Abstract
Assembly of immunoglobulin G (igG) molecules from two heavy and two light chains can be facilitated by connecting the light chain to the heavy chain by an oligopeptide linker. production of the anti-lysozyme D1.3-single chain (sc) igG1 in HeK293t cells yielded up to 8 mg/L functional scigG polypeptide. Size exclusion chromatography of material purified by protein-A affinity chromatography revealed that the majority of the D1.3-scigG1 molecules were 150 kDa monomers, with a KD of 1.8 × 10−10 M measured by surface plasmon resonance; however, significant fractions of scigG dimers and oligomers with molecular masses of 300 kDa and >600 kDa, respectively, were identified. the oligomerization resulted in an increased avidity. observed oligomerization capability may allow new approaches for the generation of bispecific/multivalent antibodies.
Key words: recombinant antibodies, single chain immunoglobulin, scIgG, single chain fragment antigen binding, scFab
In 1999, Lee et al.1 described the production of single chain IgG (scIgG) in CHO cells, pointing towards a possible way to facilitate the complex assembly process necessary for the recombinant production of immunoglobulins, where heavy (HC) and light immunoglobulin chains (LC) must be correctly assembled based on a well-balanced expression of both chains.2,3 Connection of the light chain to the heavy chain should significantly improve the yield of correctly assembled IgG-like molecules since this reduces the bimolecular reaction of heavy to light chain pairing to a much more favourable monomolecular reaction. The Fc dependent antibody effector functions of scIgG1 produced by Lee et al. were reported to be comparable to those of the corresponding IgG1.1
Following a similar strategy, a polypeptide linker between LC and Fd was used to generate single chain Fab (scFab) antibody fragments.4,5 The aim was to combine the benefits of the scFv format6,7 with the advantages seen for some Fab fragments, such as improved stability in long-term storage and lower tendency to aggregate compared to their corresponding scFv fragments. ScFab fragments were successfully produced in Gram negative bacteria (E. coli), Gram positive bacteria (Bacillus megaterium) and in yeasts (Pichia pastoris and Saccharomyces cerevisiae),4,5,8 and showed improved bacterial expression and phage display.4
Single chain Fv (scFv) antibody constructs were reported to form higher order oligomers, which can be explained by a crossover pairing of a domain of one polypeptide with that of another molecule. This oligomerization must not necessarily be seen as an unwanted artifact, but has indeed been employed to improve the properties of recombinant antibodies. For example, diabodies,9–11 as well as triabodies and tetrabodies12,13 that provide improved avidity have been constructed. By using two different scFv fragments, bispecific molecules can be formed.14 Analogous to scFvs, scFab fragments have been reported to form dimers (DiaFabodies), trimers (TriFabodies) and tetramers (TetraFabodies).4
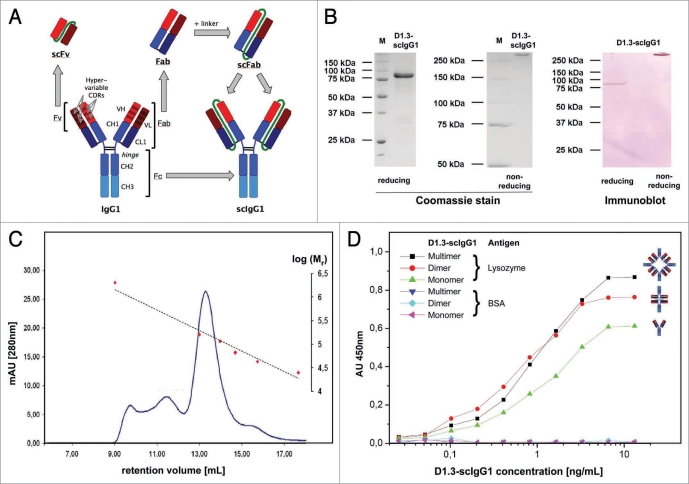
We analyzed and report here the oligomerization properties of a scIgG antibody obtained by fusion of the scFab fragment of the well characterized anti-lysozyme antibody D1.3,15 to the human IgG1 Fc moiety (Fig. 1A). This scFab fragment contains a 34 amino acid glycine-serine rich peptide linker of the sequence SGG GSG GGS EGG GSE GGG SEG GGS EGG GSG GGSG connecting the light chain with the Fd fragment of the heavy chain.4 In contrast to most scFv fragments, the linker connects the domains in the orientation light chain to heavy chain, which is dictated by the topology of the Fab in the IgG molecule.
