Abstract
Objectives
The goal of this study was to identify novel regulatory mechanisms controlling the growth and proliferation of cholesteatoma. Specifically, the potential role of microRNAs, regulators of protein translation, was studied in cholesteatoma.
Study Design
This study represents a molecular biological investigation characterizing and comparing microRNA and protein expression in cholesteatoma and normal post-auricular skin.
Methods
Cholesteatoma and normal skin were taken from patients at the time of surgery. Tissue was processed for RNA and protein extraction. Real-time RT-PCR was used to assess levels of human microRNAs, RT-PCR was used to confirm the presence of upstream regulators, and western blot analyses were used to assess levels of downstream target proteins.
Results
Among the microRNAs investigated, human microRNA-21 (hsa-mir-21) showed a 4.4-fold higher expression in cholesteatoma as compared to normal skin (p=0.0011). The downstream targets of hsa-mir-21, PTEN and PDCD4, were found to be greatly reduced in 3 of 4 cholesteatoma samples. Proposed upstream regulators of hsa-mir-21 expression (CD14, IL-6R, gp130 and STAT3) were present in all cholesteatoma tissues.
Conclusions
MicroRNAs represent powerful regulators of protein translation and their dysregulation has been implicated in many neoplastic diseases. This study specifically identified up-regulation of hsa-mir-21 concurrent with down-regulation of potent tumor suppressor proteins, PTEN and PDCD4. These proteins control aspects of apoptosis, proliferation, invasion and migration. The results of this study were used to develop a model for cholesteatoma proliferation through microRNA dysregulation. This model can serve as a template for further study into potential RNA-based therapies for the treatment of cholesteatoma.
Keywords: cholesteatoma, microRNA, hsa-mir-21, PTEN, STAT3, PDCD4
INTRODUCTION
Cholesteatomas are benign epidermally-derived temporal bone lesions that are locally destructive and frequently recurrent. Cholesteatomas cause temporal bone destruction due to mechanical pressure, enzymatically mediated bone resorption, and promotion of acute and chronic infections. Temporal bone erosion may involve the otic capsule and cause sensorineural hearing loss and dizziness. Erosion into the ossicular chain leading to conductive hearing loss is a frequent finding. Cholesteatoma may involve the facial nerve causing paralysis. Erosion of the tegmen may lead to seizures, encephaloceles, cerebrospinal fluid leaks and meningitis. Chronic infection with cholesteatoma is common and may lead to mastoiditis, bacterial labyrinthitis, sigmoid sinus thrombophlebitis, and brain abscess (1,2).
There are no medical therapies for cholesteatoma and current treatment is surgical resection. Recurrences are common and many individuals with cholesteatoma undergo multiple operations (3,4). Recurrences, and the need for multiple operations, result in progressively worse hearing deficits and an increased risk of facial nerve paralysis and other complications. Ultimate eradication of disease often requires performance of a radical mastoidectomy which removes the tympanic membrane, ossicles, mastoid and ear canal wall. This results in, at best, a maximal conductive hearing loss, an abnormal external ear canal, and a chronic need for clinic visits to clean the mastoid cavity. For the uninsured, indigent, and those without access to regular medical care, cholesteatoma and its treatment can thus lead to substantial morbidity and even mortality.
The molecular events governing cholesteatoma formation are not well established (5,6). It is clear, however, that when squamous epithelium lining the external ear canal or the tympanic membrane becomes trapped in the external or middle ear spaces a process of hyper-proliferation ensues (7). Gene expression studies have shown an up-regulation of many growth factors and signaling molecules (8–13). Similarly, protein expression studies have shown significant increases in molecules related to growth, proliferation, anti-apoptosis and bone resorption (14–20). Despite these numerous studies showing activation of similar pathways there have been no discrete pharmacological targets identified for cholesteatoma management.
We propose that master regulators of these pathways, microRNAs, may provide a better molecular target for medical management of cholesteatoma. MicroRNAs are evolutionarily conserved, 22–24 nucleotide long, non-coding RNA molecules that inhibit protein translation. They play significant roles in regulating cellular pathways including differentiation, development and apoptosis (21,22). They have also been strongly implicated in oncogenesis (23). This manuscript provides the first evidence for the potential role of microRNAs in regulating growth and proliferation in cholesteatoma. Specifically, we identified significant up-regulation of human microRNA-21 (hsa-mir-21) in cholesteatoma as compared to normal skin. Further, we investigated the expression of upstream regulators and levels of downstream targets of hsa-mir-21 to develop a model of cholesteatoma growth and proliferation. This model may identify targets for RNA and protein based therapeutic interventions for the non-surgical or adjunctive treatment of cholesteatoma.
METHODS
Tissue Collection
This study was approved by the Institutional Review Board of the Medical College of Wisconsin and Froedtert Hospital. Seven subjects undergoing cholesteatoma surgery consented to providing samples of their lesion as well as normal post-auricular skin for laboratory analysis. All subjects were over 18 years of age, had secondarily acquired cholesteatoma, and presented with chronic otitis media and cholesteatoma. In cases of revision surgery the prior scar was resected and uninvolved post-auricular skin was obtained. Cholesteatoma and normal skin samples were placed into RNase-free microcentrifuge tubes on dry ice and immediately transported to the laboratory.
