utilize more than 500 nonproteinogenic amino acid building blocks (http://bioinfo.lifl.fr/norine/, accessed October 2009). Just as with specialized PKS monomers, the genes that encode these nonproteinogenic amino acids are embedded in their respective NRPS gene clusters. For example, four of the seven amino acid building blocks for the glycopeptide antibiotic vancomycin are nonproteinogenic.64,65 Of note are β-hydroxytyrosine, which derives from the enzymatic hydroxylation of tyrosine,66 and two monomers that are constructed de novo: 4-hydroxyphenylglycine (Hpg) and 3,5-dihydroxyphenylglycine (Dpg). Hpg arises by a four-enzyme pathway that starts with prephenate,67 while Dpg is constructed by four distinct enzymes (including a small PKS) from four molecules of malonyl-CoA.68 All eight of the Hpg- and Dpg-forming enzymes are encoded in the gene cluster for vancomycin. PK and NRP building blocks are not the only ones constructed for specialized use in a natural product; another building block of vancomycin, the unusual hexose L-vancosamine, is the product of an five-step enzymatic pathway encoded by the vancomycin gene cluster.69
Much of the enzymatic machinery encoded by a natural product gene cluster can be devoted to generating a single unusual monomer. Coronatine, a hybrid NRP-PK (see Section 5),70,71 is a phytotoxin secreted by plant-associated strains of Pseudomonas syringae that mimics the phytohormome jasmonic acid, itself a natural product.72 The amino acid component of coronatine is coronamic acid, a methycyclopropyl amino acid, whose biosynthetic genes are clustered with the PKS genes that generate the coronofacic acid component. The genesis of the cyclopropane ring from allo-isoleucine is itself a fascinating story, whose deciphering led to the identification of a new class of mononuclear iron-containing halogenases that make a δ-chloroisoleucyl thioester that is an intermediate for a novel enzyme-directed cyclopropanation step.73 Some specialized building blocks are bona fide natural products themselves: a related mononuclear iron-dependent halogenase converts a tethered aminobutyrate molecule to γ,γ-dichloroaminobutyrate, a Streptomyces-produced antimetabolite.74
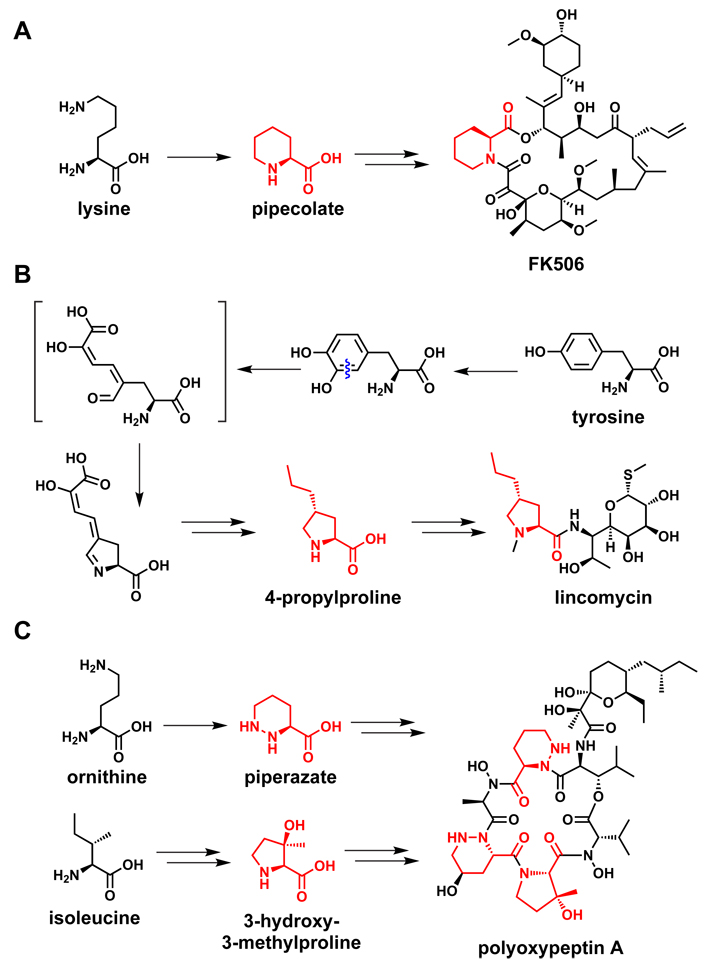
One further set of examples involves building blocks related to the imino acid proline (Figure 5). One of these building blocks is derived from proline itself: enzymes that convert a thioester-tethered proline molecule to a pyrrole-2-carboxy group by two subsequent two-electron oxidations75 are found in numerous natural product pathways. This pyrrole-2-carboxy group is an electrophilic partner for capture by nucleophilic cosubstrates, for example in prodiginine biosynthesis,76 in coumermycin assembly,75 and in many marine natural products.77
Figure 5.
Proline and its derivatives as a building block for natural products. (A) Lysine gets converted to pipecolate during FK506 biosynthesis.78 (B) Tyrosine gets converted to propylproline during lincomycin biosynthesis.80 (C) The proposed pathways to piperazate and 3-hydroxy-3-methylproline, both of which are building blocks of polyoxypeptin A.79
Other proline-like building blocks are derived from non-proline amino acids. Pipecolic acid, the six-ring homolog of proline found in FK506 and rapamycin, is generated enzymatically by cyclizing lysine in an NAD-dependent oxidation to the C6 imine with expulsion of ammonia, and reducing the resulting imine to pipecolate.78 The proposed pathways for two other proline-like building blocks come from distinct amino acids: piperazic acid, a nitrogen-containing homolog of pipecolic acid found in a variety of NRPs, is thought to come from glutamate;79 and propylproline, a component of the antibiotic lincomycin, is generated from tyrosine by a multi-step pathway.80 The wide range of pathways that convergently produce modified imino acid building blocks are a testament to the importance of these rigid monomers in natural product scaffolds.
Section 3: Assembly line enzymes are efficient, processive catalysts for iterative condensation chemistry
Long, multistep linear reaction sequences are inefficient for solution phase reactions, whether enzymes or abiotic catalysts are involved. For iterative couplings of equivalently reactive building blocks such as amino acids, Merrifield pioneered solid phase peptide synthesis half a century ago.81 Nature has arrived at the same solution: the biological equivalent of solid phase synthesis is used for building fatty acids, polyketides and nonribosomal peptides.57
By covalently tethering both the growing chain and the monomers to be incorporated at each elongation step, PKSs and NRPSs are fully processive polymerization catalysts, with no loss of intermediates to solution (Figure 6). Both systems use ~10 kDa carrier protein domains bearing a thiol-terminated phosphopantetheine moiety as the ‘resin’ to which monomers and nascent polymers are attached,82 so the (methyl)malonyl and ketidyl moieties for PKSs and the aminoacyl and peptidyl moieties for NRPSs are tethered as thermodynamically activated thioesters. These thioesters have sufficient kinetic stability at physiological pH and temperature to ensure that off-pathway hydrolysis, resulting in premature chain termination, is minimal. The logic of PKS and NRPS chain elongation is identical:56 the incoming monomer is tethered to the downstream carrier protein and acts as a nucleophile to attack the electrophilic thioester that tethers the growing chain to the upstream carrier protein. In NRPSs, the free base form of the aminoacyl thioester is the nucleophile, generating an amide as the growing chain gets transferred to the downstream carrier protein. For PKSs, decarboxylation of the (methyl)malonyl thioester generates the carbon nucleophile for the Claisen condensation, and loss of CO2 drives the equilibrium in favor of β-ketoacyl thioester formation. Because the incoming building blocks are the nucleophiles, chain growth for PKs and NRPs is unidirectional, from the amino terminus to the carboxy terminus of the assembly line enzyme.83,84
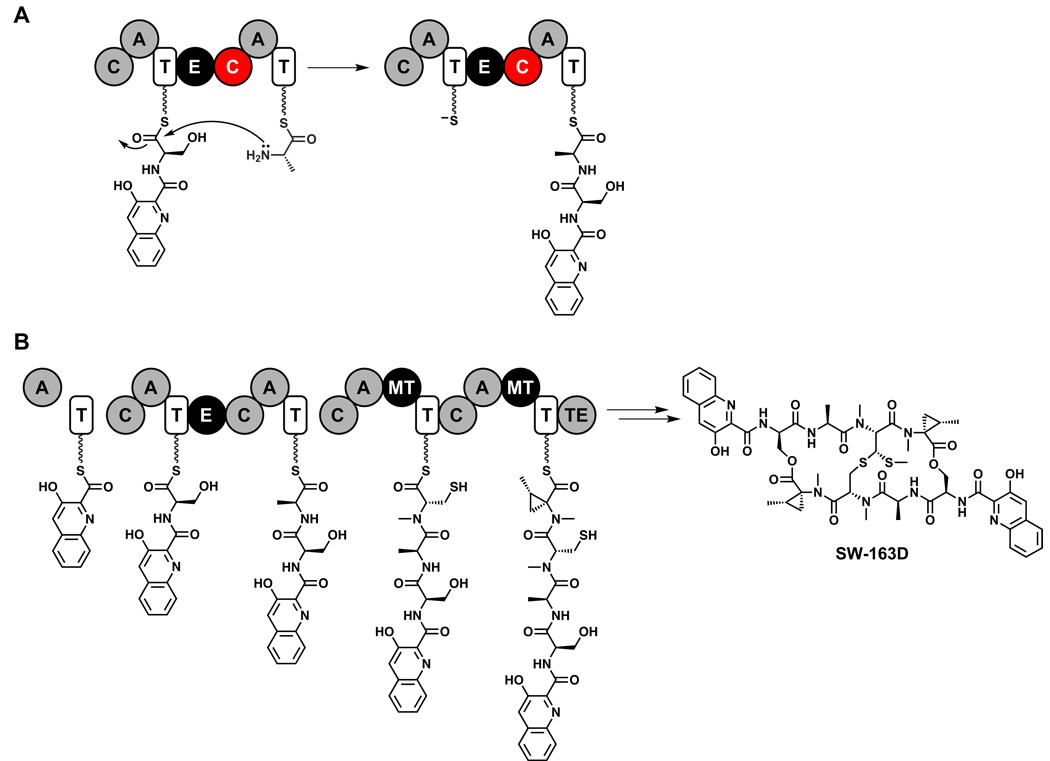
Figure 6.
Solid-phase natural product synthesis by assembly line enzymes. (A) An amide bond-forming condensation reaction during the proposed pathway for SW-163D biosynthesis.309 (B) The NRPS that constructs SW-163D, showing the nascent intermediates tethered to each thiolation domain. C = condensation, A = adenylation, T= thiolation, E = epimerization, MT = methyltransferase, TE = thioesterase.
Polyketide diversification during chain elongation: β-carbon reduction
For polyketide biosynthesis, there is almost no variation in the C-C bond-forming chain elongation step. Much of the chemical diversity in polyketide structures comes from three tailoring domains – β-keto reductase (KR), β-hydroxyacyl dehydratase (DH) and α,β-enoyl reductase (ER) – which can be present in a module in various combinations to control the oxidation state of the growing chain.40 A module with no tailoring domain carries the unreduced β-ketone forward to the next downstream module; a KR domain alone yields the β-hydroxyl; the KR+DH pair yields an olefin, while all three, KR, DH, and ER, generate a fully reduced methylene. Thus, the genetically encoded domain content of PKS module predicts the functional group at the corresponding β-carbon of its product, and conversely the functional group array of a polyketide predicts the identity and order of domains and modules that comprise its PKS.
Nonribosomal peptide diversification during chain elongation: Epimerization, α-ketone reduction, and heterocyclization
In contrast to polyketides, much of the chemical diversity in NRPs comes from the >500 different building blocks NRPSs use as monomers (http://bioinfo.lifl.fr/norine/, accessed October 2009). However, three kinds of transformations that commonly occur during chain elongation85 further diversify NRP structures: epimerization, α-ketone reduction, and heterocyclization.
D-amino acid residues are a hallmark of NRP scaffolds; for example, they comprise three of the seven residues in vancomycin. Most, but not all, NRPS assembly lines that generate products with D-amino acids load and couple the corresponding L-amino acids to make an L-L-peptidyl thioester, and then epimerize to an L-D-peptidyl thioester before the subsequent chain elongation.86
NRPs such as the emetic toxin cereulide from Bacillus cereus have alternating amide and ester linkages in their backbones. These arise by the alternate loading of amino acids and α-keto acids in adjacent NRPS modules. The tethered α-ketoacyl thioester is then reduced by an embedded reductase, and the resulting α-hydroxyl is the nucleophile for an ester-forming condensation domain.87
Heterocycles – thiazoles, oxazoles, and their reduced variants – are commonly found in NRPs, including bleomycin, epothilone, and yersiniabactin. These are formed by heterocyclization (Cy) domains, variants of the amide bond-forming condensation domain that condense an upstream peptidyl thioester with a downstream cysteinyl, seryl, or threonyl thioester. The Cy domain then catalyzes the attack of the thiolate (Cys) or hydroxyl (Ser) side chain on the newly formed amide bond. Subsequent dehydration of the tetrahedral adduct yields a thiazoline or oxazoline,88 and further dehydrogenation by embedded flavin-dependent oxidase creates the heteroaromatic thiazole and oxazole rings.89 This transformation dramatically alters a hydrolyzable peptide backbone into a nonhydrolyzable heterocycle.
Numerous exceptions to the ‘rules’ of PKS and NRPS mechanisms are now known; interested readers are encouraged to consult reviews on these (perhaps not so unusual) violations to the principles outlined above.90,91
Chain termination as an opportunity for architectural and chemical diversification
The termination machinery of assembly line enzymes offers a range of directed fates for the full-length PK or NRP chain (Figure 7).92 Most natural product assembly lines have a dedicated 35 kDa termination domain called a thioesterase (TE) domain.93 TE domains, members of the serine hydrolase superfamily, transfer the full-length acyl/peptidyl chain from the final carrier protein domain to the side chain of an active site serine, generating an acyl/peptidyl-O-enzyme intermediate as the last tethered species. Capture of the acyl/peptidyl-O-enzyme intermediate can be intermolecular or intramolecular; we consider the intramolecular fates first.
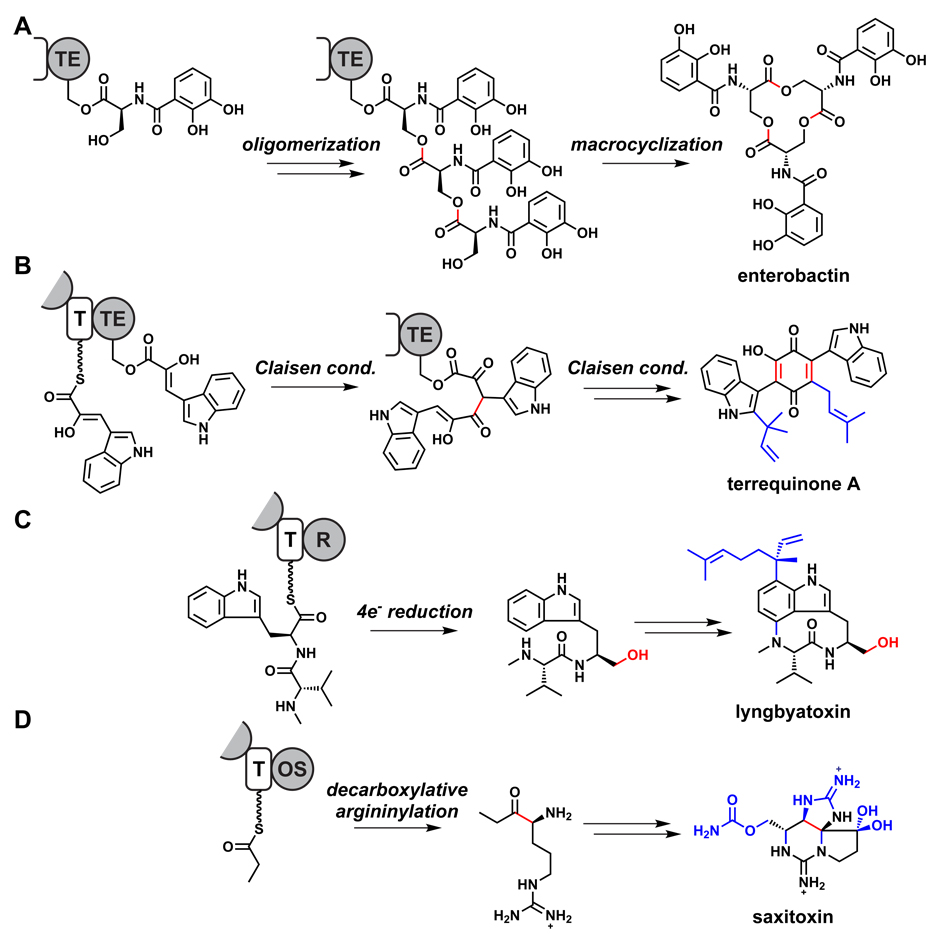
Figure 7.
Unconventional modes of chain termination. (A) Oligomerization and subsequent macrocyclication catalyzed by the enterobactin TE.95 (B) Claisen condensations catalyzed by the terrequinone TE.98 (C) Reductase-catalyzed release during lyngbyatoxin biosynthesis.182 (D) A proposed alpha-oxoamine synthase-catalyzed chain release during saxitoxin biosynthesis.111 Bonds formed or modified during chain release are colored red, and post-assembly modifications are highlighted in blue. R = reductase, OS = alpha-oxoamine synthase.