Figure 1.
(A) Schematic illustration of igG and its single chain derivatives. Variable immunoglobulin regions (VL, VH) are shown in red, constant regions (CH1, CH2, CH3) including the hinge region are shown in blue. interchain disulphide bonds are coloured black and non-sequences (linkers) are colored green. (B) SDS-pAGe and immunoblot of D1.3-scigG. After protein A affinity purification, protein samples were prepared under reducing and non-reducing conditions and separated by SDS-pAGe. Gels were stained using Coomassie Blue or blotted onto a pVDf membrane which was stained with a goat-anti-human (fc-specific) alkaline phosphate antibody conjugate (NBt-BCip substrate). (C) Size exclusion chromatography of protein A purified D1.3-scigG1. peak positions found for the molecular weight calibration standards chymotrypsinogen A (25 kDa), ovalbumin (43 kDa), albumin (67 kDa), Crp (115 kDa), igG (160 kDa) and Blue Dextran 2000 (2 MDa) are indicated in red. the dashed line indicates the deduced calibration curve. (D) Antigen eLiSA of D1.3-scigG1 SeC fractions. Serial concentrations of the D1.3-scigG1 were analysed with BSA as negative control antigen. proposed oligomerization forms suggested by the observed molecular mass are schematically illustrated.
The D1.3-scIgG1 construct was cloned into the mammalian expression vector pCMV-myc-ER. A stop codon was introduced at the 3′ terminus of the scIgG1 gene construct for secretory expression with this vector system. The pCMV-D1.3-scIgG1 vector was transiently transfected into HEK293T cells. The production yields of D1.3-scIgG1 protein were between 5 to 8 mg/L culture supernatant determined by human IgG/Fc capture ELISA. The D1.3-scIgG1 protein was purified by protein A affinity chromatography.
On SDS-PAGE under reducing conditions, a single polypeptide chain was seen, corresponding to the expected molecular mass of 75–80 kDa of the scIgG1-polypeptide including glycosylations in the human IgG1 Fc moiety (Fig. 1B). Under non-reducing conditions, the relative molecular mass of the scIgG1 preparation appeared to be more than 250 kDa, which greatly exceeded the theoretical size of about 150 kDa of the homodimeric scIgG1 molecule. The increased apparent molecular mass in non-reducing SDS-PAGE is probably due to the non-globular shape of SDS denatured scIgG1 in the presence of interchain disulphide bounds in the hinge region, as observed for other IgG1 molecules.2,16 No additional bands of lower apparent molecular mass, the presence of which would indicate unpaired polypeptide chains, were observed either in the Coomassie Blue stained gel or in the sensitive immunoblot (Fig. 1B). This suggests complete assembly into scIgG within the secreted material. Correct antigen binding function of the antibody construct was confirmed by affinity determinations.
The apparent affinity of the D1.3-scIgG1 with a KD of 1.8 × 10−10 M measured by surface plasmon resonance (SPR) analysis on an excess of antigen was 55 times higher than that of the D1.3 scFv,4 although the D1.3 scFv almost exclusively forms dimers upon production in E. coli as well. This appears to indicate that the specific functionality of the scFv antibody fragment in comparison to the scIgG is decreased, but it may also be influenced by the more constrained flexibility of the diabody format resulting from its shortened linker, restraining the mobility of the antigen binding sites. Interestingly, the KD of the D1.3-scIgG1 was 100 times better than that of the D1.3-Fab fragment and 190 times better than that of the monomeric D1.3-scFab fragment.4 As is to be expected for an avidity effect, this result was due to the low dissociation rate (kOFF of 4.62 × 10−4 s−1) rather than the association rate (kON of 2.62 × 106 M−1 s−1).