RNA/protein Extraction
Total RNA and protein were extracted from cholesteatoma and control skin using the TRIZOL reagent (Invitrogen, Carlsbad, CA) per manufacturer’s instructions. RNA was analyzed for integrity, purity and concentration by gel electrophoresis and spectrophotometry. Protein was isolated from the aqueous phase of the TRIZOL extraction and reconstituted in 1% SDS plus 8M urea. Protein concentration was determined by BCA assay and stored at −80°C until analysis.
Real-Time RT-PCR
Real-time RT-PCR was used to compare microRNA levels between cholesteatoma and normal skin. Ten nanograms of total RNA was reverse transcribed into cDNA using gene specific reverse transcription primers from the TaqMan® MicroRNA Assays (ABI, Foster City, CA) and the TaqMan® MicroRNA Reverse Transcription Kit (ABI, Foster City, CA) according to manufacturer’s instructions. Real-time PCR was performed on the iCycler iQ Multicolor Real-Time Detection System (Bio Rad Laboratories, Hercules, CA). The cDNA was amplified using TaqMan® Universal Master Mix (ABI, Foster City, CA) and PCR primers from the TaqMan® MicroRNA Assays. The thermal cycling conditions were as follows: 95°C hold for 10 minutes, followed by 50 cycles of a 15 second denaturing step at 95°C and a 60 second annealing and elongation step at 60°C. Fluorescent signal was detected by the iCycler during the elongation step and the Ct values were determined by the software. Small nuclear RNA RNU6B was used as a reference RNA. All reactions were run in triplicate and negative controls (no template) were run. For hsa-mir-21 levels each experiment was performed three times and standard errors calculated. Fold change in expression was normalized to RNU6B levels and calculated in standard fashion by 2−(avgΔΔCT). Statistical analyses of expression levels between cholesteatoma and normal skin were performed by two-tailed Student’s t-test.
Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
Confirmation of the presence of up-stream regulators of hsa-mir-21 in cholesteatoma was performed by RT-PCR. To generate cDNA for RT-PCR, one microgram aliquots of total RNA were treated with 1U DNase, Amp Grade (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. RNA was then divided into RT positive and RT negative (control) samples and reverse transcribed using random primers (Invitrogen, Carlsbad, CA) and SuperScript™ III Reverse Transcriptase (Invitrogen, Carlsbad, CA). Platinum® Blue PCR SuperMix (Invitrogen, Carlsbad, CA) was used to perform PCR on the resulting cDNA. PCR primers were designed to amplify CD14 and the membrane bound variant of both IL-6R and gp130.
CD14: 5′-TCACAAGTGTGAAGCCTGGAAG-3′ and 5′-ATGGTGCCGGTTATCTTTAGGTC-3′;
IL-6R: 5′-CTTCAGTACCACTGCCCACATTC-3′ and 5′-GAGCCCGTTTGTGTTTTATCATC-3′;
gp130: 5′-CCATCCCATACTCAAGGCTACAG-3′ and 5′-GGCTTCAATTTCTCCTTGAGCAAAC-3′.
Primers used to amplify STAT3α / STAT3β have been previously described (24).
STAT3α / STAT3β: 5′-ACCAATATCCTGGTGTCTCC-3′ and 5′-TTATTTCCAAACTGCATCAATGA-3′.
All RT-PCR products were run on a 2% agarose gel, purified and confirmed by sequencing.
Western Blot
Levels of down-stream targets of hsa-mir-21 were determined by western blot analysis. Protein from cholesteatoma and normal skin was extracted using the TRIZOL method as noted above. Samples (20ng) were resolved using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on a 10% Tris-HCl gel (Bio-Rad, Hercules, CA) and transferred to a polyvinylidene fluoride (PVDF) membrane. The membrane was probed with specific antibodies to control and target proteins including mouse anti-actin (control), rabbit anti-PTEN (Abcam Inc., Cambridge, MA: AB23694), and rabbit anti-PDCD4 (Abcam Inc., Cambridge, MA: AB51495-100). HRP-conjugated secondary antibodies (goat anti-rabbit HRP or goat anti-mouse HRP) and Luminol reagent (Santa Cruz Biotechnology (Santa Cruz, CA)) were used to detect chemiluminescence. Blots were subsequently exposed to x-ray film and developed. Bands were digitally scanned and analyzed with Adobe Photoshop. Specifically, average pixel densities and band sizes in actin control bands were used to normalize band density and size of the target proteins. Target bands from cholesteatoma and skin were then directly compared. Five measurements were taken for each sample and the results averaged. One western blot was performed for each of 4 specimens. Four specimens were used for each protein due to limitations in sample quantity.