Directed intramolecular capture of the acyl/peptidyl-O-TE intermediate is a catalytic property of many chain-terminating assembly line TEs, creating macrocyclic lactones or lactams from regiospecific attack of a hydroxyl or amine within the acyl/peptidyl moiety.94 This is the fate for the hundreds of known polyketide macrolactones, including erythromycin, epothilone, FK506/rapamycin, cyclosporine, daptomycin, and bacitracin. A spectacular variant of this outcome occurs in the cyclotrimerization that forms the 12-membered serine trilactone of the enterobacterial siderophore enterobactin;95 dimeric molecules such as echinomycin are formed by an equivalent TE-catalyzed oligomerization/cyclization mechanism.96,97 Another remarkable variant of a tandem reaction involved in chain disconnection occurs during biosynthesis of the Aspergillus terreus terrequinone scaffold. In this case, a dimerizing Claisen condensation creates the quinone ring as part of the disconnection step.98
There are a variety of natural diketopiperazines (DKPs). Some, such as thaxtomin99 and gliotoxin,100 are generated by dipeptide-cyclizing TEs. Interestingly, a recently discovered NRPS from Salinispora arenicola produces both the 7-membered macrolactam cyclomarin and the DKP cyclomarazine, the latter a truncation product comprising the first two residues of the former (but lacking a key methoxy group on Trp1 that might explain its early cyclization).101 Intriguingly, other DKPs form a newly discovered class that are synthesized in an NRPS-independent fashion,102 such as the bridged DKP recently elucidated as an important metabolite in Mycobacterium tuberculosis.103
A different set of fates result from intermolecular capture of the acyl/peptidyl-O-enzyme intermediate. If water is utilized as a nucleophile, then hydrolysis results and the chain is released as the free acid. This occurs during the biosynthesis of fatty acids, linear PKs such as bacillaene104,105 and thailandamide,106 and NRPs with linear precursors such as the β-lactam107 and glycopeptide64,65 antibiotics. Some TEs use a diffusible substrate rather than water as the nucleophile; during the biosynthesis of the anticancer agent bleomycin55 and the siderophores vibriobactin108 and pseudomonine,109 TE-catalyzed attack of a nucleophilic amine effects aminolysis, releasing the product amide. An intriguing variant involves chain termination mediated not by a TE, but by a PLP-dependent enzyme which decarboxylates an amino acid and then uses this intermediate during the chain-terminating condensation to form C-terminal oxoamines as in prodigiosin76,110 and saxitoxin.111
Bioinformatic analysis of some NRPS assembly lines indicates replacement of the C-terminal TE domain by NADH-dependent reductase domains. Indeed, these deliver a hydride ion to reduce the tethered thioester to the thiohemiacetal, which can unravel to release the peptidyl aldehyde. This is the oxidation state for the nascent product in the saframycin112 and nostocyclopeptide113,114 pathways; the aldehyde cyclizes to the hemiaminal to add further rigidification to the scaffold. If thioacetal deconvoluton and release of the nascent aldehyde is slow compared to binding and reaction of a second molecule of NADH, a further two-electron reduction to the product alcohol can occur; this is the route in lyngbyatoxin biosynthesis.115
Emphasizing the predictive (and sometimes surprising) connection between genes and molecules, a search for assembly line enzymes with C-terminal reductase domains yields tetramate-forming gene clusters from fungi. In the biosynthesis of the fungal metabolites equisetin116,117 and cyclopiazonate,118 the enolate form of the β-ketoacyl-aminoacyl thioester that accumulates on the final carrier protein domain is an intramolecular nucleophile for chain release. This yields the 3-acyltetramate (2,4-pyrrolinedione) ring found at the C-terminus of dozens of natural products and is likely the general release mechanism for such natural products. In such release steps, the reductase domain is not functioning as a redox catalyst, but instead for a Dieckmann-type cyclization.
Section 4: Nontemplated pathways: Advantages, limitations, and strategic similarities
Some classes of natural products, including oligosaccharides and isoprenoids, are produced by nontemplated pathways. These pathways are similar to those from primary metabolism, with the important exception that nontemplated pathways for natural products often produce multiple products while primary metabolic pathways generally make a single product.
Oligosaccharides
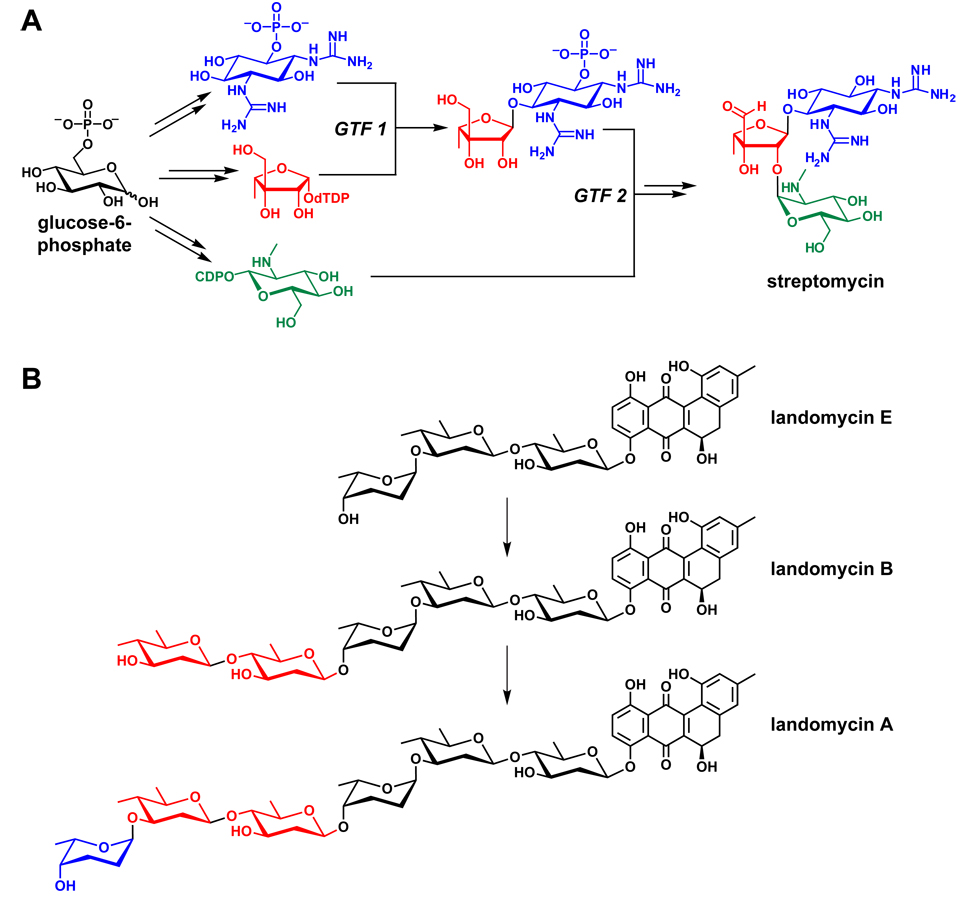
Monomers for oligosaccharide natural products are typically hexoses activated by conjugation to a nucleoside diphosphate.119,120 Glycosyltransferases,121,122 the enzymes that couple these sugars, are not organized into enzymatic assembly lines; instead, they catalyze the attack of a soluble acceptor substrate on a soluble donor substrate (NDP-sugar) (Figure 8). The aminocyclitol antibiotics exemplify two characteristics of nontemplated pathways: First, their products are generally smaller than those of templated pathways; most aminocyclitols are composed of three building blocks, despite being the products of gene clusters with several dozen genes.42 One contributing factor may be the increased challenge of channeling soluble intermediates along a nontemplated pathway, leading to diminishing yield as the number of monomer-coupling steps increases. Second, if the monomers being coupled have more than one nucleophile or electrophile, monomer-coupling steps can build branched structures. Most aminocyclitols consist of a central 2-deoxystreptamine residue that is used twice as a nucleophile, resulting in a branched trisaccharide.123
Figure 8.
Oligosaccharide pathways. (A) A schematic view of the streptomycin pathway.42 (B) Iteratively acting glycosyltransferases from the landomycin pathway.124,125 GTF = glycosyltransferase.
A third attribute of nontemplated pathways is the potential for stochastic action of sets of soluble enzymes to carry out incomplete or additional modifications on elongating oligomers to produce more scaffold diversity. This may be of use in the evolution of new scaffold variants with differentiated functions. One example is the landomycins, a family of hybrid natural products (see Section 5) in which an angular tetracyclic polyketide is found to be glycosylated with three sugars (landomycin E), five sugars (landomycin B) or six sugars (landomycin A).124,125 While this enzymatic ‘stuttering’ is more typical of nontemplated pathways, it does appear in some templated pathways.91 Since many oligosaccharides like landomycin are hybrid or tailored natural products, we will return to oligosaccharides in Sections 5 and 6.
Isoprenoids
Isoprenoid-based natural products comprise one of the largest families of natural products from plants with many thousands of molecules identified over the past decades.126 The basic chemical logic has been well established: prenyl chains are iteratively elongated by C5 units, with the incoming Δ3 isoprenyl diphosphate as the nucleophile and the elongating chain Δ2 prenyl diphosphate as the electrophile in an SN1 coupling.127,128 Chain elongations leading to polyisoprenes of C45–55 and C80–110 lengths are found in bactoprenols and dolichols, which transfer polar metabolites across bacterial and endoplasmic reticulum membranes, respectively.129 The controlled generation and capture of allylic carbocations in enzyme active sites leads to a vast range of cyclization patterns at the C10, C15, C20, C30 and C40 levels, where ‘volume control’ in the cyclase active site and the regiospecific placement of proton donors in otherwise hydrophobic active site environments controls cyclization patterns.130,131
A comprehensive review of isoprenoid biosynthesis33,44,46 is beyond the scope of this perspective, but one recent development will be covered here. Progress in identifying the genes involved in isoprenoid biosynthesis has been slow, since isoprenoids are more common among plants, and biosynthetic genes tend not to be clustered in plant genomes. Some plant isoprenoid pathways, notably those for taxol132 and gibberellin,133 have been mapped out, but identifying biosynthetic genes has been slower in plants than bacteria. However, much progress has been made recently in finding gene clusters for bacterial isoprenoids,44 and the remainder of this section is devoted to discussing these molecules.
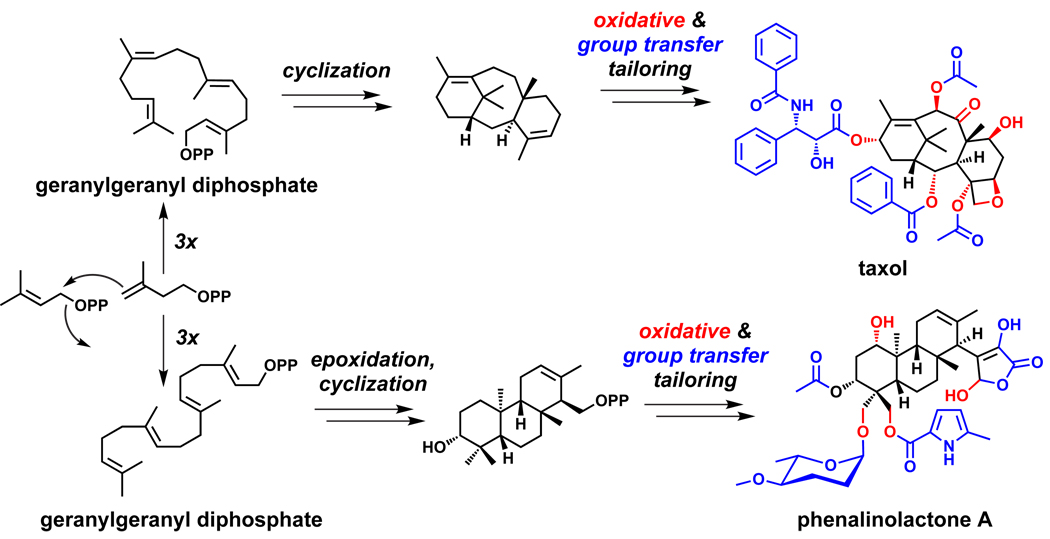
The taxol pathway (Figure 9),132 illuminated by years of pioneering work from Croteau and coworkers, established the paradigmatic logic of isoprenoid biosynthesis: a linear polyisoprenoid precursor is cyclized to a hydrophobic scaffold, which is then tailored by the addition of oxygen-based functional groups. In the taxol pathway, four isoprene building blocks are coupled to form the linear C20 precursor geranylgeranyl diphosphate, which is cyclized to taxadiene.134 Eight oxygen substituents are introduced at the periphery, some of which increase the scaffold’s solubility and serve as polar functional groups, while others undergo further tailoring by group transfer.135–141
Figure 9.
Isoprenoid biosynthesis. In the biosynthetic pathways for terpenoids such as taxol132 and phenalinolactone,151,152 a linear polyisoprenoid precursor is cyclized to a hydrophobic scaffold, which is then tailored by the addition of oxygen-based functional groups. These oxygen-based functionalities are sometimes further tailored by group transfer reactions.
Gene clusters for bacterial isoprenoids reinforce and extend this paradigm.44 They generally encode three kinds of enzymes: those that produce the Δ2 and Δ3 isopentenyl diphosphate building blocks, those that couple these monomers and cyclize the resulting polyisoprenoid intermediate, and those that catalyze group transfer and oxidative tailoring reactions.
At the C15 sesquiterpene level, farnesyl diphosphate is cyclized by epi-isozizaene synthase to the constrained tricyclic scaffold of the hydrocarbon. The adjacent gene encodes a cytochrome P450 type heme hydroxylase that oxygenates C4 first to the alcohol and then to the ketone.142–144 A second example at the C15 level is the biosynthesis of the sesquiterpene epoxide antibiotic pentalenolactone by Streptomyces avermitilis, the avermectin producer. The gene cluster that encodes the farnesyl diphosphate cyclase that generates the tricyclic pentalenene hydrocarbon also contains four putative oxygenases, the first catalyzing triple oxidation of a side chain methyl to a carboxylate, the next catalyzing a double oxygenation of one of the cyclopentane rings to an alcohol and then on to the cyclopentanone, the third carrying out a Baeyer-Villiger oxygenative ring expansion of the ketone to the six membered lactone. The final oxygenase is likely an epoxidase, reflecting four distinct routes to introduce five oxygen substituents in the mature pentalenolactone scaffold.145–150
At the C20 diterpene level, the gene cluster for the heavily oxygenated tricyclic terpenoid glycoside phenalinolactone (Figure 9) allows dissection of the chemical steps in its pathway.151,152 First, the isoprenyl diphosphate building blocks are made by genes for the methylerythritol pathway, which is more common among bacteria, rather than the mevalonate pathway, which is more common among eukaryotes but is present in some bacteria. Once four monomers have been coupled, the linear C20 precursor geranylgeranyl diphosphate is epoxidized by an FAD-containing oxidocyclase to initiate cyclization to the tricyclic scaffold. The masked aldehyde functionality in the γ-hydroxybutyrolactone ring is then elaborated and attached with participation of a nonheme oxygenase. Meanwhile, the A ring of the diterpene scaffold undergoes an oxygenation on the ring and two oxygenations on the gem-dimethyls by a suite of three P450 enzymes encoded in the gene cluster. The ring hydroxyl at C3 undergoes a tailoring acetylation, and one of the newly created pendant hydroxymethyl groups attached to C4 undergoes glycosylation by an L-amicetose residue, whose biosynthesis from TDP-D-glucose is encoded in the cluster, while the other hydroxymethyl undergoes acylation by a methylpyrrole-2-carboxyl moiety. The pyrrole unit is created from proline by a small NRPS, as noted in Section 2. The phenalinolactone gene cluster reveals the confluence of genes for required building blocks (Δ2 and Δ3 prenyl units, L-amicetose, methylpyrrole carboxylate) with the genes for isoprene chain elongation and diterpene cyclization, and a set of five oxygenases, one FAD-containing, three heme proteins, and one nonheme iron oxygenase.
Other nontemplated pathways
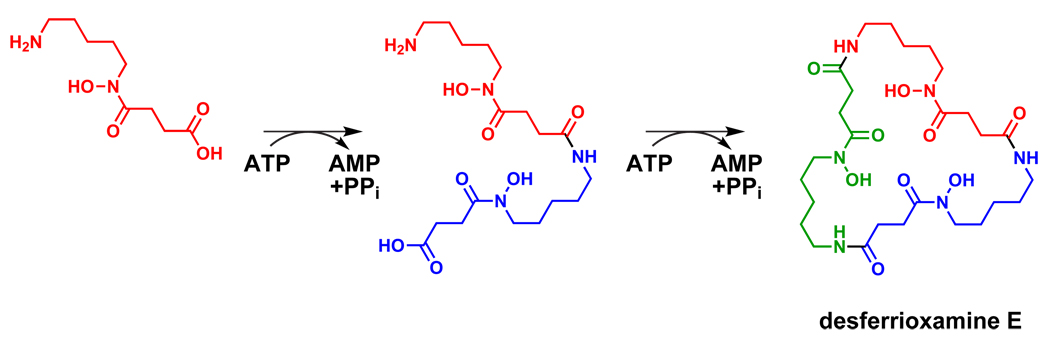
While we do not cover them in detail here, three other classes of nontemplated pathways bear mentioning. First, many natural products harbor amide bonds that derive neither from the ribosome nor from an NRPS.43 One enzyme class that synthesizes untemplated amides is the acyl-AMP ligases, homologs of NRPS adenylation domains that activate a carboxylate by adenylation for nucleophilic attack by an amine. Enzymes in this class are common in the biosynthetic pathways for hydroxamate-containing siderophores like desferrioxamine (Figure 10)153 and are also found in the pathways for molecules such as coronatine and simocyclinone.154,155 The pathway for dapdiamide, a nontemplated tripeptide, involves one amide ligation of this sort and a second by an enzyme from the ATP-grasp superfamily.156 ATP-grasp enzymes have recently been shown to catalyze the posttranslational crosslinking of microcyclamide, a cyanobacterial natural product, hinting at a wider role for these enzymes in natural product pathways.157–160
Figure 10.