To assess the potential oligomerization, the protein A purified scIgG1 fraction was examined by size exclusion chromatography (SEC) (Fig. 1C). Here, about 80% of the sample migrated corresponding to a molecular mass of about 150–160 kDa, indicating a homodimeric molecule consisting of two scIgG1 polypeptide chains. These homodimeric molecules most probably have an IgG-like conformation, and are therefore further referred to as ‘scIgG1 monomer.’ Two smaller distinct peaks of equal proportions roughly corresponded to a molar mass of about 300 kDa, and of more than 600 kDa, indicating populations of scIgG1 dimers and larger oligomers, respectively.
The resolution of the SEC system at very high molecular mass did not allow definitive determination of whether the left peak consisted of tetramers (i.e., eight identical polypeptide chains or four interacting scIgG monomers), pentamers or some higher oligomers, or a mixture thereof; however, no very high molecular mass aggregates were found. We therefore referred to this fraction as ‘oligomers.’ The formation of dimers and oligomers can be explained by the interconnection of light chain domains of one scFab moiety with the Fd domains of the scFab moiety of a second polypeptide.
The three different scIgG1 fractions obtained by gel filtration were further tested by antigen ELISA to confirm their antigen binding function (Fig. 1D). All D1.3-scIgG1 fractions specifically bound to the antigen lysozyme. The maximum absorbance measured in the antigen ELISA was slightly higher for the D1.3-scIgG1 oligomer fraction, followed by the D1.3-scIgG1 dimer and the D1.3-scIgG1 monomer fraction. The increase in total signal, despite a hardly changed EC50 of the different forms, may be explained by the increased number of Fc moieties that can be detected by the secondary antibody conjugate. The lack of a significant increase of avidity in this particular assay may be explained by the antigen density in the assays used, which does not allow significantly more re-association benefit with the increase from two to more binding arms, given the high affinity of each monovalent binding arm.
So far, the scIgG format has not been widely used, but it may help in the future to improve “whole IgG” display systems, e.g., on bacteria17 or mammalian cells,18 or other systems that allow only a single polypeptide, such as ribosomal display or yeast display. Its capability to form oligomers may be employed, in analogy to the bispecific diabody strategy,14 to generate bispecific antibody constructs with higher valency and Fc effector functions.
Material and Methods
Generation of D1.3-derived scIgG1 constructs
If not specifically indicated otherwise, all procedures were carried out according to ref. 19. The scFab gene fragment derived from the hen egg lysozyme specific antibody D1.34 was fused to the human IgG1 Fc gene fragment. The Fc moiety was amplified by polymerase chain reaction (PCR) from the vector pSH1-215,2 using the oligonucleotides CM_CH2CH3_NheI_fwd (5′-ATA TAT GCT AGC CGC TGA GCC CAA ATC TTG TGA CAA AAC TCA C-3′) and CM_Fc_Xba_rev (5′-GCC TCT CCC TGT CCC CGG GTA AAT GAT AAT CTA GAT ATA TA-3′) and cloned into the NheI and XbaI sites of the vector pPICZa-A-D1.3-scFab to obtain the vector pPICZa-A-D1.3-scIgG1. The entire D1.3-scIgG1 was re-amplified using the oligonucleotides CM_D13_BssHII_fwd (5′-AAT TAA GGC GCG CAC TCC GAC ATC GAG CTC ACC CAG TCT C-3′) and CM_Fc_Xba_rev (5′-GCC TCT CCC TGT CCC CGG GTA AAT GAT AAT CTA GAT ATA TA-3′) and cloned into the BssHII and XbaI sites of the mammalian expression vector pCMV-myc-ER (Invitrogen, Karlsruhe, Germany) to obtain the vector pCMV-D1.3-scIgG1. A stop codon was inserted at the 3′ terminus of the scIgG1 gene construct and the carboxyterminal myc tag and endoplasmic retention signal were deleted to allow secretory mammalian expression with this vector system.