RESULTS
Differential Expression of MicroRNAs in Cholesteatoma
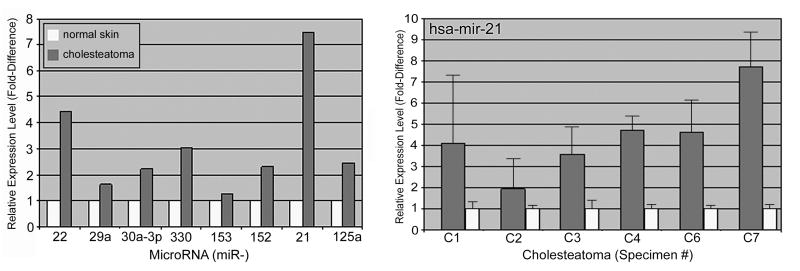
Expression levels of a series of microRNAs potentially related to tumorigenesis were quantitatively compared between cholesteatoma and normal skin using real time RT-PCR. These microRNAs included hsa-mir-21, -22, -29a, -30a-3p, -330, -153, -152 and -125a (Table 1). Several microRNAs were upregulated in cholesteatoma as compared to normal skin (Figure 1). None of the investigated microRNAs showed lower levels in cholesteatoma. The highest level of differential expression was with hsa-mir-21 which was 7.46 fold greater in cholesteatoma as compared to normal skin.
Table 1.
Raw data from real-time RT-PCR experiment examining expression levels of various microRNAs in cholesteatoma and control skin. These microRNAs were chosen as they are associated with tumor growth. CT=cycle threshold.
| MicroRNA | Cholesteatoma | Skin | U6-Cholesteatoma CT Value | U6 - Skin CT Value | Cholesteatoma to Skin | |||
|---|---|---|---|---|---|---|---|---|
| CT Value | StdDev | CT Value | StdDev | ΔΔCT | Fold Difference | |||
| 22 | 26.10 | 0.14 | 26.70 | 0.27 | 28.50 | 26.95 | 2.15 | 4.44 |
| 29a | 22.45 | 0.06 | 21.60 | 0.08 | 28.50 | 26.95 | 0.70 | 1.62 |
| 30a-3p | 27.10 | 0.00 | 26.70 | 0.00 | 28.50 | 26.95 | 1.15 | 2.22 |
| 330 | 33.85 | 0.19 | 33.90 | 0.54 | 28.50 | 26.95 | 1.60 | 3.03 |
| 153 | 35.90 | 0.71 | 34.68 | 0.26 | 28.50 | 26.95 | 0.33 | 1.26 |
| 152 | 25.68 | 0.10 | 25.32 | 0.19 | 28.50 | 26.95 | 1.19 | 2.28 |
| 21 | 19.35 | 0.06 | 20.70 | 0.00 | 28.50 | 26.95 | 2.90 | 7.46 |
| 125a | 23.63 | 0.05 | 23.38 | 0.05 | 28.50 | 26.95 | 1.30 | 2.46 |
Figure 1.
Real-time RT-PCR demonstrated increased expression of several microRNAs in a single cholesteatoma sample (C1) when compared to matched post-auricular skin (S1) (left). Subsequent real-time RT-PCR specifically for hsa-mir-21 demonstrated between two- and eightfold increased expression in cholesteatoma: avg: 4.4, p=0.0011 (right).
Hsa-mir-21 was subsequently investigated in six cholesteatomas and matched normal skin samples (Figure 1). These experiments were run four times for each sample pair. Hsa-mir-21 was up-regulated in all cholesteatomas as compared to normal skin. Expression was at least 2-fold higher in all samples and as much as 8-fold higher in one sample. The average fold increase in expression in cholesteatoma was 4.4 (p=0.0011).
Presence of Up-Stream Regulators of Hsa-mir-21 in Cholesteatoma
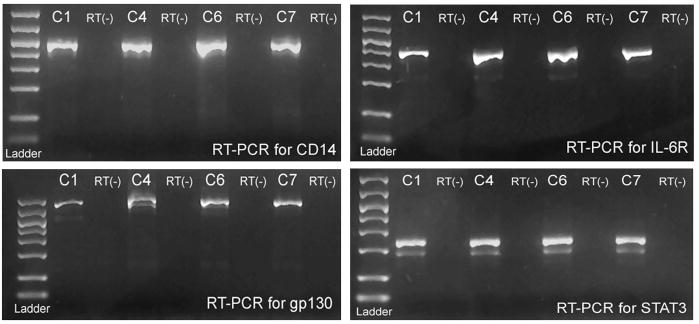
In light of prior studies, we sought to demonstrate the presence of key elements of signaling pathways that lead to the expression of hsa-mir-21. These include cell membrane receptors and transcription factors. Specifically, RT-PCR was performed on cholesteatoma to show the presence of mRNA for the lipopolysaccharide receptor CD14, the IL-6 receptor components IL-6R and gp130, and the transcription factor STAT3. These components were identified in all samples studied (Figure 2). Sequencing of the amplicons confirmed their identities. RT(−) controls demonstrated no evidence of DNA contamination.
Figure 2.
RT-PCR demonstrated mRNA expression of gp130, IL-6R, STAT3, and CD14 in cholesteatoma tissue. These proteins are integral components in pathways that may promote hsa-mir-21 expression in response to inflammation and infection. STAT3 shows a lighter second band which reflects the ϐ-isoform of STAT3.