Amide ligases form the amide bonds of hydroxamate siderophores such as desferrioxamine E.153
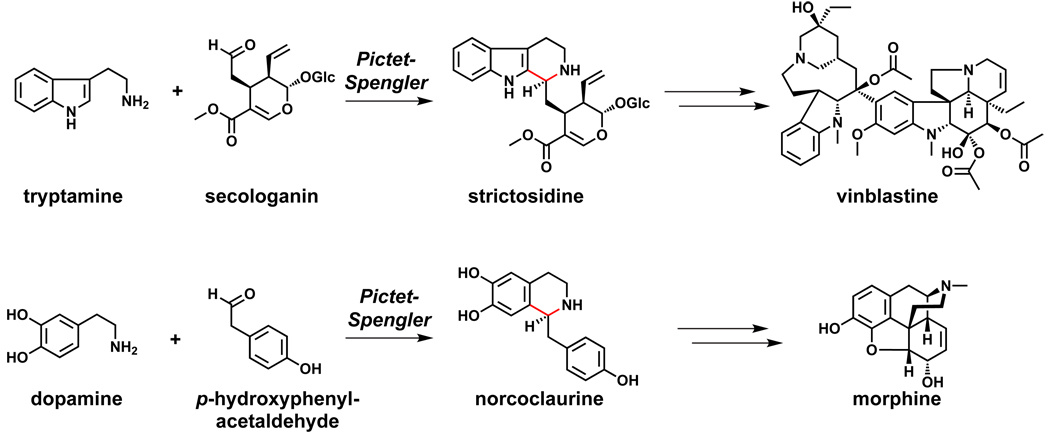
Two widely distributed plant natural product pathways involve an enzyme-catalyzed Pictet-Spengler reaction. The condensation of tryptamine and secologanin forms strictosidine, the precursor to a variety of alkaloids including vinblastine.161 The Pictet-Spengler product of dopamine and p-hydroxyphenylacetaldehyde, norcoclaurine, is the precursor to another series of alkaloids including morphine (Figure 11).162 Oxidative tailoring steps divert strictosidine and norcoclaurine down many distinct pathways, and recent efforts have shown that modified precursors can be fed into these pathways in planta to produce unnatural alkaloids.163,164
Figure 11.
Pictet-Spengler reactions in widely distributed plant pathways.161,162
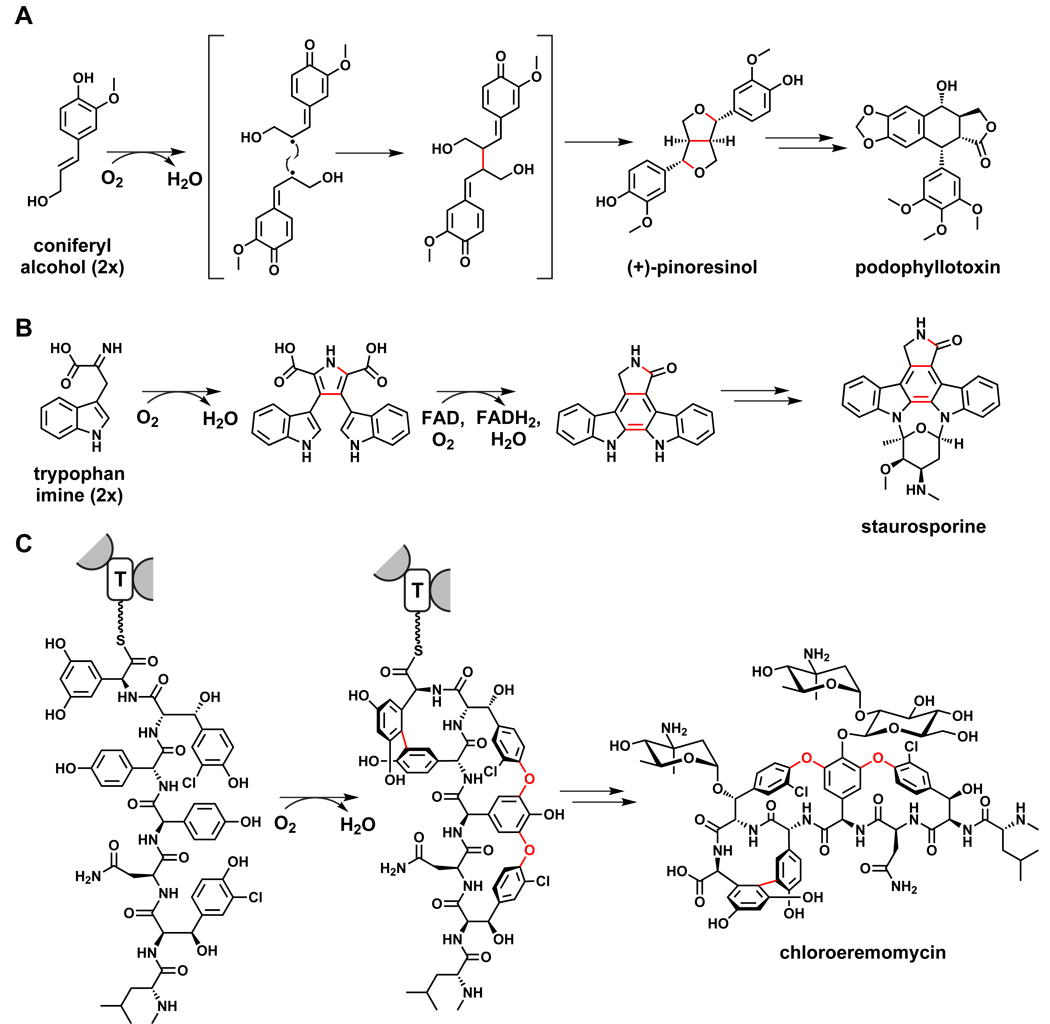
Another widely distributed plant pathway is the one from which lignins and lignans derive: the one-electron coupling of the phenylpropanoid monomer coniferyl alcohol.165 Directed coupling products such as pinoresinol166 undergo largely oxidative tailoring to become lignans like podophyllotoxin,167 or polymerize to yield lignins,168 the second most abundant organic polymer behind the oligosaccharide cellulose (Figure 12). Some bacterial pathways also involve the oxidative coupling of aromatic monomers: indolocarbazoles such as staurosporine are formed by the oxidative dimerization of two tryptophan-derived monomers,169–171 and the cyanobacterial pigment scytonemin comes from the oxidative coupling of indole-3-pyruvate and p-hydroxyphenylpyruvate.172,173
Figure 12.
Oxidative coupling reactions in biosynthetic pathways. (A) Lignin and lignan pathways in plants begin with the oxidative coupling of coniferyl alcohol.165 (B) The pathway to indolocarbazoles such as staurosporine involves the coupling of two tryptophan-derived monomers.169,171 (C) Intramolecular crosslinking of aryl monomers forms the cup-like shape of glycopeptides such as chloroeremomycin and vancomycin.226–229
Section 5: Where chemistry allows, hybrid pathways emerge
Two examples from the previous section – landomycin,124,125 a polyketide-oligosaccharide hybrid, and phenalinolactone,151,152 an isoprenoid bearing NRP and hexose functional groups – serve as a reminder that many natural products have a mixed pedigree. These hybrid molecules, which represent evolutionary opportunism in mixing genes to create chemical diversity, are synthesized collaboratively by distinct but chemically compatible biosynthetic systems. A central focus in understanding the biosynthetic origins of hybrid molecules is identifying enzymes that conjugate one class of building block to another; for example, NRPS condensation domains that accept a PK substrate, or glycosyltransferases that recognize a lipid substrate.
PK-NRP hybrids
Some medicinally important products of assembly line enzymes are PK-NRP hybrids, including the immunosuppressants FK506/FK520 and rapamycin50–53 and the anticancer agents bleomycin55 and epothilone54 (Figure 13). While the building blocks and monomer coupling chemistries of PKSs and NRPSs are quite different, some fundamental similarities have allowed hybrid pathways to emerge.56
Figure 13.
Hybrid natural products. (A) Examples of hybrid natural products. Nonribosomal peptide-derived monomers are colored blue, polyketide monomers are colored red, oligosaccharide monomers are colored green, and isoprenoid monomers are colored pink. (B) A condensation reaction links a nonribosomal peptide monomer to a polyketide monomer during the proposed pathway for salinosporamide.
Most notably, both PKS and NRPS assembly lines carry out acyl transfer chemistry and tether monomers and growing polymers as pantetheinyl thioesters to carrier protein domains.57 As noted in Section 3, the logic of PKS and NRPS chain elongation is identical: the incoming monomer is tethered to the downstream carrier protein and acts as a nucleophile (enolate or amine) to attack the electrophilic thioester that tethers the growing chain to the upstream carrier protein.
For hybrid chain elongation to proceed, two protein-based recognition problems have to be solved. The first is a catalytic loosening of stringency. At an NRPS-PKS interface, the KS domain must be able to catalyze a Claisen condensation with an upstream peptidyl thioester chain; at a PKS-NRPS junction, the condensation domain must catalyze amide bond formation to an upstream ketidyl thioester chain. The second is a protein-protein recognition problem: NRPS and PKS modules must form a productive interface with enough affinity to allow hybrid chain elongation to proceed. Much progress has been made in defining and swapping recognition domains at the N-and C-termini of modules to direct/redirect partner module recognition.174–179
One well-studied example is the epothilone synthase. Epothilone is almost entirely a polyketide with one pharmacophoric thiazole ring appended to the macrocycle. As expected, the epothilone assembly line54 has eight PKS modules distributed over five separate proteins, and only a single NRPS module. The beginning of the assembly line is composed of three separate proteins, EpoA-C. EpoA is a starter PKS module, decarboxylating malonyl-S-EpoA to acetyl-S-EpoA. EpoB is a four domain NRPS module which begins by activating and loading cysteine as a pantetheinyl thioester on its carrier protein domain; it then condenses the cysteinyl thioester with the upstream acetyl thioester, across the PKS-NRPS interface, to give N-acetyl-S-EpoB; and finally it cyclizes, dehydrates, and dehydrogenates this intermediate to yield methylthiazolyl-S-EpoB. 180 EpoC, a methylmalonyl-CoA-utilizing PKS module, mediates chain transfer of the methylthiazolyl moiety across the NRPS-PKS interface by C-C bond formation and then reduces and dehydrates the initial β-ketoacyl-S-EpoC to the methylthiazolyl-enoyl-S-EpoC.181 EpoA-C thus traverse two hybrid module interfaces, and the remainder of the assembly line enzyme, including the TE-mediated macrocyclization, is PKS chemistry.
Other hybrid natural products
There are a variety of natural products that undergo prenylation. Both lyngbyatoxin,182 which incorporates a C10 geranyl group, and cyclopiazonate,118 which incorporates a C5 dimethylallyl group at C4 of the indole ring (subsequently used to create the pentacyclic framework), are NRP examples. Perhaps more intriguing is the de novo construction of prenyl groups during polyketide chain extension.183 This occurs to introduce beta methyl and higher alkyl branches in such molecules as bacillaene,184 myxovirescin,185 rhizoxin,186 leinamycin, and a cyclopropyl group in jamaicamide.187 This merger of isoprene-building chemistry and Claisen chemistry on an assembly line enzyme can now be predicted from a subset of five genes in PK gene clusters.
In contrast to the landomycin pathway124,125 (Section 4), in which a PK scaffold is synthesized and then elaborated by the addition of multiple sugars, the pathway for orthosomycin antibiotics such as avilamycin188 and everninomicin189 begins with the construction of a heptasaccharide chain and ends with its conjugation to a PK aglycone, dichloroisoeverninic acid. Avilamycin bears unusual sugars such as D-olivose, D-fucose, 2-deoxy-evalose, and L-xylose, so its biosynthetic machinery must convert the primary/core metabolite TDP-D-glucose into all of these NDP-sugar variants for oligosaccharide chain elongation, and then connect each to the next in the proper order. A 54-gene cluster has been described that has not only the anticipated glycosyltransferases, but all the genes encoding the TDP-sugar-tailoring enzymes to create those needed building blocks.
Remarkably, some hybrid natural products consist of building blocks from four or more sources. The enediyne C-1027190 (Figure 2) consists of NRP, PK, hexose, and shikimate pathway monomers, while leupyrrin191 (Figure 13) consists of PK, NRP, isoprenoid, and dicarboxylic acid monomers.
Section 6: Scaffold Tailoring is Inevitable and Advantageous
Enzymatic tailoring of a nascent scaffold is a hallmark of almost all natural product classes. Tailored and hybrid natural products are two faces of the same phenomenon: evolutionary opportunism in mixing genes to create chemical diversity by juxtaposing distinct but chemically compatible biosynthetic systems. For our purposes, tailoring reactions are generally peripheral – the pathway could proceed in their absence and related non-tailored molecules are known – and they serve to functionalize the core scaffold, often occurring later in the pathway. Clearly, the distinction between tailored and hybrid natural products can be blurry; the landomycin hexasaccharide124 (landomycin A) could reasonably be considered either a hybrid PK-oligosaccharide or a PK tailored by glycosylation. As with hybrid pathways, a central focus in studying tailoring is identifying enzymes (and their encoding genes) that conjugate one class of building block to a scaffold from a different class (e.g., glycosyltransferases specific for PK scaffolds).
We have already noted a few examples of tailoring, such as the oxidative tailoring of reduced isoprenoid intermediates described in Section 4. Tailoring enzyme chemistries can be grouped into two broad categories: group transfer reactions and oxidative transformations. Many tailoring enzymes from both categories are homologous to enzymes from primary/core metabolism, consistent with the premise that contemporary natural product tailoring enzymes have been conscripted from that enzyme pool.
Tailoring by group transfer enzymes
Nearly all group transfers involve coupling an electrophilic fragment of a cosubstrate or primary metabolite to a nucleophilic N, O, or S in the natural product scaffold. These cosubstrates include NDP-sugars as glycosyl donors, S-adenosylmethionine as a CH3+ donor, acyl-CoA as an acyl donor, ATP as a phosphoryl donor, phosphoadenosine phosphosulfate as a SO3 donor, and dimethylallyl diphosphate as a prenyl donor. For example, the aglycones of glycopeptides like vancomycin and teicoplanin can undergo N-methylation,192 O-sulfation, 193 and glycosylation by UDP-D-glucose and TDP-L-vancosamine;194 a pendant sugar can be further tailored by acylation with a decanoyl group.195 None of these group transfer steps involve oxygen, so this scaffold decoration chemistry could have evolved early in anaerobic organisms.
Glycosylation196–198 is the most extensive category of group transfer tailoring. The catalyzed addition of monosaccharides and their iterative elongation into oligosaccharide chains diversifies natural product scaffolds and often increases water solubility. Pendant sugars can be essential for biological activity; the erythromycin aglycone, for example, lacks antibacterial activity since its two sugars make specific contacts to the 50S ribosomal subunit.199,200 The sugars added are occasionally common hexoses such as glucose, N-acetylglucosamine, and mannose, but more often are specially constructed deoxy- and deoxyaminosugars119,120 to control the balance of hydrophobicity/hydrophilicity and provide additional sites for hydrogen bonding or further tailoring of the hydroxyl or amine groups. These auxiliary sugars are constructed from UDP- or TDP-glucose by enzymes encoded within the biosynthetic gene cluster, allowing de novo bioinformatic predictions of sugar-bearing natural products. The genes responsible for constructing sugars and transferring them to nascent scaffolds can be moved between gene clusters to provide new variants of related scaffolds, both naturally and in the laboratory.196,201,202
Recent studies have shown that natural product glycosyltransferases have equilibria not far from unity, such that a glycosyl moiety on one mature natural product can be transferred back to a nucleoside diphosphate and then onto a new scaffold in vitro.203 If this could happen in vivo, it would be another route, at the protein rather than the gene level, to construct novel glycosylated natural products. Most scaffold glycosylations occur by attack of PK or NRP hydroxyl groups on the electrophilic C1 of an NDP-sugar. In addition, there are examples of N-glycosylation on indolocarbazole204 and ansamitocin205 scaffolds, and C-glycosylation at carbons ortho or para to phenolate oxygens in the salmochelin siderophores206 and in anthracycline polyketides such as urdamycin207–210 and hedamycin.211
Much recent effort has gone into glycodiversification efforts, building on the sometimes-relaxed specificity of tailoring glycosyltransferases, the lack of templating, and the occasional capacity for a glycosyltransferase to act iteratively.196,198,201,202 In vivo studies in various anathracycline systems have shown that replacing endogenous glycosyltransferases with variant homologs, co-expression with new glycosyltransferases, and mutational analysis can lead to products with novel glycosyl tailoring outcomes, including replacing an O-glycosyltransferase with a C-glycosyltransferase.212,213 Construction of operons containing collections of glycosyltransferases under the control of inducible promoters has yielded new glycovariants of elloramycins, mithramycins, and indolocarbazoles.212,214,215
Two types of embedded tailoring domains in NRPS assembly lines were noted in Section 3: epimerization and heterocyclization. Perhaps the most common tailoring domain contained within NRPS modules is the N-methyltransferase, which uses S-adenosylmethionine as donor of a CH3+ equivalent to the amine of the aminoacyl thioester before condensation.85 In cyclosporine synthetase, 7 of the 11 modules contain such N-methyltransferase domains, and accordingly the residues they insert into the growing chain are N-methylated in cyclosporine.216,217 N-methylation alters the resonance stabilization in NRP amide bonds, allowing them to adopt a distinct set of conformations, increasing their hydrophobicity, and promoting their stability to proteolytic cleavage.