Expression of scIgG1 in HEK293T cells
A total of 6 × 106 HEK293T cells (ATCC No: CRL-11268) were seeded into a 10 cm tissue culture plate (Sarstedt, Nümbrecht, Germany) containing 15 mL Dulbecco’s modified Eagle’s medium (DMEM with 4.5 g/L glucose, PAA, Parching, Germany) supplemented with 8% (v/v) fetal calf serum (FCS, PAA) and 1% penicillin/streptomycin (P/S, PAA). After one day cells were transfected with the plasmid pCMV-D1.3-scIgG1 using HEKfectin (BioRad, Munich, Germany) according to the manufacturer’s description. One day after transfection medium was exchanged with DMEM containing 4% (v/v) “FCS ultra low IgG” (PAA). Culture supernatant was harvested and medium was exchanged every day for 7 to 10 days.
Protein A purification of scIgG1
The scIgG1 protein from the production supernatants was purified by protein A affinity chromatography using Protein A HiTrap FF columns and the Äkta Prime fast performance liquid chromatography (FPLC) device (GE Healthcare, Munich, Germany) according to the manufacturer’s protocols (binding buffer: 20 mM Na2HPO4 × 2H2O, pH 7.0; washing buffer: 100 mM citric acid, pH 5.0; elution buffer: 100 mM citric acid, pH 2.5). Elution fractions were immediately neutralized with the adequate amount of 2 M TrisHCl [Tris-(hydroxymethyl)-aminomethan-HCl, pH 9.0]. The scIgG1-Protein was quantified by a human IgG/Fc capture ELISA in both purified and non-purified samples in comparison to the N protein SL human serum protein standard (Dade Behring, now Siemens Healthcare, Erlangen, Germany). Purified scIgG1 was quantified by a Bradford total protein assay (BioRad, Munich, Germany) using bovine serum albumin (BSA, Sigma) as protein standard.
Sodium dodecyl sulphate (SDS) polyacrylamide gel electrophoresis (PAGE), coomassie-blue and silver stain and immunoblot
Samples of scIgG1 protein A purification were prepared under reducing and non-reducing conditions. After SDS-PAGE gels were either stained with Coomassie-blue (staining solution: 25% (v/v) isopropanol, 10% (v/v) acetic acid, 0.5% (w/v) Coomassie blue G-250; destaining solution: 25% (v/v) isopropanol and 10% (v/v) acetic acid) or transferred onto PVDF membranes (Roth, Karlsruhe, Germany) for western blotting. Blots were blocked in Phosphate buffered saline (PBS, 10x stock solution contains 1.37 M NaCl, 26 mM KCl, 80 mM Na2HPO4, 15 mM KH2PO4) supplemented with 2% (w/v) skim milk powder (Roth) and 0.05% (v/v) Tween-20 (Serva) (2% MPBST) for one hour and stained with an α-human-IgG (Fc specific) antibody-alkaline phosphatase conjugate (Sigma, Germany) which was diluted 1:10,000 in 2% MPBST. After three washing steps in PBS and one washing step in substrate buffer (100 mM TrisHCl pH 9.5, 0.5 mM MgCl2) the blot was stained with 0.03% (w/v) nitro-blue-tetrazolium (NBT, Roth) chloride and 0.015% (w/v) 5-bromo-4-Chloro-3-indolylphosphate (BCIP, Roth) in substrate buffer.