Down-Regulation of Down-Stream Targets of Hsa-mir-21
Hsa-mir-21 has been well-studied due to its role in tumorigenesis. Among the downstream targets of hsa-mir-21 are PTEN and PDCD4. PTEN is considered the most highly mutated tumor-suppressor gene found in neoplasms since the identification of p53 (25). PTEN suppresses several pathways involved in cell proliferation, anti-apoptosis, and cell migration (25,26). Reduced PTEN expression has been identified in approximately half of all tumors investigated (26). Our hypothesis is that the increased levels of hsa-mir-21 would inhibit PTEN translation and similarly reduce levels of this protein in cholesteatoma.
While PTEN is the most prominent of hsa-mir-21 targets, other transcript targets have also been associated with tumorigenesis. Among these is PDCD4: programmed cell death 4. In a mouse cancer model PDCD4 was shown to suppress benign and malignant skin tumor formation and progression (27). PDCD4 levels have been clearly shown to be down-regulated by hsa-mir-21 (28). PDCD4 down-regulation would release restraints on cell growth and proliferation in cholesteatoma. Thus, this experiment investigated whether the increased levels of hsa-mir-21 are associated with decreased levels of the tumor suppressor PDCD4.
Western blot was used to compare levels of PTEN and PDCD4 protein in cholesteatoma as compared to normal skin (Figure 3). Expression of PTEN was reduced in 3 of 4 cholesteatoma samples. In two of these experiments there was an absence of detectable PTEN in cholesteatoma. Similarly, cholesteatoma expression of PDCD4 was also reduced in 3 of 4 patients when compared to matched normal skin controls. Absence of PDCD4 was noted in one sample. There was no direct correlation between protein levels and hsa-mir-21 levels (see figure 1) within each sample.
Figure 3.
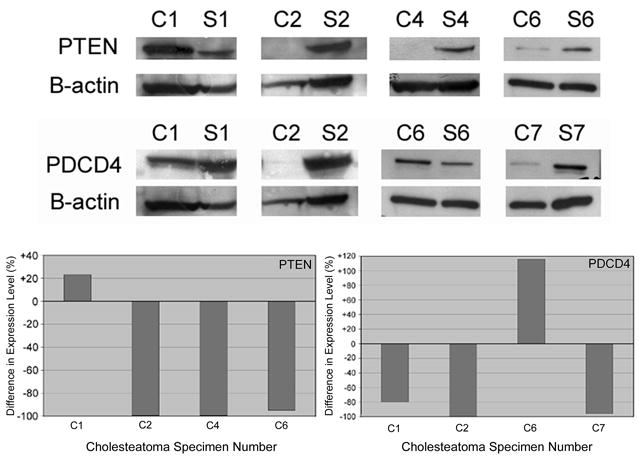
Cholesteatoma and matched skin samples were analyzed by western blot for levels of downstream hsa-mir-21 targets (top). These analyses demonstrated significant reductions in PTEN and PDCD4 protein levels in cholesteatoma as compared to normal skin in 3 of 4 specimens studied for each protein (bottom). The y-axis represents percent difference in expression in cholesteatoma than in skin (i.e., negative values equate to less expression in cholesteatoma).
DISCUSSION
MicroRNAs represent powerful regulators of protein translation and have been implicated in neoplastic and hyper-proliferative diseases. This study specifically identified up-regulation of hsa-mir-21 in cholesteatoma as compared to normal skin. Increased levels of hsa-mir-21 have been identified in many solid tumors including breast, liver, colorectal and brain (28–32). Hsa-mir-21 up-regulation has also been seen in non-malignant lesions such as vestibular schwannoma (33). Hsa-mir-21 inhibits the translation of several tumor suppressor proteins and thus represents a common entry-point into pathways regulating growth and proliferation.
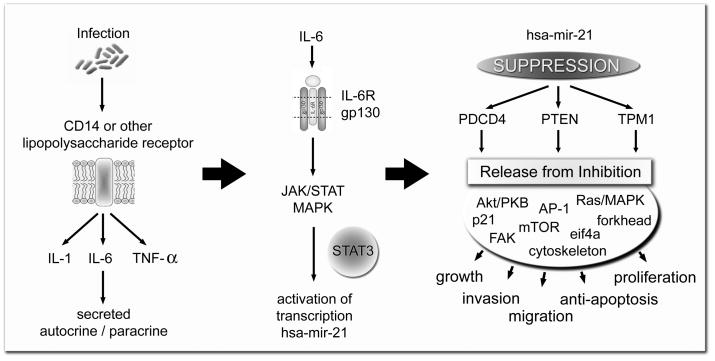
Based upon the results of this study and evidence in the literature a model for hsa-mir-21 regulated cholesteatoma growth and proliferation was developed. This model proposes that infection and lipopolysaccharide receptor activation is the initial stimulus for cholesteatoma growth. Subsequent IL-6 production activates the JAK/STAT/MAPK pathway through the IL-6 receptor composed of IL-6R and gp130. The subsequent phosphorylation of STAT3 causes increased expression of hsa-mir-21 which suppresses the translation of PTEN and PDCD4 leading to keratinocyte proliferation, migration, growth, and invasion (Figure 4).