Tailoring by oxidative enzymes
The other major type of tailoring reaction involves O2 as a cosubstrate and would have been conscripted from oxygenases of primary metabolism after aerobic organisms gained hold. To overcome the kinetic barrier to the favorable thermodynamics of O2 reduction, nearly all oxygenases provide cofactors or coenzymes that can perform a one-electron transfer to a ground state triplet O2 molecule. Two strategies predominate: one is to use redox-active transition metals, most commonly iron, both in heme218 and nonheme219 microenvironments in oxygenase active sites. The second (independently evolved) strategy is to use the riboflavin-based coenzymes FMN and FAD, in which the dihydro oxidation state can engage in kinetically and thermodynamically accessible one-electron chemistry with O2.220
Heme iron-containing enzymes of the cytochrome P450 oxygenase superfamily are widespread tailoring enzymes for natural products. Among the best studied have been the regiospecific hydroxylases that hydroxylate the product of the erythromycin PKS, 6-deoxyerythronolide B (6dEB), first at C6 and then at C12. 221,222 The crystal structure of the 6dEB 6-hydroxylase, EryF, with bound 6dEB gives molecular insight into regioselectivity,223,224 as does the crystal structure of the P450 that converts epothilone D to epothilone B by a regioselective epoxidation.225 A set of three cytochromes P450 from the vancomycin gene cluster, OxyABC, are the catalysts that sequentially crosslink side chains 2 and 4, 4 and 6 and 5 and 7 of the heptapeptidyl chain while it is tethered at the last carrier protein of the vancomcyin synthetase assembly line (Figure 12).226–229 These three cross-links create the cup-shaped architecture of vancomycin and are the essential conformational constraints for high-affinity recognition of the D-Ala-D-Ala target on peptidoglycan.
A large number of nonheme iron oxygenases219 tailor natural product scaffolds by hydroxylation, using high valent oxo-iron intermediates and carbon centered radicals at the sites to be hydroxylated. Among the most fascinating of this enzyme class are the diverse transformations in which O2 is reduced and split but none of the oxygen atoms end up in the resculpted product. Four examples from β-lactam pathways show the chemical range of these iron-based scaffold maturation enzymes.
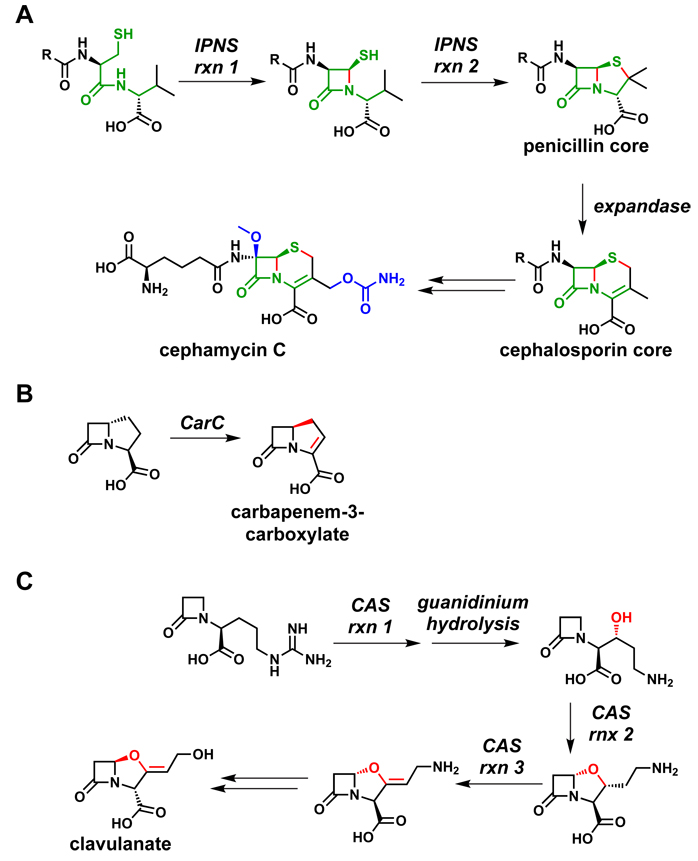
The best-known pair are isopenicillin N synthase (IPNS)230 and deacetoxycephalosporin synthase (DAOCS)231 (Figure 14), which function sequentially to convert the acyclic tripeptide aminoadipoyl-Cys-Val, the product of a three-module NRPS, to isopenicillin N and then to the cephalosporin cephamycin C. The IPNS reaction involves a four-electron reduction of cosubstrate O2 to two molecules of H2O while the tripeptide undergoes a four electron oxidation in two 2-electron steps. The first step constructs the four-membered β-lactam ring, and the second step constructs the thiazolidine in the 4,5-ring system of penicillin. Following an epimerization in the acyl chain, DAOCS (known colloquially as expandase) converts the 5-membered thiazolidine ring of the penicillin core to the 6-membered dihydrothiazine ring of the cephalopsorin scaffold. Both enzymes use one-electron chemistry and control the flux of the carbon and sulfur-centered intermediates coordinated to the active site iron.
Figure 14.
Oxidative tailoring reactions from β-lactam pathways. (A) IPNS230 catalyzes two successive two-electron oxidations to form the 4,5-ring system of the penicillins. Following an epimerization reaction in the acyl chain, DAOCS231 (expandase) catalyzes the conversion of the 5-membered ring to a 6-membered ring to form the 4,6-ring system of the cephalosporins. The portion of the aminoadipoyl-Cys-Val tripeptide that becomes the β-lactam core is highlighted in green, the bonds formed by oxidation are colored red, and post-assembly modifications are shown in blue. (B) CarC catalyzes both the epimerization and the desaturation of the carbapenem core; both modifications are shown in red.232 (C) CAS catalyzes three different oxidative transformations during clavulanate biosynthesis, each of which is shown in red.233
The two enzymes in this superfamily that participate in clavulanate and carbapenem assembly perform equally intriguing chemistry.232,233 The carbapenem synthase takes a (3S,5S)-carbapenam substrate and, in an O2-dependent reaction, epimerizes the unactivated C5 center and installs the 2,3-double bond in the five-membered ring. Even more remarkably, the clavaminate synthase uses three O2 molecules for a sequential six-electron oxidation of its monocyclic β-lactam for sequential hydroxylation and ring closure to generate the fused 4,5-ring system of clavulanate, and then carries out a dehydrogenation to form the exocyclic double bond of the enol ether moiety.
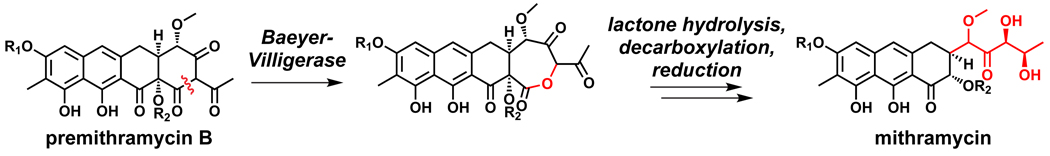
An independent and parallel strategy is found in oxygenases that are iron-free but use flavin-bound coenzymes. While they are typically found in gene clusters where activated aromatic substrate moieties undergo capture of an electrophilic oxygen from flavin-hydroperoxy intermediates, the flavin-hydroperoxides can also utilize the distal oxygen as a nucleophile to attack an electrophilic ketone or aldehyde in a natural product scaffold. By converting ketones into lactones that undergo hydrolysis, Baeyer-Villiger chemistry can effect regioselective and stereoselective C-C bond cleavages;234 two pathways in which this chemistry carves away and remodels a portions of a polycyclic framework are those for aflatoxin235,236 and mithramycin (Figure 15).237
Figure 15.
Baeyer-Villigerase action remodels the mithramycin scaffold by effecting C-C bond cleavage.237
Section 7: Ribosomally synthesized peptides can become natural products by complexity-generating posttranslational modifications
Normally, we do not think of ribosomally synthesized peptides as being precursors to natural products. However, genomic and bioinformatic studies have recently revealed that heterocycle-containing cyclic peptides from cyanobacteria such as the patellamides, ulithiacyclamides, and lissoclinamides are derived from ribosomally synthesized peptide precursors.238 Almost a hundred variants of these peptides are generated by the posttranslational cyclodehydration and dehydrogenation of Cys, Ser, and Thr residues, with protease-mediated excision of these highly modified octapeptides from a preprotein backbone. Combinatorial diversity arises from mutations in the preprotein octapeptide sequences.239
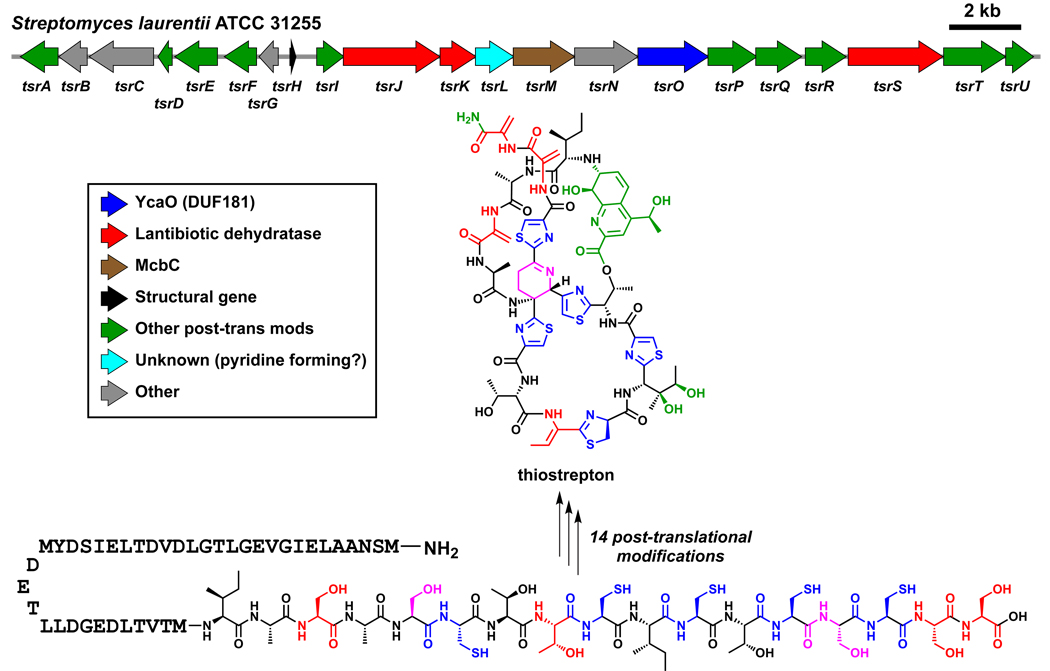
An analogous combination of bionformatics and genetics has led to the recent finding that thiomuracin, thiocillin, thiostrepton, and nosiheptide, representing the class of thiazolylpeptide antibiotics240 targeting the 50S subunit of the bacterial ribosome, are also generated by the posttranslational tailoring of Cys, Ser, and Thr-rich peptide sequences found at the C-termini of precursor proteins (Figure 16).241–245 More than 80 members of this antibiotic class are currently known240 but microbial genome scanning suggests that many more exist. Of special note in molecules such as thiocillins and thiomuracin is the trithiazolyl pyridine ring system at the center of these antibiotics. Crystal structures of these molecules bound to the 50S ribosome246 show that the trithiazolyl pyridine has a propeller structure, directing the pendant peptide chains in an architectural array that creates the three dimensional contacts necessary for high affinity binding and blockade of bacterial protein synthesis. Among the many questions of biosynthetic interest is the construction of the central pyridine ring. As indicated by the existence of thiostrepton and other congeners with six-membered nitrogen heterocycles in reduced oxidation states, the ring-forming step may be an aza [4+2] cyclization process, similar to strategies utilized in the total synthesis of several members of this antibiotic class.247,248 It is not yet known whether this reaction is concerted or stepwise, nor if it involves active or passive participation by one of the proteins encoded in these gene clusters. In such a mechanism, the initial dihydropyridine ring could be reduced to the tetrahydro state found in thiostrepton or the upstream peptide chain could be oxidatively eliminated to give the heteraromatic pyridine found in thiocillin and nosiheptide. This chemistry has analogy to the proposed carba [4+2] cyclizations noted in the next section for lovastatin,249 kijanimicin,250 and indanomycin.251
Figure 16.
Converting a ribosomally synthesized peptide into a natural product. A schematic view of the thiostrepton pathway is shown.241,244 The C-terminal 17 amino acids of the structural peptide TsrH undergo 14 posttranslational modifications, including cleavage of the leader peptide, to become thiostrepton.
Section 8: Oxidation control and cascade reactions are important components of biosynthetic logic
We have noted the extensive use of redox chemistry by tailoring enzymes to introduce oxygen atoms and to carry out oxidative cyclizations of nascent scaffolds. More generally, PK and terpenoid pathways rely on oxidation control of the functional group inventory to generate dramatic product structural diversity from simple building blocks. As discussed in Section 4, terpenoid pathways exert oxidation control by first generating a reduced, unreactive polycyclic intermediate and then tailoring it with oxygen-based functionality. Some PKSs employ a different form of oxidation control: generating a reactive polyketone or polyene intermediate and then steering its reactivity toward markedly divergent structural outcomes.
Fused aromatic scaffolds: Polyketone cyclization steered by regiospecific reduction
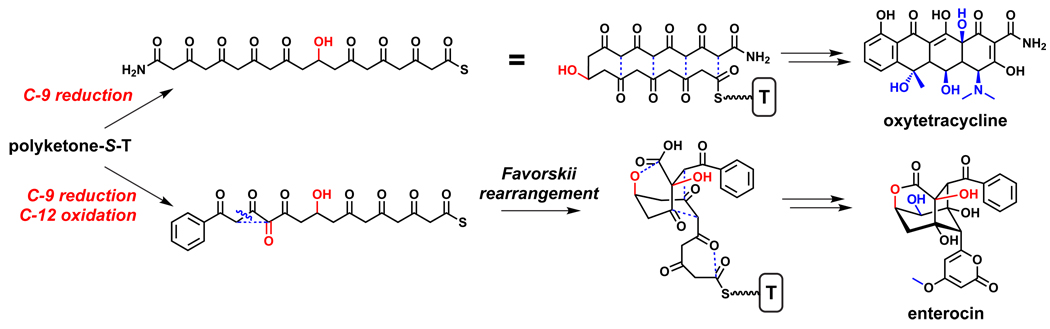
While modular PKSs like the erythromycin synthase have a separate module for each elongation cycle, most iterative PKSs have a single KS, AT, and ACP domain for elongation cycles, so they generate polyketone chains bound to an ACP domain.40 In contrast to the polyethers discussed below, none of the oxygens in these PK intermediates derive from molecular oxygen. The fates of these polyketone chains are controlled by reductase, cyclase, and aromatase domains as in the assembly of tetracycline (Figure 17).252,253 Generation of alcohols by KR domains at various points in the chains helps establish the regiochemistry of cyclization. Other forms of oxidative intervention at the polyketone stage can divert product structure dramatically: in the enterocin pathway, oxidation of an α-methylene in a thioester-tethered polyketone chain sets up a Favorskii rearrangement to generate the polycyclic scaffold of enterocin (Figure 17).254,255
Figure 17.
Oxidation control in iterative PKS pathways. The regiochemistry of cyclization is controlled by regiospecific reduction or oxidation reactions, leading to widely divergent outcomes.253,254
Enediynes: A conjugated polyene intermediate
Gene clusters for multiple enediynes have revealed that the core is assembled by an iterative PKS which harbors KR and DH domains but no ER.190,256–258 Two of these PKSs were recently shown to generate conjugated polyenes and/or polyene methyl ketones, presumably derived from decarboxylation of an initial β-keto or β-hydroxy carboxylic acid (Figure 18).259–261 It is not yet known whether these intermediates are on pathway, but it appears likely that the pathway to the enediyne core involves oxidative tailoring of a conjugated polyene intermediate.
Figure 18.
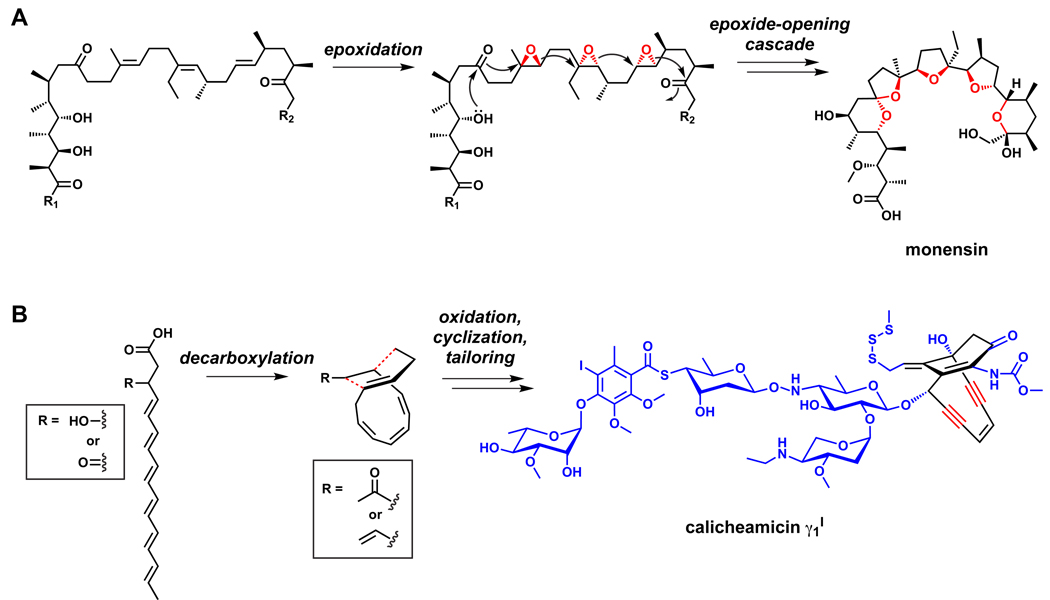
Oxidative chemistry that converts polyenes into polycyclic scaffolds. (A) The pathways to polyether polyketides involve the epoxidation of a polyolefinic precursor, followed by an epoxide-opening cascade that constructs the tetrahydropyran and tetrahydrofuran rings.262,263 (B) Polyenes are the precursors of enediynes.259–261 Additional oxidation reactions may occur out of the sequence shown in the figure.