Size exclusion chromatography
SEC was performed using an Äkta-Purifier-FPLC system (GE Healthcare) according to the manufacturer’s recommendations. Briefly, analytic SEC was performed using a Superdex 200 10/30 GL column (GE-Healthcare). Preparative scales were performed using a Superdex 200 HiLoad16/60 column (GE-Healthcare). PBS (pH 7.2) was used as diluent and flow rate was adjusted to 1 mL/min. Molecular weight calibration was performed with a Gelfiltration LMW Calibration Kit (GE Healthcare; chymotrypsinogen A—25 kDa, ovalbumin—43 kDa, albumin—67 kDa and Blue Dextran 2000—2 MDa) and additional protein standards human C-reactive protein (115 kDa, BiosPacific, Emeryville, CA) and human IgG (160 kDa) before and after analysis of the samples. Optical extinction was monitored at 280 nm. Samples were fractionated and stored for further ELISA analysis at 4°C.
Comparative antigen ELISA
Functionality of the D1.3-scIgG1 antibody fractions obtained from SEC containing scIgG1 monomers, dimers and oligomers were analyzed in an antigen ELISA as described.4 Briefly, 100 µL/well of 1 µg lysozyme diluted in 50 mM sodium bicarbonate buffer (pH 9.6) was coated in 96-well Maxisorp plates (Nunc, Thermo Fisher Scientific, Langenselbold, Germany) overnight at 4°C. Plates were blocked by addition of 300 µL 10% (v/v) FCS in PBS and incubation for 1 hour at 37°C. After three washing steps with PBST (PBS containing 0.05% (v/v) Tween-20) using an ELISA washer (TECAN, Crailsheim, Germany) each well was incubated for 1 hour at 37°C with 100 µL of a goat human IgG (Fc specific) antibody horseradish peroxidase (HRP) conjugate (1:60,000 in blocking buffer). After additional three washing steps, 100 µL/well of a TMB peroxidase substrate solution were added. The color reaction was stopped with 100 µL/well of 1N H2SO4. Absorbance was measured at 450 nm (absorbance at 620 nm was substracted) in a microtitre plate ELISA reader (Sunrise, TECAN).
Surface plasmon resonance analysis (SPR)
The dissociation constant (KD) and association and dissociation rates (kon and koff) were measured using a BIAcore® 2000 as previously described.4 Briefly, the antigen lysozyme was covalently bound to the CM5 chip by amino coupling with 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), N-hydroxysuccinimide (NHS) and ethanolamine according to the manufacturer’s description (BIAcore, GE Healthcare). In a parallel flow cell BSA was coupled as control antigen to substract unspecific signals. Measurements were performed at 25°C with a constant flow (25 µL/min) in a HEPES buffer (10 mM HEPES pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.005% (v/v) Nonidet P20). After measurement the chip was regenerated with 20 µL 10 mM NaOH. For affinity determination, at least four different scIgG1 concentrations in duplicate experiments were measured and analyzed using the BIAevaluation software.
Acknowledgements
This work was supported by the Land Niedersachsen. Original DNA encoding D1.3 VH and VL was provided by Arne Skerra.
Abbreviations
- BSA
bovine serum albumin
- IgG
immunoglobulin
- G
scFab, single chain fragment antigen binding
- scFv
single chain fragment variable
- scIgG
single chain immunoglobulin G
- SPR
surface plasmon resonance
- SEC
size exclusion chromatography
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/10784
References
- 1.Lee HS, Shu L, De Pascalis R, Giuliano M, Zhu M, Padlan EA, et al. Generation and characterization of a novel single-gene-encoded single-chain immunoglobulin molecule with antigen binding activity and effector functions. Mol Immunol. 1999;36:61–71. doi: 10.1016/s0161-5890(98)00109-6. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Menzel C, Meier D, Zhang C, Dübel S, Jostock T. A comparative study of different vector designs for the mammalian expression of recombinant IgG antibodies. J Immunol Methods. 2007;318:113–124. doi: 10.1016/j.jim.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Zhang C, Jostock T, Dübel S. Analysis of IgG heavy chain to light chain ratio with mutant Encephalomyocarditis virus internal ribosome entry site. Protein Eng Des Sel. 2007;20:491–496. doi: 10.1093/protein/gzm038. [DOI] [PubMed] [Google Scholar]