Figure 4.
These data help to formulate our model of hsa-mir-21 up-regulation in epithelium, which leads to PTEN and PDCD4 suppression and subsequent cholesteatoma formation. We postulate that hsa-mir-21 up-regulation is secondary to infection and resultant cytokine production.
The most prominent target of hsa-mir-21 is PTEN, whose expression has been clearly shown to be mediated by hsa-mir-21 (30,31). PTEN is a phosphatase that maintains low levels of phosphatidylinositol (3,4,5)-triphosphate (PIP-3) by conversion to PIP-2. If PTEN fails to maintain this homeostasis, PIP-3 levels increase and activate the Akt/PKB pathway. Akt/PKB pathway activation has many effects including inhibition of apoptosis by effects on BAD, caspase-9, and Forkhead transcription factors (25,34). The Akt/PKB pathway also promotes cell growth and proliferation by activating the mTOR pathway and by effects on p27Kip1 and GSK-3(35,36). Also seen is activation of Ras/MAPK signaling through release of inhibition on Shc (37).
This study demonstrated decreased levels of PTEN concurrent with increased levels of hsa-mir-21. If this is a functional relationship we would expect cholesteatoma to show alterations in the targets of PTEN, namely Akt/PKB mediated pathways. Indeed, Akt/PKB and Ras/MAPK pathways have been identified as up-regulated in cholesteatoma as compared to normal skin (38,39). Further, PTEN has been found to be reduced in cholesteatoma concurrent with increased Akt phosphorylation (39). The Akt/PKB and Ras/MAPK pathways also show increased activation in keratinocytes of a PTEN-deficient mouse model (26). This mouse develops epidermal hyperplasia and hyperkeratosis and has keratinocytes which are hyper-proliferative, resistant to apoptosis, and show increased migration.
This study demonstrated that PDCD4, another target of hsa-mir-21, is also reduced in cholesteatoma as compared to normal skin. PDCD4 suppresses several targets which regulate translation and cell proliferation: eukaryotic initiation factor 4a, AP-1-mediated trans-activation, and the kinase inhibitor p21 (29,40). Several studies have shown that PDCD4 levels are down-regulated by hsa-mir-21 (28,29,40). These reduced levels of PDCD4 have been implicated in tissue invasion and proliferation (29,40). In a mouse cancer model, the presence of PDCD4 was shown to suppress benign and malignant skin tumor formation and progression (27). Our model is consistent with these findings and proposes that hsa-mir-21 inhibits PTEN and PDCD4 translation leading to keratinocyte proliferation.
One theory of cholesteatoma proliferation is that activation of growth and proliferation is secondary to infection and resultant cytokine production (5,41). Lipopolysaccharides from bacterial infections are postulated to stimulate the bone resorption caused by cholesteatoma through activation of cytokines and proteinases (42). Lipopolysaccharides activate IL-1, IL-6 and TNF-α production as well as induce increased levels of matrix-metalloproteinases (42). These factors have been shown to be over-expressed in cholesteatoma (10,14,15,43–46). Most notable among these activators is IL-6 which, through a gp130 mediated receptor, activates STAT3, a critical signal transducer and activator of transcription associated with malignant transformation and tumorigenesis (47). STAT3 has been identified as a transcription factor definitively promoting hsa-mir-21 transcription and expression (48). We showed that these components of the pathway promoting hsa-mir-21 expression (i.e., IL-6R, GP130, CD14 and STAT3) are present in cholesteatoma. This would provide a mechanism by which infection can lead to microRNA expression and subsequent suppression of PTEN and PDCD4 production.
An experimental consideration is the use of post-auricular skin as a control for cholesteatoma. Some studies have suggested that canal skin provides a better control for cholesteatoma as it has greater similarity in protein expression (14,49). Those studies, however, investigated only a limited number of proteins as opposed to a comprehensive genetic profile. Further, canal skin is typically within the infected field and may not be representative of the normal skin condition. For example, exposure of the canal skin to chronic purulent otorrhea may stimulate receptors and cytokine expression leading to changes in baseline gene/protein expression. Post-auricular skin, on the other hand, is outside the infected field and is widely used and continues to serve as a control in many cholesteatoma studies (14,39). It is also abundant and allows enough material for RNA and protein studies. Further, the genetic background provided by post-auricular skin (i.e., epithelial) remains a good template for interpreting cholesteatoma findings.
Another consideration in this study is the heterogeneity of the tissues studied. Cholesteatoma may include bacteria as well as immune response cells such as neutrophils and lymphocytes. Skin controls may inadvertently include subcutaneous fat or have different proportions of keratinocytes and fibroblasts. Inflammatory and fat cells are a distinct minority of the total cell mass in these tissues and are unlikely to be the source of differential expression. Keratinocytes are the predominant cell in both these tissues and are the likely source of differing levels of microRNA and protein; however, fibroblasts may also be implicated in this pathology. There is some evidence that fibroblasts may serve as a “feeder” for keratinocytes with regards to the cholesteatoma phenotype (50,51). If this is the case, the heterogeneity inherent in this study is advantageous and a better reflection of the pathologic condition. Current experiments in our laboratory looking at isolated keratinocytes in culture may help to distinguish the independent roles of these cell types.