Polyethers: Oxidation sets up a cascade reaction
Polyene chains, both conjugated and unconjugated, can also be generated by modular PKSs. The precursor to polyethers is a carrier protein-tethered polyene chain devoid of oxygens at the ring precursor positions, but oxygen can be re-incorporated by tailoring, this time by O2-utilizing epoxidases (Figure 18).262,263 These transformations are the key steps that enable ether bond formations as the epoxides undergo reaction and closure to the tetrahydrofuran and tetrahydropyran rings found in the large class of polyether natural products. The gene clusters for a handful of polyethers have been identified,264–269 and they encode two key enzyme classes: flavoprotein epoxidases to epoxidize the olefins, and epoxide hydrolases that are thought to guide the cascade of epoxide-opening events. As yet, none of the PKS gene clusters for the giant polyether toxins such as maitotoxin, ciguatoxin, and brevetoxin have been cloned, but similar mechanisms have been proposed for their biosyntheses.270
Other intramolecular cyclizations that form polycyclic ring systems
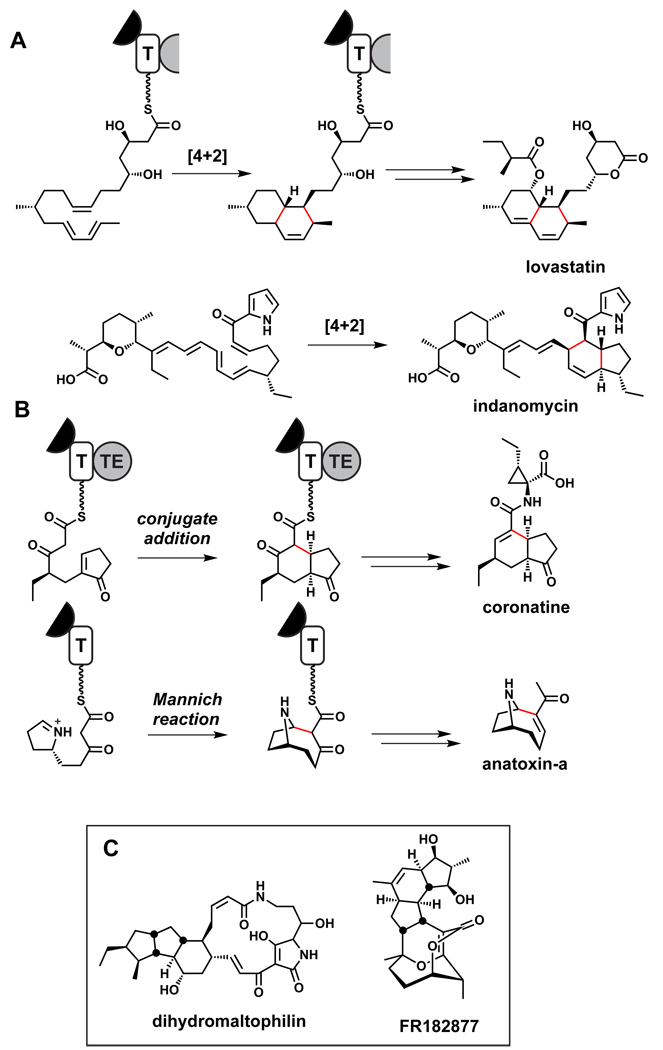
There are less well understood and more complex tailoring events that lead to the formation of polycyclic ring systems by certain PKSs. Among the most notable are cyclizations that have the attributes of [4+2] cyclizations, although it is still debated whether they proceed by stepwise or concerted mechanisms, or if they are catalyzed by Diels-Alderase enzymes.271,272 The most famous PKS in this class is the lovastatin nonaketide synthase,273 and others include the recently described PKSs for the octahydronapthalene ring in kijanimicin250 and the indane bicyclic system in indanomycin (Figure 19).251 The availability of more gene clusters and encoded enzymes makes it likely that conclusive mechanistic analyses of these natural [4+2]-type ring-forming reactions will be conducted. Note that three PKS modules in apposition must contain functional KR and DH domains but lack functional ER domains to set up the tri-olefinic precursors for the [4+2] cyclizations. In the pathway to kijanimicin’s octahydronapthalene ring system, the PKS would generate a chain with olefins in a 1,3,9 relationship prior to the [4+2]-type cyclization.250 Similar cyclizations may play a role in forming the polycyclic ring systems of FR182877,274 dihydromaltophilin,275 and related molecules.
Figure 19.
Cascade reactions for polycyclic ring systems. (A) Proposed [4+2]-like cyclizations during the biosynthesis of lovastatin273 and indanomycin.251 (B) Proposed intramolecular cyclizations during the coronatine276 and anatoxin-a277 pathways.
An intriguing variant of a cascade reaction occurs during the biosynthesis of the coronofacic acid, the polyketide moiety of the phytotoxin coronatine. Studies with a purified PKS protein indicate that a cycolepentenone-β-ketoacyl thioester arising from an assembly line-mediated Claisen condensation undergoes an intramolecular endo-trig cyclization to give a 5,6-hydrindane ring system as a precursor to coronofacic acid.276 Similar on-assembly-line intramolecular cyclizations may play a role in the biosynthesis of anatoxin,277 spinosyn,278 and tetronomycin.265
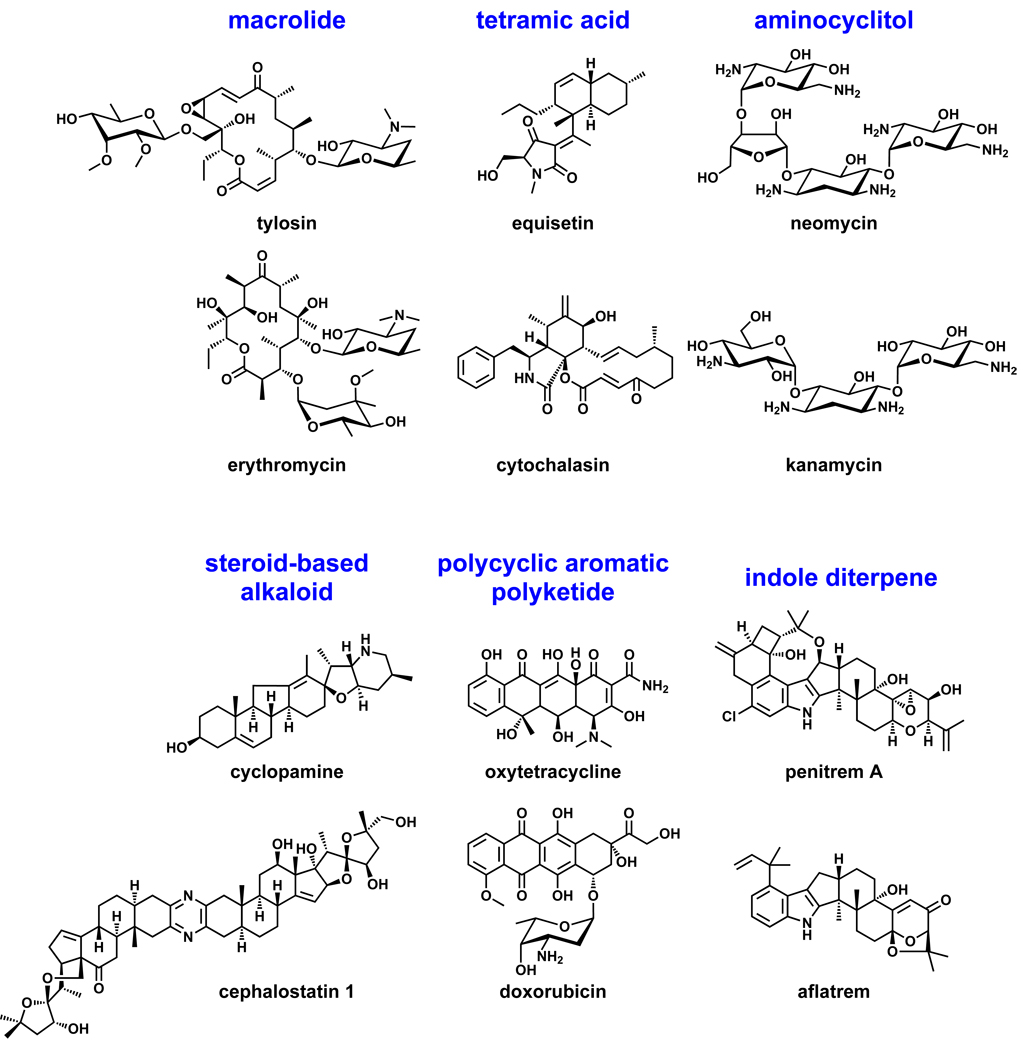
Section 9: What would it take to find all natural product scaffolds?
Given that natural products are genetically encoded and gene pools are finite, can we hope to discover most (if not all) of the natural product scaffold classes (Figure 20)? Chemists have been isolating natural products in earnest for much of the past century, in part to catalog Nature’s molecules and in part by bioactivity-guided extraction and purification for medicinal purposes. During the third quarter of the 20th century plant extracts from more than 3000 species were extensively assayed for anticancer activity alone.279 Of the 250,000–500,000 plant species, estimates are that ~10% have been studied for natural products.280,281
Figure 20.
Examples of common natural product scaffolds.
The isolation of natural products from microbes, mostly bacteria and fungi, followed from their utility as antibiotics; in a golden era from the 1930s–1950s, most of the natural antibiotic classes that have seen wide clinical use were discovered.282,283 During the past two decades, however, natural product discovery efforts in the pharmaceutical industry have decreased almost to zero.15,16 Several factors have contributed to the decline, including rediscovery of known molecules; difficulties with stability, formulation and resupply of molecules with complex architectural scaffolds; the incompatibility of natural products with HTS screens; and a preference for reduced stereochemical complexity in screening decks. Even after 50 years of intensively screening terrestrial actinomycetes, it is estimated that less than 10% of their natural product inventory has been sampled.14 Rather than discussing proposals for increased screening to improve discovery rates for new natural product scaffolds,284 we turn instead to genomics and bioinformatics as predictors of new molecules.
The ~1000 sequenced bacterial genomes harbor thousands of predicted biosynthetic gene clusters.24,26 As large as the potential currently seems, there is an important reason to think it might even be understated: the set of sequenced bacterial genomes is biased toward pathogens. Pathogenic bacteria like enterobacteria, staphylococci, and streptococci have very limited capacity for natural product biosynthesis; one contributing factor is that many of their genomes are ~3 Mb, which is a size breakpoint for genomes with significant auxiliary metabolic capacity. In general, above 3 Mb, the larger the genome, the greater the coding capacity devoted to biosynthetic gene clusters.285 For genomes as large as Streptomyces (8–9 Mb), 25–400 kb of DNA can be devoted to such gene clusters, consonant with historical experience that streptomycetes are prolific antibiotic producers.
Sequenced streptomycetes such as S. avermitilis and S. griseus have been observed to make ~2–3 natural products but harbor ~25–30 predicted biosynthetic gene clusters.23,27,286 To date, we are missing 90% of the natural product biosynthetic capacity of even the workhorse producers. If even 20% of the 20–25 cryptic molecules were novel, the current knowledge base from streptomycetes would double. Various efforts to activate these ‘cryptic’ gene clusters are underway, including placing them under the control of strong promoters.29
One test of the power of bacterial genomics and bioinformatics would be to sequence 1000 new bacterial genomes from genera such as Streptomyces and Myxobacteria to determine whether the average of 30 clusters per genome holds. If so, and 20% of the 30,000 encoded molecules, once expressed, isolated, and characterized were novel, 6,000 new molecules would be available for screening. While the fraction that would have new scaffolds and/or biological activities is unknown, the odds of novelty would increase if bacteria from underexplored niches were emphasized, including those from marine sediments21,287 and symbionts of plants and insects,17 since novel molecules18,19,288,289 have recently been characterized by this route.
The power of bioinformatics would allow the evaluation of whether gene cluster saturation were being approached. Such a sequencing/bioinformatics effort would also determine whether an ambitious goal of ~90% coverage of all the encoded scaffolds (perhaps beginning with the more bioinformatically accessible PK and NRP scaffolds) could be approached, and how many genomes would be required to get there. This project would be analogous to the ongoing protein structure initiative (http://www.structuralgenomics.org) in which high-throughput x-ray structures of proteins are being solved for a ten-year period to approach the point where most protein folds have a solved structure, in part as a prerequisite for protein design and functional re-engineering. If scientists similarly knew the universe of PKSs, NRPSs, and the associated enzymes that make novel building blocks, a gene-based mix and match strategy should enable the generation and testing of new scaffold variants.
In parallel, there are ongoing efforts in enzyme evolution in many laboratories around the world to evolve enzymes with engineered catalytic abilities for transformations of industrial synthetic interest. All of those principles and methodologies would be transferable to modulate enzyme specificity and gatekeeper roadblocks in natural product pathways. In that sense, the 90% of all microbial natural products from the contemporary global microbial communities would be a starting point to connect genotype evolution to chemotype evolution of useful new molecules.
Section 10: Which disciplines will merge with natural product research in the future?
Natural product research has been dominated by chemists and biochemists, and for good reason: isolation, structural characterization, total synthesis, diverted synthesis for analoging, and biochemical characterization of pathways are all chemical pursuits. But as the connection between natural products and the genes that encode them grows stronger, the tools of modern genetics will increasingly be brought to bear on natural product research, leading to its likely merger with four disciplines:
Genomics and bioinformatics
The connection with genomics and bioinformatics has its roots in the discovery of the erythromycin synthase by Leadlay and Katz,47,48 grew stronger with the revelation of abundant cryptic gene clusters in the Streptomyces coelicolor and Streptomyces avermitilis genomes,23,286 and continues to expand as the database of sequenced bacterial genomes exceeds 1000.24,26 One long-term goal of this branch of chemoinformatics is to automate the identification of gene clusters in sequenced genomes and the prediction of their small molecule products, and significant progress toward this goal has been made with efforts to predict the structures of nonribosomal peptides and polyketides.106,290,291
Microbial ecology
While the natural roles of natural products are only beginning to be understood, it appears likely that many natural products mediate interactions among microbes or between microbes and larger organisms.292–295 The nexus of natural product research and microbial ecology will involve studying, inter alia, the global distribution of classes of gene clusters;296 how differences in the complement of encoded natural products allow related microbes to adapt to distinct ecological niches; and how the set of natural products produced by a microbe influences the other organisms in its niche. Insights from this line of study will guide efforts to stimulate the production of cryptic natural products in microbial genomes.
Synthetic biology
One important contribution from the decades of research into natural product biosynthesis is a large ‘parts list’ of enzymes that catalyze specific transformations relevant to the construction and tailoring of molecular scaffolds.297 Synthetic biologists have proven adept at compiling parts lists and using them to understand existing processes and design new ones.298 The reconstitution of complex terpenoid,299,300 polyketide,301 and nonribosomal peptide302 pathways in hosts like Saccharomyces cerevisiae and Escherichia coli likely presage more widespread efforts to reconstitute existing pathways and design new pathways by constructing artificial operons.
Systems biology
Researchers are beginning to use the tools of systems biology to answer important questions in natural product research: How do core metabolic networks link to auxiliary pathways and adapt to their conditional expression?303 How do global and pathway-specific regulatory networks govern the production of natural products?304,305 Can transcriptomic and proteomic responses to a novel natural product reveal its mechanism of action?306 Metabolomics, a direct merger between systems biology and small molecule research, will play an increasingly important role in the coming decades.37
Acknowledgments
We are indebted to our current and former coworkers for countless discussions and insights featured in this perspective. We are particularly indebted to Jon Clardy for ideas and discussions about connecting natural products to the genes that encode them. Research in the authors’ laboratories is supported by NIH grants GM20011, GM49338, AI42738 (C.T.W.); NERCE award AI057159 (C.T.W. and M.A.F.); the Department of Bioengineering and Therapeutic Sciences at UCSF (M.A.F.); and the California Institute for Quantitative Biosciences (M.A.F.).
Contributor Information
Christopher T. Walsh, Email: christopher_walsh@hms.harvard.edu.
Michael A. Fischbach, Email: fischbach@fischbachgroup.org.