- 4.Hust M, Jostock T, Menzel C, Voedisch B, Mohr A, Brenneis M, et al. Single chain Fab (scFab) fragment. BMC Biotechnol. 2007;7:14. doi: 10.1186/1472-6750-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jordan E, Al-Halabi L, Schirrmann T, Hust M, Dübel S. Production of single chain Fab (scFab) fragments in Bacillus megaterium. Microb Cell Fact. 2007;6:38. doi: 10.1186/1475-2859-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird RE, Hardman KD, Jacobson JW, Johnson S, Kaufman BM, Lee SM, et al. Single-chain antigen-binding proteins. Science. 1988;242:423–426. doi: 10.1126/science.3140379. [DOI] [PubMed] [Google Scholar]
- 7.Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotny J, Margolies MN, et al. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci USA. 1988;85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker LM, Bowley DR, Burton DR. Efficient recovery of high-affinity antibodies from a single-chain Fab yeast display library. J Mol Biol. 2009;389:365–375. doi: 10.1016/j.jmb.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holliger P, Prospero T, Winter G. “Diabodies“: small bivalent and bispecific antibody fragments. Proc Natl Acad Sci USA. 1993;90:6444–6448. doi: 10.1073/pnas.90.14.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arndt MA, Krauss J, Rybak SM. Antigen binding and stability properties of non-covalently linked anti-CD22 single-chain Fv dimers. FEBS Lett. 2004;578:257–261. doi: 10.1016/j.febslet.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Hudson PJ, Kortt AA. High avidity scFv multimers; diabodies and triabodies. J Immunol Methods. 1999;231:177–189. doi: 10.1016/s0022-1759(99)00157-x. [DOI] [PubMed] [Google Scholar]
- 12.Iliades P, Kortt AA, Hudson PJ. Triabodies: single chain Fv fragments without a linker form trivalent trimers. FEBS Lett. 1997;409:437–441. doi: 10.1016/s0014-5793(97)00475-4. [DOI] [PubMed] [Google Scholar]
- 13.Schmiedl A, Zimmermann J, Scherberich JE, Fischer P, Dübel S. Recombinant variants of antibody 138H11 against human gamma-glutamyltransferase for targeting renal cell carcinoma. Hum Antibodies. 2006;15:81–94. [PubMed] [Google Scholar]
- 14.Kontermann RE, Wing MG, Winter G. Complement recruitment using bispecific diabodies. Nat Biotechnol. 1997;15:629–631. doi: 10.1038/nbt0797-629. [DOI] [PubMed] [Google Scholar]
- 15.Boulot G, Eisele JL, Bentley GA, Bhat TN, Ward ES, Winter G, et al. Crystallization and preliminary X-ray diffraction study of the bacterially expressed Fv from the monoclonal anti-lysozyme antibody D1.3 and of its complex with the antigen, lysozyme. J Mol Biol. 1990;213:617–619. doi: 10.1016/S0022-2836(05)80248-7. [DOI] [PubMed] [Google Scholar]
- 16.Simmons LC, Reilly D, Klimowski L, Raju TS, Meng G, Sims P, et al. Expression of full-length immunoglobulins in Escherichia coli: rapid and efficient production of aglycosylated antibodies. J Immunol Methods. 2002;263:133–147. doi: 10.1016/s0022-1759(02)00036-4. [DOI] [PubMed] [Google Scholar]
- 17.Mazor Y, Van Blarcom T, Mabry R, Iverson BL, Georgiou G. Isolation of engineered, full-length antibodies from libraries expressed in Escherichia coli. Nat Biotechnol. 2007;25:563–565. doi: 10.1038/nbt1296. [DOI] [PubMed] [Google Scholar]
- 18.Akamatsu Y, Pakabunto K, Xu Z, Zhang Y, Tsurushita N. Whole IgG surface display on mammalian cells: Application to isolation of neutralizing chicken monoclonal anti-IL-12 antibodies. J Immunol Methods. 2007;327:40–52. doi: 10.1016/j.jim.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]