Surgical resection is the only treatment option currently available for primary, secondary, and recurrent cholesteatoma. The proposed model for cholesteatoma pathogenesis provides novel targets for pharmacological therapies. Of particular interest would be targeting hsa-mir-21 with a microRNA inhibitor. Indeed, RNA-based therapies have shown promising results for the suppression of biological pathways. For example, systemic administration of an anti-microRNA toward mir-122 in monkeys successfully reduced microRNA levels and affected down-stream cholesterol production (52). Interfering RNAs applied trans-tympanically or topically at the time of surgery may be capable of suppressing cholesteatoma growth or recurrence. Such pharmacological therapies may need to target multiple components of the hsa-mir-21 pathway. For example, STAT3 inhibitors have demonstrated the ability to decrease cell proliferation in breast cancer and would add to the inhibition of hsa-mir-21 activity (47). We are currently characterizing a keratinocyte cell culture model in which to test potential inhibitors of this pathway.
CONCLUSION
This study proposes a novel microRNA-based model to explain the growth and proliferation of cholesteatoma. We have demonstrated the presence and expression levels of many of the components of this model within cholesteatoma. Ongoing studies are characterizing additional proteins related to hsa-mir-21 expression and activity. These may reveal critical roles of other targets of hsa-mir-21 such as TPM1, an enzyme down-regulated in breast carcinoma and able to inhibit tumorigenesis when over-expressed (53–55). The relatively easy access of the middle ear to intra-tympanic drug delivery makes an RNA-based topical inhibitor of cholesteatoma growth an attractive mode of primary or adjunctive therapy. This model provides an initial template to begin to explore such new leads and novel medical treatments for cholesteatoma.
Acknowledgments
Supported by NIDCD K08DC006227
References
- 1.Vikram BK, Udayashankar SG, Naseeruddin K, Venkatesha BK, Manjunath D, Savantrewwa IR. Complications in primary and secondary acquired cholesteatoma: a prospective comparative study of 62 ears. Am J Otolaryngol. 2008;29:1–6. doi: 10.1016/j.amjoto.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 2.Friedland DR, Pensak ML, Kveton JF. Cranial and Intracranial Complications of Acute and Chronic Otitis Media. In: Wackym PA, Snow J, editors. Ballenger’s Otorhinolaryngology Head and Neck Surgery. Hamilton ON: BC Decker; 2008. in press. [Google Scholar]
- 3.Mukherjee P, Saunders N, Liu R, Fagan P. Long-term outcome of modified radical mastoidectomy. J Laryngol Otol. 2004;118:612–6. doi: 10.1258/0022215041917970. [DOI] [PubMed] [Google Scholar]
- 4.Yung M, Jacobsen NL, Vowler SL. A 5-year observational study of the outcome in pediatric cholesteatoma surgery. Otol Neurotol. 2007;28:1038–40. doi: 10.1097/mao.0b013e318159e799. [DOI] [PubMed] [Google Scholar]
- 5.Semaan MT, Megerian CA. The pathophysiology of cholesteatoma. Otolaryngol Clin North Am. 2006;39:1143–59. doi: 10.1016/j.otc.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Olszewska E, Wagner M, Bernal-Sprekelsen M, et al. Etiopathogenesis of cholesteatoma. Eur Arch Otorhinolaryngol. 2004;261:6–24. doi: 10.1007/s00405-003-0623-x. [DOI] [PubMed] [Google Scholar]
- 7.Kim HJ, Chole RA. Experimental models of aural cholesteatomas in Mongolian gerbils. Ann Otol Rhinol Laryngol. 1998;107:129–34. doi: 10.1177/000348949810700208. [DOI] [PubMed] [Google Scholar]
- 8.Kim CS, Lee CH, Chung JW, Kim CD. Interleukin-1 alpha, interleukin-1 beta and interleukin-8 gene expression in human aural cholesteatomas. Acta Otolaryngol. 1996;116:302–6. doi: 10.3109/00016489609137846. [DOI] [PubMed] [Google Scholar]
- 9.Hwang SJ, Kang HJ, Song JJ, et al. Up-regulation of peroxidase proliferator-activated receptor gamma in cholesteatoma. Laryngoscope. 2006;116:58–61. doi: 10.1097/01.mlg.0000184507.49254.f9. [DOI] [PubMed] [Google Scholar]
- 10.Bujia J, Kim C, Boyle D, Hammer C, Firestein G, Kastenbauer E. Quantitative analysis of interleukin-1-alpha gene expression in middle ear cholesteatoma. Laryngoscope. 1996;106:217–20. doi: 10.1097/00005537-199602000-00021. [DOI] [PubMed] [Google Scholar]