References
- 1.Newman DJ, Cragg GM. Journal of natural products. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 2.Wender PA, Miller BL. Nature. 2009;460:197–201. doi: 10.1038/460197a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies HM. Nature. 2009;459:786–787. doi: 10.1038/459786a. [DOI] [PubMed] [Google Scholar]
- 4.Feher M, Schmidt JM. J Chem Inf Comput Sci. 2003;43:218–227. doi: 10.1021/ci0200467. [DOI] [PubMed] [Google Scholar]
- 5.Shang S, Tan DS. Curr Opin Chem Biol. 2005;9:248–258. doi: 10.1016/j.cbpa.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Chen MS, White MC. Science (New York, N.Y. 2007;318:783–787. doi: 10.1126/science.1148597. [DOI] [PubMed] [Google Scholar]
- 7.Chen K, Baran PS. Nature. 2009;459:824–828. doi: 10.1038/nature08043. [DOI] [PubMed] [Google Scholar]
- 8.Davie EA, Mennen SM, Xu Y, Miller SJ. Chemical reviews. 2007;107:5759–5812. doi: 10.1021/cr068377w. [DOI] [PubMed] [Google Scholar]
- 9.Li JW, Vederas JC. Science. 2009;325:161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 10.Inoki K, Corradetti MN, Guan KL. Nat Genet. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- 11.Tamaoki T, Nomoto H, Takahashi I, Kato Y, Morimoto M, Tomita F. Biochem Biophys Res Commun. 1986;135:397–402. doi: 10.1016/0006-291x(86)90008-2. [DOI] [PubMed] [Google Scholar]
- 12.Capdeville R, Buchdunger E, Zimmermann J, Matter A. Nat Rev Drug Discov. 2002;1:493–502. doi: 10.1038/nrd839. [DOI] [PubMed] [Google Scholar]
- 13.Lamb J, et al. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 14.Watve MG, Tickoo R, Jog MM, Bhole BD. Archives of microbiology. 2001;176:386–390. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- 15.Baltz RH. SIM News. 2005;55:186–196. [Google Scholar]
- 16.Baltz RH. J Ind Microbiol Biotechnol. 2006;33:507–513. doi: 10.1007/s10295-005-0077-9. [DOI] [PubMed] [Google Scholar]
- 17.Piel J. Natural product reports. 2009;26:338–362. doi: 10.1039/b703499g. [DOI] [PubMed] [Google Scholar]
- 18.Bister B, Bischoff D, Strobele M, Riedlinger J, Reicke A, Wolter F, Bull AT, Zahner H, Fiedler HP, Sussmuth RD. Angewandte Chemie (International ad. 2004;43:2574–2576. doi: 10.1002/anie.200353160. [DOI] [PubMed] [Google Scholar]
- 19.Scott JJ, Oh DC, Yuceer MC, Klepzig KD, Clardy J, Currie CR. Science (New York, N.Y. 2008;322:63. doi: 10.1126/science.1160423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Partida-Martinez LP, Hertweck C. Nature. 2005;437:884–888. doi: 10.1038/nature03997. [DOI] [PubMed] [Google Scholar]
- 21.Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Proc Natl Acad Sci U S A. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wenzel SC, Muller R. Current opinion in drug discovery & development. 2009;12:220–230. [PubMed] [Google Scholar]
- 23.Bentley SD, et al. Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 24.Challis GL. Journal of medicinal chemistry. 2008;51:2618–2628. doi: 10.1021/jm700948z. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S. Nature biotechnology. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 26.McAlpine JB. Journal of natural products. 2009;72:566–572. doi: 10.1021/np800742z. [DOI] [PubMed] [Google Scholar]
- 27.Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S. Journal of bacteriology. 2008;190:4050–4060. doi: 10.1128/JB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF. Nature biotechnology. 2007;25:447–453. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- 29.Scherlach K, Hertweck C. Org Biomol Chem. 2009;7:1753–1760. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- 30.Schneiker S. Nat Biotechnol. 2007;25:1281–1289. doi: 10.1038/nbt1354. [DOI] [PubMed] [Google Scholar]
- 31.Goldman BS, et al. Proc Natl Acad Sci U S A. 2006;103:15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernhardt P, O'Connor SE. Curr Opin Chem Biol. 2009;13:35–42. doi: 10.1016/j.cbpa.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirby J, Keasling JD. Annu Rev Plant Biol. 2009;60:335–355. doi: 10.1146/annurev.arplant.043008.091955. [DOI] [PubMed] [Google Scholar]
- 34.Leonard E, Runguphan W, O'Connor S, Prather KJ. Nat Chem Biol. 2009;5:292–300. doi: 10.1038/nchembio.160. [DOI] [PubMed] [Google Scholar]
- 35.Shendure J, Ji H. Nat Biotechnal. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 36.Vinayavekhin N, Saghatelian A. ACS Chem Biol. 2009;4:617–623. doi: 10.1021/cb900075n. [DOI] [PubMed] [Google Scholar]
- 37.Rochfort S. J Nat Prod. 2005;68:1813–1820. doi: 10.1021/np050255w. [DOI] [PubMed] [Google Scholar]
- 38.Schroeder FC, Gibson DM, Churchill AC, Sojikul P, Wursthorn EJ, Krasnoff SB, Clardy J. Angew Chem Int Ed Engl. 2007;46:901–904. doi: 10.1002/anie.200603821. [DOI] [PubMed] [Google Scholar]
- 39.Molinski TF. Curr Opin Drug Discov Devel. 2009;12:197–206. [PubMed] [Google Scholar]
- 40.Hertweck C. Angewandte Chemie (International ad. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 41.Marahiel MA. J Pept Sci. 2009;15:799–807. doi: 10.1002/psc.1183. [DOI] [PubMed] [Google Scholar]
- 42.Flatt PM, Mahmud T. Natural product reports. 2007;24:358–392. doi: 10.1039/b603816f. [DOI] [PubMed] [Google Scholar]
- 43.Challis GL. Chembiochem. 2005;6:601–611. doi: 10.1002/cbic.200400283. [DOI] [PubMed] [Google Scholar]
- 44.Daum M, Herrmann S, Wilkinson B, Bechthold A. Curr Opin Chem Biol. 2009;13:180–188. doi: 10.1016/j.cbpa.2009.02.029. [DOI] [PubMed] [Google Scholar]
- 45.Buchanan BB, Gruissem W, Jones RL. Biochemistry & molecular biology of plants. Rockville, Md: American Society of Plant Physiologists; 2000. [Google Scholar]
- 46.Christianson DW. Curr Opin Chem Biol. 2008;12:141–150. doi: 10.1016/j.cbpa.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donadio S, Staver MJ, McAlpine JB, Swanson SJ, Katz L. Science (New York, N.Y. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 48.Cortes J, Haydock SF, Roberts GA, Bevitt DJ, Leadlay PF. Nature. 1990;348:176–178. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- 49.Marahiel MA, Stachelhaus T, Mootz HD. Chem Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 50.Schwecke T, Aparicio JF, Molnar I, Konig A, Khaw LE, Haydock SF, Oliynyk M, Caffrey P, Cortes J, Lester JB, et al. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aparicio JF, Molnar I, Schwecke T, Konig A, Haydock SF, Khaw LE, Staunton J, Leadlay PF. Gene. 1996;169:9–16. doi: 10.1016/0378-1119(95)00800-4. [DOI] [PubMed] [Google Scholar]
- 52.Motamedi H, Shafiee A. Eur J Biochem. 1998;256:528–534. doi: 10.1046/j.1432-1327.1998.2560528.x. [DOI] [PubMed] [Google Scholar]
- 53.Wu K, Chung L, Revill WP, Katz L, Reeves CD. Gene. 2000;251:81–90. doi: 10.1016/s0378-1119(00)00171-2. [DOI] [PubMed] [Google Scholar]
- 54.Tang L, Shah S, Chung L, Carney J, Katz L, Khosla C, Julien B. Science (New York, N.Y. 2000;287:640–642. doi: 10.1126/science.287.5453.640. [DOI] [PubMed] [Google Scholar]
- 55.Du L, Sanchez C, Chen M, Edwards DJ, Shen B. Chem Biol. 2000;7:623–642. doi: 10.1016/s1074-5521(00)00011-9. [DOI] [PubMed] [Google Scholar]
- 56.Cane DE, Walsh CT. Chem Biol. 1999;6:R319–R325. doi: 10.1016/s1074-5521(00)80001-0. [DOI] [PubMed] [Google Scholar]
- 57.Fischbach MA, Walsh CT. Chemical reviews. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 58.Boucher Y, Nesbo CL, Doolittle WF. Curr Opin Microbiol. 2001;4:285–289. doi: 10.1016/s1369-5274(00)00204-6. [DOI] [PubMed] [Google Scholar]
- 59.Chan YA, Podevels AM, Kevany BM, Thomas MG. Nat Prod Rep. 2009;26:90–114. doi: 10.1039/b801658p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moore BS, Hertweck C. Nat Prod Rep. 2002;19:70–99. doi: 10.1039/b003939j. [DOI] [PubMed] [Google Scholar]
- 61.Lowden PA, Wilkinson B, Bohm GA, Handa S, Floss HG, Leadlay PF, Staunton J. Angew Chem Int Ed Engl. 2001;40:777–779. [PubMed] [Google Scholar]
- 62.Eustaquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G, Alhamadsheh MM, Lechner A, Kale AJ, Kobayashi Y, Reynolds KA, Moore BS. Proc Natl Acad Sci U S A. 2009;106:12295–12300. doi: 10.1073/pnas.0901237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wenzel SC, Williamson RM, Grunanger C, Xu J, Gerth K, Martinez RA, Moss SJ, Carroll BJ, Grond S, Unkefer CJ, Muller R, Floss HG. J Am Chem Soc. 2006;128:14325–14336. doi: 10.1021/ja064408t. [DOI] [PubMed] [Google Scholar]
- 64.Hubbard BK, Walsh CT. Angewandte Chemie (International ed. 2003;42:730–765. doi: 10.1002/anie.200390202. [DOI] [PubMed] [Google Scholar]
- 65.Donadio S, Sosio M, Stegmann E, Weber T, Wohlleben W. Mol Genet Genomics. 2005;274:40–50. doi: 10.1007/s00438-005-1156-3. [DOI] [PubMed] [Google Scholar]
- 66.Puk O, Bischoff D, Kittel C, Pelzer S, Weist S, Stegmann E, Sussmuth RD, Wohlleben W. J Bacteriol. 2004;186:6093–6100. doi: 10.1128/JB.186.18.6093-6100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hubbard BK, Thomas MG, Walsh CT. Chem Biol. 2000;7:931–942. doi: 10.1016/s1074-5521(00)00043-0. [DOI] [PubMed] [Google Scholar]
- 68.Chen H, Tseng CC, Hubbard BK, Walsh CT. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:14901–14906. doi: 10.1073/pnas.221582098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen H, Thomas MG, Hubbard BK, Losey HC, Walsh CT, Burkart MD. Proc Natl Acad Sci U S A. 2000;97:11942–11947. doi: 10.1073/pnas.210395097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Penfold CN, Bender CL, Turner JG. Gene. 1996;183:167–173. doi: 10.1016/s0378-1119(96)00550-1. [DOI] [PubMed] [Google Scholar]
- 71.Ullrich M, Bender CL. J Bacteriol. 1994;176:7574–7586. doi: 10.1128/jb.176.24.7574-7586.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weiler EW, Kutchan TM, Gorba T, Brodschelm W, Niesel U, Bublitz F. FEBS Lett. 1994;345:9–13. doi: 10.1016/0014-5793(94)00411-0. [DOI] [PubMed] [Google Scholar]
- 73.Vaillancourt FH, Yeh E, Vosburg DA, O'Connor SE, Walsh CT. Nature. 2005;436:1191–1194. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- 74.Ueki M, Galonic DP, Vaillancourt FH, Garueau-Tsodikova S, Yeh E, Vosburg DA, Schroeder FC, Osada H, Walsh CT. Chem Biol. 2006;13:1183–1191. doi: 10.1016/j.chembiol.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 75.Garneau S, Dorrestein PC, Kelleher NL, Walsh CT. Biochemistry. 2005;44:2770–2780. doi: 10.1021/bi0476329. [DOI] [PubMed] [Google Scholar]
- 76.Garneau-Tsodikova S, Dorrestein PC, Kelleher NL, Walsh CT. J Am Chem Soc. 2006;128:12600–12601. doi: 10.1021/ja063611l. [DOI] [PubMed] [Google Scholar]
- 77.O'Hagan D. Nat Prod Rep. 2000;17:435–446. doi: 10.1039/a707613d. [DOI] [PubMed] [Google Scholar]
- 78.Gatto GJ, Jr, Boyne MT, 2nd, Kelleher NL, Walsh CT. J Am Chem Soc. 2006;128:3838–3847. doi: 10.1021/ja0587603. [DOI] [PubMed] [Google Scholar]
- 79.Umezawa K, Ikeda Y, Kawase O, Naganawa H, Kondo S. J Chem Soc, Perkin Trans 1. 2001:1550–1553. [Google Scholar]
- 80.Neusser D, Schmidt H, Spizek J, Novotna J, Peschke U, Kaschabeck S, Tichy P, Piepersberg W. Archives of microbiology. 1998;169:322–332. doi: 10.1007/s002030050578. [DOI] [PubMed] [Google Scholar]
- 81.Merrifield RB. Science. 1965;150:178–185. doi: 10.1126/science.150.3693.178. [DOI] [PubMed] [Google Scholar]
- 82.Lai JR, Koglin A, Walsh CT. Biochemistry. 2006;45:14869–14879. doi: 10.1021/bi061979p. [DOI] [PubMed] [Google Scholar]
- 83.Tanovic A, Samel SA, Essen LO, Marahiel MA. Science. 2008;321:659–663. doi: 10.1126/science.1159850. [DOI] [PubMed] [Google Scholar]
- 84.Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. Proc Natl Acad Sci U S A. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walsh CT, Chen H, Keating TA, Hubbard BK, Losey HC, Luo L, Marshall CG, Miller DA, Patel HM. Curr Opin Chem Biol. 2001;5:525–534. doi: 10.1016/s1367-5931(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 86.Luo L, Kohli RM, Onishi M, Linne U, Marahiel MA, Walsh CT. Biochemistry. 2002;41:9184–9196. doi: 10.1021/bi026047+. [DOI] [PubMed] [Google Scholar]
- 87.Magarvey NA, Ehling-Schulz M, Walsh CT. J Am Chem Soc. 2006;128:10698–10699. doi: 10.1021/ja0640187. [DOI] [PubMed] [Google Scholar]
- 88.Kelly WL, Hillson NJ, Walsh CT. Biochemistry. 2005;44:13385–13393. doi: 10.1021/bi051124x. [DOI] [PubMed] [Google Scholar]
- 89.Schneider TL, Shen B, Walsh CT. Biochemistry. 2003;42:9722–9730. doi: 10.1021/bi034792w. [DOI] [PubMed] [Google Scholar]
- 90.Shen B. Curr Opin Chem Biol. 2003;7:285–295. doi: 10.1016/s1367-5931(03)00020-6. [DOI] [PubMed] [Google Scholar]
- 91.Wenzel SC, Muller R. Curr Opin Chem Biol. 2005;9:447–458. doi: 10.1016/j.cbpa.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 92.Keating TA, Ehmann DE, Kohli RM, Marshall CG, Trauger JW, Walsh CT. Chembiochem. 2001;2:99–107. doi: 10.1002/1439-7633(20010202)2:2<99::AID-CBIC99>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 93.Koglin A, Walsh CT. Natural product reports. 2009;26:987–1000. doi: 10.1039/b904543k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tseng CC, Bruner SD, Kohli RM, Marahiel MA, Walsh CT, Sieber SA. Biochemistry. 2002;41:13350–13359. doi: 10.1021/bi026592a. [DOI] [PubMed] [Google Scholar]
- 95.Gehring AM, Mori I, Walsh CT. Biochemistry. 1998;37:2648–2659. doi: 10.1021/bi9726584. [DOI] [PubMed] [Google Scholar]
- 96.Angell S, Bench BJ, Williams H, Watanabe CM. Chemistry & biology. 2006;13:1349–1359. doi: 10.1016/j.chembiol.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 97.Robbel L, Hoyer KM, Marahiel MA. FEBS J. 2009;276:1641–1653. doi: 10.1111/j.1742-4658.2009.06897.x. [DOI] [PubMed] [Google Scholar]
- 98.Balibar CJ, Howard-Jones AR, Walsh CT. Nat Chem Biol. 2007;3:584–592. doi: 10.1038/nchembio.2007.20. [DOI] [PubMed] [Google Scholar]
- 99.Loria R, Bignell DR, Moll S, Huguet-Tapia JC, Joshi MV, Johnson EG, Seipke RF, Gibson DM. Antonie Van Leeuwenhoek. 2008;94:3–10. doi: 10.1007/s10482-008-9240-4. [DOI] [PubMed] [Google Scholar]
- 100.Balibar CJ, Walsh CT. Biochemistry. 2006;45:15029–15038. doi: 10.1021/bi061845b. [DOI] [PubMed] [Google Scholar]
- 101.Schultz AW, Oh DC, Carney JR, Williamson RT, Udwary DW, Jensen PR, Gould SJ, Fenical W, Moore BS. J Am Chem Soc. 2008;130:4507–4516. doi: 10.1021/ja711188x. [DOI] [PubMed] [Google Scholar]
- 102.Gondry M, et al. Nat Chem Biol. 2009;5:414–420. doi: 10.1038/nchembio.175. [DOI] [PubMed] [Google Scholar]
- 103.Belin P, Le Du MH, Fielding A, Lequin O, Jacquet M, Charbonnier JB, Lecoq A, Thai R, Courcon M, Masson C, Dugave C, Genet R, Pernodet JL, Gondry M. Proc Natl Acad Sci U S A. 2009;106:7426–7431. doi: 10.1073/pnas.0812191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moldenhauer J, Chen XH, Borriss R, Piel J. Angew Chem Int Ed Engl. 2007;46:8195–8197. doi: 10.1002/anie.200703386. [DOI] [PubMed] [Google Scholar]
- 105.Butcher RA, Schroeder FC, Fischbach MA, Straight PD, Kolter R, Walsh CT, Clardy J. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1506–1509. doi: 10.1073/pnas.0610503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nguyen T, Ishida K, Jenke-Kodama H, Dittmann E, Gurgui C, Hochmuth T, Taudien S, Platzer M, Hertweck C, Piel J. Nature biotechnology. 2008;26:225–233. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]
- 107.Liras P, Martin JF. Int Microbiol. 2006;9:9–19. [PubMed] [Google Scholar]
- 108.Keating TA, Marshall CG, Walsh CT. Biochemistry. 2000;39:15522–15530. doi: 10.1021/bi0016523. [DOI] [PubMed] [Google Scholar]
- 109.Sattely ES, Walsh CT. J Am Chem Soc. 2008;130:12282–12284. doi: 10.1021/ja804499r. [DOI] [PubMed] [Google Scholar]
- 110.Cerdeno AM, Bibb MJ, Challis GL. Chem Bio1. 2001;8:817–829. doi: 10.1016/s1074-5521(01)00054-0. [DOI] [PubMed] [Google Scholar]
- 111.Kellmann R, Mihali TK, Jeon YJ, Pickford R, Pomati F, Neilan BA. Appl Environ Microbio1. 2008;74:4044–4053. doi: 10.1128/AEM.00353-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li L, Deng W, Song J, Ding W, Zhao QF, Peng C, Song WW, Tang GL, Liu W. J Bacteriol. 2008;190:251–263. doi: 10.1128/JB.00826-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kopp F, Mahlert C, Grunewald J, Marahiel MA. J Am Chem Soc. 2006;128:16478–16479. doi: 10.1021/ja0667458. [DOI] [PubMed] [Google Scholar]
- 114.Becker JE, Moore RE, Moore BS. Gene. 2004;325:35–42. doi: 10.1016/j.gene.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 115.Read JA, Walsh CT. J Am Chem Soc. 2007;129:15762–15763. doi: 10.1021/ja077374d. [DOI] [PubMed] [Google Scholar]
- 116.Sims JW, Fillmore JP, Warner DD, Schmidt EW. Chem Commun (Camb) 2005:186–188. doi: 10.1039/b413523g. [DOI] [PubMed] [Google Scholar]
- 117.Sims JW, Schmidt EW. J Am Chem Soc. 2008;130:11149–11155. doi: 10.1021/ja803078z. [DOI] [PubMed] [Google Scholar]
- 118.Liu X, Walsh CT. Biochemistry. 2009;48:8746–8757. doi: 10.1021/bi901123r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.He XM, Liu HW. Annu Rev Biochem. 2002;71:701–754. doi: 10.1146/annurev.biochem.71.110601.135339. [DOI] [PubMed] [Google Scholar]
- 120.Thibodeaux CJ, Melancon CE, 3rd, Liu HW. Angew Chem Int Ed Eng1. 2008;47:9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lairson LL, Henrissat B, Davies GJ, Withers SG. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 122.Liu J, Mushegian A. Protein Sci. 2003;12:1418–1431. doi: 10.1110/ps.0302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Busscher GF, Rutjes FP, van Delft FL. Chem Rev. 2005;105:775–791. doi: 10.1021/cr0404085. [DOI] [PubMed] [Google Scholar]
- 124.Luzhetskyy A, Fedoryshyn M, Durr C, Taguchi T, Novikov V, Bechthold A. Chem Bio1. 2005;12:725–729. doi: 10.1016/j.chembiol.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 125.Westrich L, Domann S, Faust B, Bedford D, Hopwood DA, Bechthold A. FEMS Microbial Lett. 1999;170:381–387. doi: 10.1111/j.1574-6968.1999.tb13398.x. [DOI] [PubMed] [Google Scholar]
- 126.McGarvey DJ, Croteau R. Plant Cell. 1995;7:1015–1026. doi: 10.1105/tpc.7.7.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Thulasiram HV, Erickson HK, Poulter CD. J Am Chem Soc. 2008;130:1966–1971. doi: 10.1021/ja0771282. [DOI] [PubMed] [Google Scholar]
- 128.Thulasiram HV, Erickson HK, Poulter CD. Science. 2007;316:73–76. doi: 10.1126/science.1137786. [DOI] [PubMed] [Google Scholar]
- 129.Swiezewska E, Dauikiewicz W. Prog Lipid Res. 2005;44:235–258. doi: 10.1016/j.plipres.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 130.Austin MB, O'Maille PE, Noel JP. Nat Chem Bio1. 2008;4:217–222. doi: 10.1038/nchembio0408-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yoshikuni Y, Ferrin TE, Keasling JD. Nature. 2006;440:1078–1082. doi: 10.1038/nature04607. [DOI] [PubMed] [Google Scholar]
- 132.Jennewein S, Croteau R. Appl Microbiol Biotechnol. 2001;57:13–19. doi: 10.1007/s002530100757. [DOI] [PubMed] [Google Scholar]
- 133.Bomke C, Tudzynski B. Phytochemistry. 2009 doi: 10.1016/j.phytochem.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 134.Lin X, Hezari M, Koepp AE, Floss HG, Croteau R. Biochemistry. 1996;35:2968–2977. doi: 10.1021/bi9526239. [DOI] [PubMed] [Google Scholar]
- 135.Long RM, Lagisetti C, Coates RM, Croteau RB. Arch Biochem Biophys. 2008;477:384–389. doi: 10.1016/j.abb.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chau M, Croteau R. Arch Biochem Biophys. 2004;427:48–57. doi: 10.1016/j.abb.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 137.Chau M, Jennewein S, Walker K, Croteau R. Chem Biol. 2004;11:663–672. doi: 10.1016/j.chembiol.2004.02.025. [DOI] [PubMed] [Google Scholar]
- 138.Jennewein S, Long RM, Williams RM, Croteau R. Chem Bio1. 2004;11:379–387. doi: 10.1016/j.chembiol.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 139.Jennewein S, Rithner CD, Williams RM, Croteau R. Arch Biochem Biophys. 2003;413:262–270. doi: 10.1016/s0003-9861(03)00090-0. [DOI] [PubMed] [Google Scholar]
- 140.Jennewein S, Rithner CD, Williams RM, Croteau RB. Proc Natl Acad Sci U S A. 2001;98:13595–13600. doi: 10.1073/pnas.251539398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schoendorf A, Rithner CD, Williams RM, Croteau RB. Proc Natl Acad Sci U S A. 2001;98:1501–1506. doi: 10.1073/pnas.98.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lin X, Cane DE. J Am Chem Soc. 2009;131:6332–6333. doi: 10.1021/ja901313v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhao B, Lin X, Lei L, Lamb DC, Kelly SL, Waterman MR, Cane DE. J Biol Chem. 2008;283:8183–8189. doi: 10.1074/jbc.M710421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lin X, Hopson R, Cane DE. J Am Chem Soc. 2006;128:6022–6023. doi: 10.1021/ja061292s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jiang J, Tetzlaff CN, Takamatsu S, Iwatsuki M, Komatsu M, Ikeda H, Cane DE. Biochemistry. 2009;48:6431–6440. doi: 10.1021/bi900766w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.You Z, Omura S, Ikeda H, Cane DE, Jogl G. J Biol Chem. 2007;282:36552–36560. doi: 10.1074/jbc.M706358200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.You Z, Omura S, Ikeda H, Cane DE. Arch Biochem Biophys. 2007;459:233–240. doi: 10.1016/j.abb.2006.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Quaderer R, Omura S, Ikeda H, Cane DE. J Am Chem Soc. 2006;128:13036–13037. doi: 10.1021/ja0639214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.You Z, Omura S, Ikeda H, Cane DE. J Am Chem Soc. 2006;128:6566–6567. doi: 10.1021/ja061469i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tetzlaff CN, You Z, Cane DE, Takamatsu S, Omura S, Ikeda H. Biochemistry. 2006;45:6179–6186. doi: 10.1021/bi060419n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Binz TM, Wenzel SC, Schnell HJ, Bechthold A, Muller R. Chembiochem. 2008;9:447–454. doi: 10.1002/cbic.200700549. [DOI] [PubMed] [Google Scholar]
- 152.Durr C, Schnell HJ, Luzhetskyy A, Murillo R, Weber M, Welzel K, Vente A, Bechthold A. Chem Biol. 2006;13:365–377. doi: 10.1016/j.chembiol.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 153.Kadi N, Oves-Costales D, Barona-Gomez F, Challis GL. Nat Chem Biol. 2007;3:652–656. doi: 10.1038/nchembio.2007.23. [DOI] [PubMed] [Google Scholar]
- 154.Pacholec M, Freel Meyers CL, Oberthur M, Kahne D, Walsh CT. Biochemistry. 2005;44:4949–4956. doi: 10.1021/bi047303g. [DOI] [PubMed] [Google Scholar]
- 155.Liyanage H, Penfold C, Turner J, Bender CL. Gene. 1995;153:17–23. doi: 10.1016/0378-1119(94)00661-b. [DOI] [PubMed] [Google Scholar]
- 156.Hollenhorst MA, Clardy J, Walsh CT. Biochemistry. 2009 doi: 10.1021/bi9013165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Philmus B, Guerrette JP, Hemscheidt TK. ACS Chem Biol. 2009;4:429–434. doi: 10.1021/cb900088r. [DOI] [PubMed] [Google Scholar]
- 158.Philmus B, Christiansen G, Yoshida WY, Hemscheidt TK. Chembiochem. 2008;9:3066–3073. doi: 10.1002/cbic.200800560. [DOI] [PubMed] [Google Scholar]
- 159.Ziemert N, Ishida K, Liaimer A, Hertweck C, Dittmann E. Angew Chem Int Ed Engl. 2008;47:7756–7759. doi: 10.1002/anie.200802730. [DOI] [PubMed] [Google Scholar]
- 160.Moore BS. Angewandte Chemie (International ed. 2008;47:9386–9388. doi: 10.1002/anie.200803868. [DOI] [PubMed] [Google Scholar]
- 161.Maresh JJ, Giddings LA, Friedrich A, Loris EA, Panjikar S, Trout BL, Stockigt J, Peters B, O'Connor SE. Journal of the American Chemical Society. 2008;130:710–723. doi: 10.1021/ja077190z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Luk LY, Bunn S, Liscombe DK, Facchini PJ, Tanner ME. Biochemistry. 2007;46:10153–10161. doi: 10.1021/bi700752n. [DOI] [PubMed] [Google Scholar]
- 163.Runguphan W, Maresh JJ, O'Connor SE. Proc Natl Acad Sci U S A. 2009;106:13673–13678. doi: 10.1073/pnas.0903393106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Runguphan W, O'Connor SE. Nat Chem Biol. 2009;5:151–153. doi: 10.1038/nchembio.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pan JY, Chen SL, Yang MH, Wu J, Sinkkonen J, Zou K. Nat Prod Rep. 2009;26:1251–1292. doi: 10.1039/b910940d. [DOI] [PubMed] [Google Scholar]
- 166.Davin LB, Wang HB, Crowell AL, Bedgar DL, Martin DM, Sarkanen S, Lewis NG. Science. 1997;275:362–366. doi: 10.1126/science.275.5298.362. [DOI] [PubMed] [Google Scholar]
- 167.Xia ZQ, Costa MA, Proctor J, Davin LB, Lewis NG. Phytochemistry. 2000;55:537–549. doi: 10.1016/s0031-9422(00)00242-9. [DOI] [PubMed] [Google Scholar]
- 168.Whetten RW, MacKay JJ, Sederoff RR. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:585–609. doi: 10.1146/annurev.arplant.49.1.585. [DOI] [PubMed] [Google Scholar]
- 169.Howard-Jones AR, Walsh CT. Journal of the American Chemical Society. 2007;129:11016–11017. doi: 10.1021/ja0743801. [DOI] [PubMed] [Google Scholar]
- 170.Howard-Jones AR, Kruger RG, Lu W, Tao J, Leimkuhler C, Kahne D, Walsh CT. J Am Chem Soc. 2007;129:10082–10083. doi: 10.1021/ja0735857. [DOI] [PubMed] [Google Scholar]
- 171.Howard-Jones AR, Walsh CT. Biochemistry. 2005;44:15652–15663. doi: 10.1021/bi051706e. [DOI] [PubMed] [Google Scholar]
- 172.Balskus EP, Walsh CT. J Am Chem Soc. 2009;131:14648–14649. doi: 10.1021/ja906752u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Balskus EP, Walsh CT. J Am Chem Soc. 2008;130:15260–15261. doi: 10.1021/ja807192u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Richter CD, Nietlispach D, Broadhurst RW, Weissman KJ. Nat Chem Biol. 2008;4:75–81. doi: 10.1038/nchembio.2007.61. [DOI] [PubMed] [Google Scholar]
- 175.Weissman KJ. Chembiochem. 2006;7:485–494. doi: 10.1002/cbic.200500435. [DOI] [PubMed] [Google Scholar]
- 176.Kumar P, Li Q, Cane DE, Khosla C. J Am Chem Soc. 2003;125:4097–4102. doi: 10.1021/ja0297537. [DOI] [PubMed] [Google Scholar]
- 177.Hahn M, Stachelhaus T. Proc Natl Acad Sci U S A. 2006;103:275–280. doi: 10.1073/pnas.0508409103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hahn M, Stachelhaus T. Proc Natl Acad Sci U S A. 2004;101:15585–15590. doi: 10.1073/pnas.0404932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Menzella HG, Carney JR, Santi DV. Chem Biol. 2007;14:143–151. doi: 10.1016/j.chembiol.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 180.Chen H, O'Connor S, Cane DE, Walsh CT. Chem Biol. 2001;8:899–912. doi: 10.1016/s1074-5521(01)00064-3. [DOI] [PubMed] [Google Scholar]
- 181.O'Connor SE, Chen H, Walsh CT. Biochemistry. 2002;41:5685–5694. doi: 10.1021/bi020006w. [DOI] [PubMed] [Google Scholar]
- 182.Edwards DJ, Gerwick WH. Journal of the American Chemical Society. 2004;126:11432–11433. doi: 10.1021/ja047876g. [DOI] [PubMed] [Google Scholar]
- 183.Calderone CT. Nat Prod Rep. 2008;25:845–853. doi: 10.1039/b807243d. [DOI] [PubMed] [Google Scholar]
- 184.Calderone CT, Kowtoniuk WE, Kelleher NL, Walsh CT, Dorrestein PC. Proc Natl Acad Sci U S A. 2006;103:8977–8982. doi: 10.1073/pnas.0603148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Calderone CT, Iwig DF, Dorrestein PC, Kelleher NL, Walsh CT. Chem Biol. 2007;14:835–846. doi: 10.1016/j.chembiol.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kusebauch B, Busch B, Scherlach K, Roth M, Hertweck C. Angew Chem Int Ed Engl. 2009;48:5001–5004. doi: 10.1002/anie.200900277. [DOI] [PubMed] [Google Scholar]
- 187.Gu L, Wang B, Kulkarni A, Geders TW, Grindberg RV, Gerwick L, Hakansson K, Wipf P, Smith JL, Gerwick WH, Sherman DH. Nature. 2009;459:731–735. doi: 10.1038/nature07870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Weitnauer G, Muhlenweg A, Trefzer A, Hoffmeister D, Sussmuth RD, Jung G, Welzel K, Vente A, Girreser U, Bechthold A. Chem Biol. 2001;8:569–581. doi: 10.1016/s1074-5521(01)00040-0. [DOI] [PubMed] [Google Scholar]
- 189.Hosted TJ, Wang TX, Alexander DC, Horan AC. J Ind Microbiol Biotechnol. 2001;27:386–392. doi: 10.1038/sj.jim.7000189. [DOI] [PubMed] [Google Scholar]
- 190.Liu W, Christenson SD, Standage S, Shen B. Science (New York, N.Y. 2002;297:1170–1173. doi: 10.1126/science.1072110. [DOI] [PubMed] [Google Scholar]
- 191.Bode HB, Wenzel SC, Irschik H, Hofle G, Muller R. Angew Chem Int Ed Engl. 2004;43:4163–4167. doi: 10.1002/anie.200454240. [DOI] [PubMed] [Google Scholar]
- 192.Shi R, Lamb SS, Zakeri B, Proteau A, Cui Q, Sulea T, Matte A, Wright GD, Cygler M. Chem Biol. 2009;16:401–410. doi: 10.1016/j.chembiol.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 193.Lamb SS, Patel T, Koteva KP, Wright GD. Chem Biol. 2006;13:171–181. doi: 10.1016/j.chembiol.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 194.Losey HC, Peczuh MW, Chen Z, Eggert US, Dong SD, Pelczer I, Kahne D, Walsh CT. Biochemistry. 2001;40:4745–4755. doi: 10.1021/bi010050w. [DOI] [PubMed] [Google Scholar]
- 195.Kruger RG, Lu W, Oberthur M, Tao J, Kahne D, Walsh CT. Chem Bio1. 2005;12:131–140. doi: 10.1016/j.chembiol.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 196.Salas JA, Mendez C. Trends Microbiol. 2007;15:219–232. doi: 10.1016/j.tim.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 197.Blanchard S, Thorson JS. Curr Opin Chem Biol. 2006;10:263–271. doi: 10.1016/j.cbpa.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 198.Luzhetskyy A, Bechthold A. Appl Microbiol Biotechnol. 2008;80:945–952. doi: 10.1007/s00253-008-1672-2. [DOI] [PubMed] [Google Scholar]
- 199.Weymouth-Wilson AC. Nat Prod Rep. 1997;14:99–110. doi: 10.1039/np9971400099. [DOI] [PubMed] [Google Scholar]
- 200.Tu D, Blaha G, Moore PB, Steitz TA. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 201.Mendez C, Salas JA. Trends Biotechnol. 2001;19:449–456. doi: 10.1016/s0167-7799(01)01765-6. [DOI] [PubMed] [Google Scholar]
- 202.Griffith BR, Langenhan JM, Thorson JS. Curr Opin Biotechnol. 2005;16:622–630. doi: 10.1016/j.copbio.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 203.Zhang C, Griffith BR, Fu Q, Albermann C, Fu X, Lee IK, Li L, Thorson JS. Science. 2006;313:1291–1294. doi: 10.1126/science.1130028. [DOI] [PubMed] [Google Scholar]
- 204.Salas AP, Zhu L, Sanchez C, Brana AF, Rohr J, Mendez C, Salas JA. Mol Microbiol. 2005;58:17–27. doi: 10.1111/j.1365-2958.2005.04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zhao P, et al. Chem Biol. 2008;15:863–874. doi: 10.1016/j.chembiol.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 206.Fischbach MA, Lin H, Liu DR, Walsh CT. Proc Natl Acad Sci U S A. 2005;102:571–576. doi: 10.1073/pnas.0408463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mittler M, Bechthold A, Schulz GE. J Mol Biol. 2007;372:67–76. doi: 10.1016/j.jmb.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 208.Hoffmeister D, Drager G, Ichinose K, Rohr J, Bechthold A. J Am Chem Soc. 2003;125:4678–4679. doi: 10.1021/ja029645k. [DOI] [PubMed] [Google Scholar]