- 11.Ishibashi T, Shinogami M, Kaga K, Fukaya T. Keratinocyte growth factor and receptor mRNA expression in cholesteatoma of the middle ear. Acta Otolaryngol. 1997;117:714–8. doi: 10.3109/00016489709113465. [DOI] [PubMed] [Google Scholar]
- 12.Kojima H, Matsuhisa A, Shiwa M, et al. Expression of messenger RNA for keratinocyte growth factor in human cholesteatoma. Arch Otolaryngol Head Neck Surg. 1996;122:157–60. doi: 10.1001/archotol.1996.01890140043009. [DOI] [PubMed] [Google Scholar]
- 13.Song JJ, Chae SW, Woo JS, Lee HM, Jung HH, Hwang SJ. Differential expression of human beta defensin 2 and human beta defensin 3 in human middle ear cholesteatoma. Ann Otol Rhinol Laryngol. 2007;116:235–40. doi: 10.1177/000348940711600312. [DOI] [PubMed] [Google Scholar]
- 14.Mehta D, Daudia A, Birchall JP, Banerjee AR. The localization of matrix metalloproteinases-8 and -13 in cholesteatoma, deep-meatal and post-auricular skin: a comparative analysis. Acta Otolaryngol. 2007;127:138–42. doi: 10.1080/00016480600781807. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt M, Grunsfelder P, Hoppe F. Up-regulation of matrix metalloprotease-9 in middle ear cholesteatoma–correlations with growth factor expression in vivo? Eur Arch Otorhinolaryngol. 2001;258:472–6. doi: 10.1007/s004050100359. [DOI] [PubMed] [Google Scholar]
- 16.Cho JG, Lim HW, Woo JS, Hwang SJ, Lee HM, Chae SW. Overexpression of placenta growth factor in human middle ear cholesteatoma. Acta Otolaryngol. 2006;126:900–4. doi: 10.1080/00016480500546334. [DOI] [PubMed] [Google Scholar]
- 17.Sakamoto T, Kondo K, Yamasoba T, Suzuki M, Sugasawa M, Kaga K. Overexpression of ErbB-2 protein in human middle ear cholesteatomas. Laryngoscope. 2004;114:1988–91. doi: 10.1097/01.mlg.0000147934.21638.d8. [DOI] [PubMed] [Google Scholar]
- 18.Miyao M, Shinoda H, Takahashi S. Caspase-3, caspase-8, and nuclear factor-kappaB expression in human cholesteatoma. Otol Neurotol. 2006;27:8–13. doi: 10.1097/01.mao.0000180482.34545.b8. [DOI] [PubMed] [Google Scholar]
- 19.Jeong JH, Park CW, Tae K, et al. Expression of RANKL and OPG in middle ear cholesteatoma tissue. Laryngoscope. 2006;116:1180–4. doi: 10.1097/01.mlg.0000224345.59291.da. [DOI] [PubMed] [Google Scholar]
- 20.Huisman MA, De Heer E, Grote JJ. Sustained extracellular signal-regulated kinase1/2 mitogen-activated protein kinase signalling is related to increased p21 expression in cholesteatoma epithelium. Acta Otolaryngol. 2005;125:134–40. doi: 10.1080/00016480410022813. [DOI] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 22.Fabbri M, Croce CM, Calin GA. MicroRNAs. Cancer J. 2008;14:1–6. doi: 10.1097/PPO.0b013e318164145e. [DOI] [PubMed] [Google Scholar]
- 23.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 24.Maritano D, Sugrue ML, Tininini S, et al. The STAT3 isoforms alpha and beta have unique and specific functions. Nat Immunol. 2004;5:401–9. doi: 10.1038/ni1052. [DOI] [PubMed] [Google Scholar]
- 25.Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387–90. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki A, Nakano T, Mak TW, Sasaki T. Portrait of PTEN: messages from mutant mice. Cancer Sci. 2008;99:209–13. doi: 10.1111/j.1349-7006.2007.00670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jansen AP, Camalier CE, Colburn NH. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res. 2005;65:6034–41. doi: 10.1158/0008-5472.CAN-04-2119. [DOI] [PubMed] [Google Scholar]
- 28.Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 29.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–33. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 30.Meng F, Henson R, Lang M, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–29. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 31.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 33.Cioffi JA, Mendolia-Loffredo S, Massey B, Wackym PA. MicroRNA-21 over-expression may contibute to tumor growth in vestibular schwannomaa. Abstracts of the Association for Otolaryngology. 2008;31:264. [Google Scholar]
- 34.Paez JG, Sellers WR. PI3K/PTEN/AKT Pathway: A Critical Mediator of Oncogenic Signaling. In: Frank DA, editor. Signal Transduction in Cancer. New York: Springer; 2002. pp. 145–68. [Google Scholar]
- 35.Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455–64. doi: 10.1038/sj.onc.1209085. [DOI] [PubMed] [Google Scholar]
- 36.Hlobilkova A, Knillova J, Bartek J, Lukas J, Kolar Z. The mechanism of action of the tumour suppressor gene PTEN. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2003;147:19–25. [PubMed] [Google Scholar]
- 37.Gu J, Tamura M, Pankov R, et al. Shc and FAK differentially regulate cell motility and directionality modulated by PTEN. J Cell Biol. 1999;146:389–403. doi: 10.1083/jcb.146.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huisman MA, De Heer E, Grote JJ. Survival signaling and terminal differentiation in cholesteatoma epithelium. Acta Otolaryngol. 2007;127:424–9. doi: 10.1080/00016480600868430. [DOI] [PubMed] [Google Scholar]
- 39.Yune TY, Byun JY. Expression of PTEN and phosphorylated Akt in human cholesteatoma epithelium. Acta Otolaryngol. 2008:1–6. doi: 10.1080/00016480802258802. [DOI] [PubMed] [Google Scholar]
- 40.Lu Z, Liu M, Stribinskis V, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008 doi: 10.1038/onc.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sudhoff H, Tos M. Pathogenesis of attic cholesteatoma: clinical and immunohistochemical support for combination of retraction theory and proliferation theory. Am J Otol. 2000;21:786–92. [PubMed] [Google Scholar]
- 42.Peek FA, Huisman MA, Berckmans RJ, Sturk A, Van Loon J, Grote JJ. Lipopolysaccharide concentration and bone resorption in cholesteatoma. Otol Neurotol. 2003;24:709–13. doi: 10.1097/00129492-200309000-00002. [DOI] [PubMed] [Google Scholar]
- 43.Kato A, Ohashi Y, Masamoto T, Sakamoto H, Uekawa M, Nakai Y. Interleukin-6 and tumour necrosis factor alpha synthesized by cholesteatoma cells affect mucociliary function in the eustachian tube. Acta Otolaryngol Suppl. 1998;538:90–7. doi: 10.1080/00016489850182792. [DOI] [PubMed] [Google Scholar]
- 44.Wilmoth JG, Schultz GS, Antonelli PJ. Tympanic membrane metalloproteinase inflammatory response. Otolaryngol Head Neck Surg. 2003;129:647–54. doi: 10.1016/S0194-59980301388-3. [DOI] [PubMed] [Google Scholar]
- 45.Akimoto R, Pawankar R, Yagi T, Baba S. Acquired and congenital cholesteatoma: determination of tumor necrosis factor-alpha, intercellular adhesion molecule-1, interleukin-1-alpha and lymphocyte functional antigen-1 in the inflammatory process. ORL J Otorhinolaryngol Relat Spec. 2000;62:257–65. doi: 10.1159/000027756. [DOI] [PubMed] [Google Scholar]
- 46.Bujia J, Kim C, Ostos P, Sudhoff H, Kastenbauer E, Hultner L. Interleukin 1 (IL-1) and IL-1-receptor antagonist (IL-1-RA) in middle ear cholesteatoma: an analysis of protein production and biological activity. Eur Arch Otorhinolaryngol. 1996;253:252–5. doi: 10.1007/BF00171137. [DOI] [PubMed] [Google Scholar]
- 47.Siddiquee K, Zhang S, Guida WC, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci U S A. 2007;104:7391–6. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loffler D, Brocke-Heidrich K, Pfeifer G, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–3. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 49.Lee RJ, Mackenzie IC, Hall BK, Gantz BJ. The nature of the epithelium in acquired cholesteatoma. Clin Otolaryngol Allied Sci. 1991;16:168–73. doi: 10.1111/j.1365-2273.1991.tb01970.x. [DOI] [PubMed] [Google Scholar]
- 50.Yoshikawa M, Kojima H, Wada K, et al. Identification of specific gene expression profiles in fibroblasts derived from middle ear cholesteatoma. Arch Otolaryngol Head Neck Surg. 2006;132:734–42. doi: 10.1001/archotol.132.7.734. [DOI] [PubMed] [Google Scholar]
- 51.Laeeq S, Faust R. Modeling the cholesteatoma microenvironment: coculture of HaCaT keratinocytes with WS1 fibroblasts induces MMP-2 activation, invasive phenotype, and proteolysis of the extracellular matrix. Laryngoscope. 2007;117:313–8. doi: 10.1097/01.mlg.0000251164.26405.1a. [DOI] [PubMed] [Google Scholar]
- 52.Elmen J, Lindow M, Schutz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 53.Mahadev K, Raval G, Bharadwaj S, et al. Suppression of the transformed phenotype of breast cancer by tropomyosin-1. Exp Cell Res. 2002;279:40–51. doi: 10.1006/excr.2002.5583. [DOI] [PubMed] [Google Scholar]
- 54.Raval GN, Bharadwaj S, Levine EA, et al. Loss of expression of tropomyosin-1, a novel class II tumor suppressor that induces anoikis, in primary breast tumors. Oncogene. 2003;22:6194–203. doi: 10.1038/sj.onc.1206719. [DOI] [PubMed] [Google Scholar]
- 55.Shah V, Braverman R, Prasad GL. Suppression of neoplastic transformation and regulation of cytoskeleton by tropomyosins. Somat Cell Mol Genet. 1998;24:273–80. doi: 10.1023/b:scam.0000007130.08611.fc. [DOI] [PubMed] [Google Scholar]