- 209.Hoffmeister D, Wilkinson B, Foster G, Sidebottom PJ, Ichinose K, Bechthold A. Chem Biol. 2002;9:287–295. doi: 10.1016/s1074-5521(02)00114-x. [DOI] [PubMed] [Google Scholar]
- 210.Decker H, Haag S. J Bacteriol. 1995;177:6126–6136. doi: 10.1128/jb.177.21.6126-6136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Bililign T, Hynn CG, Williams JS, Czisny AM, Thorson JS. Chem Biol. 2004;11:959–969. doi: 10.1016/j.chembiol.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 212.Trefzer A, Blanco G, Remsing L, Kunzel E, Rix U, Lipata F, Brana AF, Mendez C, Rohr J, Bechthold A, Salas JA. J Am Chem Soc. 2002;124:6056–6062. doi: 10.1021/ja017385l. [DOI] [PubMed] [Google Scholar]
- 213.Zhu L, Luzhetskyy A, Luzhetska M, Mattingly C, Adams V, Bechthold A, Rohr J. Chembiochem. 2007;8:83–88. doi: 10.1002/cbic.200600360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Salas JA, Mendez C. Curr Opin Chem Biol. 2009;13:152–160. doi: 10.1016/j.cbpa.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 215.Perez M, Lombo F, Zhu L, Gibson M, Brana AF, Rohr J, Salas JA, Mendez C. Chem Commun (Comb) 2005:1604–1606. doi: 10.1039/b417815g. [DOI] [PubMed] [Google Scholar]
- 216.Velkov T, Lawen A. J Biol Chem. 2003;278:1137–1148. doi: 10.1074/jbc.M209719200. [DOI] [PubMed] [Google Scholar]
- 217.Lawen A, Zocher R. J Biol Chem. 1990;265:11355–11360. [PubMed] [Google Scholar]
- 218.Ortiz de Montellano PR. Cytochrome P450: structure, mechanism, and biochemistry. 3rd ed. New York: Kluwer Academic/Plenum Publishers; 2005. [Google Scholar]
- 219.Costas M, Mehn MP, Jensen MP, Que L., Jr Chem Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 220.Fraaije MW, Mattevi A. Trends Biochem Sci. 2000;25:126–132. doi: 10.1016/s0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
- 221.Andersen JF, Tatsuta K, Gunji H, Ishiyama T, Hutchinson CR. Biochemistry. 1993;32:1905–1913. doi: 10.1021/bi00059a004. [DOI] [PubMed] [Google Scholar]
- 222.Lambalot RH, Cane DE, Aparicio JJ, Katz L. Biochemistry. 1995;34:1858–1866. doi: 10.1021/bi00006a006. [DOI] [PubMed] [Google Scholar]
- 223.Cupp-Vickery J, Anderson R, Hatziris Z. Proc Natl Acad Sci U S A. 2000;97:3050–3055. doi: 10.1073/pnas.050406897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Cupp-Vickery JR, Poulos TL. Nat Struct Biol. 1995;2:144–153. doi: 10.1038/nsb0295-144. [DOI] [PubMed] [Google Scholar]
- 225.Nagano S, Li H, Shimizu H, Nishida C, Ogura H, Ortiz de Montellano PR, Poulos TL. J Biol Chem. 2003;278:44886–44893. doi: 10.1074/jbc.M308115200. [DOI] [PubMed] [Google Scholar]
- 226.Woithe K, Geib N, Meyer 0, Wortz T, Zerbe K, Robinson JA. Org Biomol Chem. 2008;6:2861–2867. doi: 10.1039/b805956j. [DOI] [PubMed] [Google Scholar]
- 227.Zerbe K, Pylypenko O, Vitali F, Zhang W, Rouset S, Heck M, Vrijbloed JW, Bischoff D, Bister B, Sussmuth RD, Pelzer S, Wohlleben W, Robinson JA, Schlichting I. J Biol Chem. 2002;277:47476–47485. doi: 10.1074/jbc.M206342200. [DOI] [PubMed] [Google Scholar]
- 228.Pylypenko O, Vitali F, Zerbe K, Robinson JA, Schlichting I. J Biol Chem. 2003;278:46727–46733. doi: 10.1074/jbc.M306486200. [DOI] [PubMed] [Google Scholar]
- 229.Stegmann E, Pelzer S, Bischoff D, Puk O, Stockert S, Butz D, Zerbe K, Robinson J, Sussmuth RD, Wohlleben W. J Biotechnol. 2006;124:640–653. doi: 10.1016/j.jbiotec.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 230.Roach PL, Clifton IJ, Hensgens CM, Shibata N, Schofield CJ, Hajdu J, Baldwin JE. Nature. 1997;387:827–830. doi: 10.1038/42990. [DOI] [PubMed] [Google Scholar]
- 231.Valegard K, van Scheltinga AC, Lloyd MD, Hara T, Ramaswamy S, Perrakis A, Thompson A, Lee HJ, Baldwin JE, Schofield CJ, Hajdu J, Andersson I. Nature. 1998;394:805–809. doi: 10.1038/29575. [DOI] [PubMed] [Google Scholar]
- 232.Kershaw NJ, Caines ME, Sleeman MC, Schofield CJ. Chem Commun (Camb) 2005:4251–4263. doi: 10.1039/b505964j. [DOI] [PubMed] [Google Scholar]
- 233.Townsend CA. Curr Opin Chem Biol. 2002;6:583–589. doi: 10.1016/s1367-5931(02)00392-7. [DOI] [PubMed] [Google Scholar]
- 234.Fraaije MW, Kamerbeek NM, van Berkel WJ, Janssen DB. FEES Lett. 2002;518:43–47. doi: 10.1016/s0014-5793(02)02623-6. [DOI] [PubMed] [Google Scholar]
- 235.Ehrlich KC, Montalbano B, Boue SM, Bhatnagar D. Appl Environ Microbiol. 2005;71:8963–8965. doi: 10.1128/AEM.71.12.8963-8965.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Watanabe CM, Townsend CA. J Org Chem. 1996;61:1990–1993. [Google Scholar]
- 237.Gibson M, Nur-e-alam M, Lipata F, Oliveira MA, Rohr J. Journal of the American Chemical Society. 2005;127:17594–17595. doi: 10.1021/ja055750t. [DOI] [PubMed] [Google Scholar]
- 238.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Proc Natl Acad Sci U S A. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Nat Chem Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 240.Bagley MC, Dale JW, Merritt EA, Xiong X. Chemical reviews. 2005;105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 241.Liao R, Duan L, Lei C, Pan H, Ding Y, Zhang Q, Chen D, Shen B, Yu Y, Liu W. Chem Biol. 2009;16:141–147. doi: 10.1016/j.chembiol.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wieland Brown LC, Acker MG, Clardy J, Walsh CT, Fischbach MA. Proc Natl Acad Sci U S A. 2009;106:2549–2553. doi: 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Morris RP, Leeds JA, Naegeli HU, Oberer L, Memmert K, Weber E, LaMarche MJ, Parker CN, Burrer N, Esterow S, Hein AE, Schmitt EK, Krastel P. J Am Chem Soc. 2009;131:5946–5955. doi: 10.1021/ja900488a. [DOI] [PubMed] [Google Scholar]
- 244.Kelly WL, Pan L, Li C. J Am Chem Soc. 2009;131:4327–4334. doi: 10.1021/ja807890a. [DOI] [PubMed] [Google Scholar]
- 245.Yu Y, Duan L, Zhang Q, Liao R, Ding Y, Pan H, Wendt-Pienkowski E, Tang G, Shen B, Liu W. ACS Chem Biol. 2009;4:855–864. doi: 10.1021/cb900133x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CM, Fucini P. Mol Cell. 2008;30:26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 247.Nicolaou KC, Safina BS, Zak M, Lee SH, Nevalainen M, Bella M, Estrada AA, Funke C, Zecri FJ, Bulat S. J Am Chem Soc. 2005;127:11159–11175. doi: 10.1021/ja0529337. [DOI] [PubMed] [Google Scholar]
- 248.Moody CJ, Hughes RA, Thompson SP, Alcaraz L. Chem Commun (Camb) 2002:1760–1761. doi: 10.1039/b204868j. [DOI] [PubMed] [Google Scholar]
- 249.Sutherland A, Auclair K, Vederas JC. Curr Opin Drug Discov Devel. 2001;4:229–236. [PubMed] [Google Scholar]
- 250.Zhang H, White-Phillip JA, Melancon CE, 3rd, Kwon HJ, Yu WL, Liu HW. J Am Chem Soc. 2007;129:14670–14823. doi: 10.1021/ja0744854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Li C, Roege KE, Kelly WL. Chembiochem. 2009;10:1064–1072. doi: 10.1002/cbic.200800822. [DOI] [PubMed] [Google Scholar]
- 252.Zakeri B, Wright GD. Biochem Cell Biol. 2008;86:124–136. doi: 10.1139/O08-002. [DOI] [PubMed] [Google Scholar]
- 253.Zhang W, Watanabe K, Wang CC, Tang Y. J Biol Chem. 2007;282:25717–25725. doi: 10.1074/jbc.M703437200. [DOI] [PubMed] [Google Scholar]
- 254.Cheng Q, Xiang L, Izumikawa M, Meluzzi D, Moore BS. Nature chemical biology. 2007;3:557–558. doi: 10.1038/nchembio.2007.22. [DOI] [PubMed] [Google Scholar]
- 255.Xiang L, Kalaitzis JA, Moore BS. Proc Natl Acad Sci U S A. 2004;101:15609–15614. doi: 10.1073/pnas.0405508101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Van Lanen SG, Oh TJ, Liu W, Wendt-Pienkowski E, Shen B. J Am Chem Soc. 2007;129:13082–13094. doi: 10.1021/ja073275o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Liu W, Nonaka K, Nie L, Zhang J, Christenson SD, Bae J, Van Lanen SG, Zazopoulos E, Farnet CM, Yang CF, Shen B. Chem Dial. 2005;12:293–302. doi: 10.1016/j.chembiol.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 258.Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang K, Fonstein L, Czisny A, Whitwam RE, Farnet CM, Thorson JS. Science. 2002;297:1173–1176. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- 259.Belecki K, Crawford JM, Townsend CA. Journal of the American Chemical Society. 2009;131:12564–12566. doi: 10.1021/ja904391r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Kong R, Goh LP, Liew CW, Ho QS, Murugan E, Li B, Tang K, Liang ZX. Journal of the American Chemical Society. 2008;130:8142–8143. doi: 10.1021/ja8019643. [DOI] [PubMed] [Google Scholar]
- 261.Zhang J, Van Lanen SG, Ju J, Liu W, Dorrestein PC, Li W, Kelleher NL, Shen B. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1460–1465. doi: 10.1073/pnas.0711625105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Gallimore AR, Stark CB, Bhatt A, Harvey BM, Demydchuk Y, Bolanos-Garcia V, Fowler DJ, Staunton J, Leadlay PF, Spencer JB. Chem Biol. 2006;13:453–460. doi: 10.1016/j.chembiol.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 263.Bhatt A, Stark CB, Harvey BM, Gallimore AR, Demydchuk YA, Spencer JB, Staunton J, Leadlay PF. Angewandte Chemie (International ed. 2005;44:7075–7078. doi: 10.1002/anie.200501757. [DOI] [PubMed] [Google Scholar]
- 264.Migita A, Watanabe M, Hirose Y, Watanabe K, Tokiwano T, Kinashi H, Oikawa H. Biosci Biotechnol Biochem. 2009;73:169–176. doi: 10.1271/bbb.80631. [DOI] [PubMed] [Google Scholar]
- 265.Demydchuk Y, Sun Y, Hong H, Staunton J, Spencer JB, Leadlay PF. Chembiochem. 2008;9:1136–1145. doi: 10.1002/cbic.200700715. [DOI] [PubMed] [Google Scholar]
- 266.Harvey BM, Mironenko T, Sun Y, Hong H, Deng Z, Leadlay PF, Weissman KJ, Haydock SF. Chem Biol. 2007;14:703–714. doi: 10.1016/j.chembiol.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 267.Izumikawa M, Murata M, Tachibana K, Ebizuka Y, Fujii I. Bioorg Med Chem. 2003;11:3401–3405. doi: 10.1016/s0968-0896(03)00337-7. [DOI] [PubMed] [Google Scholar]
- 268.Sun Y, Zhou X, Dong H, Tu G, Wang M, Wang B, Deng Z. Chem Biol. 2003;10:431–441. doi: 10.1016/s1074-5521(03)00092-9. [DOI] [PubMed] [Google Scholar]
- 269.Leadlay PF, et al. J Ind Microbial Biotechnol. 2001;27:360–367. doi: 10.1038/sj.jim.7000204. [DOI] [PubMed] [Google Scholar]
- 270.Gallimore AR. Nat Prod Rep. 2009;26:266–280. doi: 10.1039/b807902c. [DOI] [PubMed] [Google Scholar]
- 271.Oikawa H, Tokiwano T. Nat Prod Rep. 2004;21:321–352. doi: 10.1039/b305068h. [DOI] [PubMed] [Google Scholar]
- 272.Kelly WL. Org Biomol Chem. 2008;6:4483–4493. doi: 10.1039/b814552k. [DOI] [PubMed] [Google Scholar]
- 273.Auclair K, Sutherland A, Kennedy J, Witter DJ, Van Den Heever JP, Hutchinson CR, Vederas JC. Journal of the American Chemical Society. 2000;122:11519–11520. [Google Scholar]
- 274.Sorensen EJ. Bioorg Med Chem. 2003;11:3225–3228. doi: 10.1016/s0968-0896(03)00185-8. [DOI] [PubMed] [Google Scholar]
- 275.Yu F, Zaleta-Rivera K, Zhu X, Huffman J, Millet JC, Harris SD, Yuen G, Li XC, Du L. Antimicrob Agents Chemother. 2007;51:64–72. doi: 10.1128/AAC.00931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Strieter ER, Koglin A, Aron ZD, Walsh CT. J Am Chem Soc. 2009;131:2113–2115. doi: 10.1021/ja8077945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Mejean A, Mann S, Maldiney T, Vassiliadis G, Lequin O, Ploux O. J Am Chem Soc. 2009;131:7512–7513. doi: 10.1021/ja9024353. [DOI] [PubMed] [Google Scholar]
- 278.Kim HJ, Pongdee R, Wu Q, Hong L, Liu HW. J Am Chem Soc. 2007;129:14582–14584. doi: 10.1021/ja076580i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Cragg GM, Newman DJ. J Ethnopharmacol. 2005;100:72–79. doi: 10.1016/j.jep.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 280.Harvey A. Drug Discov Today. 2000;5:294–300. doi: 10.1016/s1359-6446(00)01511-7. [DOI] [PubMed] [Google Scholar]
- 281.Rates SM. Toxicon. 2001;39:603–613. doi: 10.1016/s0041-0101(00)00154-9. [DOI] [PubMed] [Google Scholar]
- 282.Walsh C. Antibiotics: Actions, Origins, Resistance. Washington, DC: ASM Press; 2003. [Google Scholar]
- 283.Fischbach MA, Walsh CT. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Singh SB, Phillips JW, Wang J. Curr Opin Drug Discov Devel. 2007;10:160–166. [PubMed] [Google Scholar]
- 285.Donadio S, Monciardini P, Sosio M. Nat Prod Rep. 2007;24:1073–1109. doi: 10.1039/b514050c. [DOI] [PubMed] [Google Scholar]
- 286.Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Penn K, Jenkins C, Nett M, Udwary DW, Gontang EA, McGlinchey RP, Foster B, Lapidus A, Podell S, Allen EE, Moore BS, Jensen PR. ISME J. 2009;3:1193–1203. doi: 10.1038/ismej.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Angewandte Chemie (International ed. 2003;42:355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- 289.Piel J. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Stachelhaus T, Mootz HD, Marahiel MA. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 291.Challis GL, Ravel J, Townsend CA. Chem Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 292.Bassler BL, Losick R. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 293.Dudler R, Eberl L. Current opinion in biotechnology. 2006;17:268–273. doi: 10.1016/j.copbio.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 294.Long SR. The Plant cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Shank EA, Kolter R. Current opinion in microbiology. 2009;12:205–214. doi: 10.1016/j.mib.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Krug D, Zurek G, Revermann O, Vos M, Velicer GJ, Muller R. Appl Environ Microbiol. 2008;74:3058–3068. doi: 10.1128/AEM.02863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Sattely ES, Fischbach MA, Walsh CT. Nat Prod Rep. 2008;25:757–793. doi: 10.1039/b801747f. [DOI] [PubMed] [Google Scholar]
- 298.Ferber D. Science. 2004;303:158–161. doi: 10.1126/science.303.5655.158. [DOI] [PubMed] [Google Scholar]
- 299.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, Shiba Y, Sarpong R, Keasling JD. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 300.Tsuruta H, Paddon CJ, Eng D, Lenihan JR, Horning T, Anthony LC, Regentin R, Keasling JD, Renninger NS, Newman JD. PLoS One. 2009;4:e4489. doi: 10.1371/journal.pone.0004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C. Science (New York, N.Y. 2001;291:1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- 302.Watanabe K, Hotta K, Praseuth AP, Koketsu K, Migita A, Boddy CN, Wang CC, Oguri H, Oikawa H. Nature chemical biology. 2006;2:423–428. doi: 10.1038/nchembio803. [DOI] [PubMed] [Google Scholar]
- 303.Oksman-Caldentey KM, Inze D, Oresic M. Proc Natl Acad Sci U S A. 2004;101:9949–9950. doi: 10.1073/pnas.0403636101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Yu JH, Keller N. Annu Rev Phytopathol. 2005;43:437–458. doi: 10.1146/annurev.phyto.43.040204.140214. [DOI] [PubMed] [Google Scholar]
- 305.Bibb MJ. Curr Opin Microbiol. 2005;8:208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 306.Freiberg C, Brotz-Oesterhelt H, Labischinski H. Curr Opin Microbial. 2004;7:451–459. doi: 10.1016/j.mib.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 307.Wilson MK, Abergel RJ, Raymond KN, Arceneaux JE, Byers BR. Biochem Biophys Res Commun. 2006;348:320–325. doi: 10.1016/j.bbrc.2006.07.055. [DOI] [PubMed] [Google Scholar]
- 308.Kim CG, et al. Arch Microbiol. 2008;189:463–473. doi: 10.1007/s00203-007-0337-3. [DOI] [PubMed] [Google Scholar]
- 309.Watanabe K, Hotta K, Nakaya M, Praseuth AP, Wang CC, Inada D, Takahashi K, Fukushi E, Oguri H, Oikawa H. J Am Chem Soc. 2009;131:9347–9353. doi: 10.1021/ja902261a. [DOI] [PMC free article] [PubMed] [Google Scholar]