Abstract
OBJECTIVE
Twist1 is a transcription factor that is highly expressed in murine brown and white adipose tissue (WAT) and negatively regulates fatty acid oxidation in mice. The role of twist1 in WAT is not known and was therefore examined.
RESEARCH DESIGN AND METHODS
The expression of twist1 was determined by quantitative real-time PCR in different tissues and in different cell types within adipose tissue. The effect of twist1 small interfering RNA on fatty acid oxidation, lipolysis, adipokine secretion, and mRNA expression was determined in human adipocytes. The interaction between twist1 and specific promoters in human adipocytes was investigated by chromatin immunoprecipitation (ChIP) and reporter assays.
RESULTS
Twist1 was highly expressed in human WAT compared with a set of other tissues and found predominantly in adipocytes. Twist1 levels increased during in vitro differentiation of human preadipocytes. Gene silencing of twist1 in human white adipocytes had no effect on lipolysis or glucose transport. Unexpectedly, and in contrast with results in mice, twist1 RNA interference reduced fatty acid oxidation. Furthermore, the expression and secretion of the inflammatory factors tumor necrosis factor-α, interleukin-6, and monocyte chemoattractant protein-1 were downregulated by twist1 silencing. ChIP and reporter assays confirmed twist1 interaction with the promoters of these genes.
CONCLUSIONS
Twist1 may play a role in inflammation of human WAT because it can regulate the expression and secretion of inflammatory adipokines via direct transcriptional effects in white adipocytes. Furthermore, twist1 may, in contrast to findings in mice, be a positive regulator of fatty acid oxidation in human white adipocytes.
Twist1 and twist2 are well-conserved basic helix-loop-helix transcription factors (1,2). They dimerize with other basic helix-loop-helix proteins and bind to E-boxes in the promoter regions of their target genes (3). The human twist proteins are highly homologous, although twist2 (previously Dermo1) is shorter than twist1 due to an NH2-terminal truncation. Twist1 and twist2 have partly overlapping functions but twist1 has been more extensively studied. Twist1-null mice die during embryogenesis due to failed neural tube fusion, whereas Twist2−/− mice, although growth retarded, survive, implying that the two twist isoforms also have unique effects (4). Twist2−/− animals display alterations in the morphology and function of several tissues, including adipose tissue, indicating an important role in organ development (5). Moreover, twist1 and twist2 inhibit osteogenesis and myogenesis by blocking the activity of the transcription factors Runx-2 and MyoD, respectively, which are essential for differentiation (6–9). Increased expression of twist1 is associated with tumor progression and metastasis (10).
Local low-grade inflammation is an important factor linking white adipose tissue (WAT) to insulin resistance and ultimately type 2 diabetes (11). Twist2−/− and Twist1+/−/Twist2+/− mice have increased circulating levels of the inflammatory cytokines tumor necrosis factor-α (TNF-α), interleukin (IL)-1β, and IL-6, implying a role in inflammation (5). Twist1 expression in T helper 1 lymphocytes and bone marrow–derived macrophages attenuates the expression of interferon-γ, IL-2, and TNF-α (12,13), further implicating twist1 in the regulation of cytokine expression. Moreover, twist1 inhibits the activity of cytokine promoters in COS cells, partly through an interaction with nuclear factor-κB (5).
Twist1 was recently shown to be highly, selectively, and similarly expressed in murine brown adipose tissue (BAT) and WAT (14). Furthermore, twist1 was found to inhibit the transcriptional activity of peroxisome proliferator–activated receptor-γ coactivator-1α (PGC-1α), which has a central role in brown adipocytes. Knockdown of twist1 in murine brown adipocytes induced the expression of genes involved in oxidative metabolism and fatty acid (FA) oxidation, for example, uncoupling protein 1 (UCP-1) and carnitine palmitoyl transferase 1 (CPT-1), whereas twist1 overexpression in C2C12 myotubes resulted in a reduction in PGC-1α–stimulated FA oxidation (14). This identifies a novel role for twist1 in energy homeostasis by regulating PGC-1α–induced energy expenditure in BAT.
Taken together, in vitro as well as animal studies suggest that twist1 regulates pathways involved in inflammation and energy homeostasis, processes that are closely involved in the development of obesity and its associated disorders, for example, insulin resistance and type 2 diabetes (11). However, the function of twist1 in WAT is not known. Because of the selective expression of twist1 in murine WAT and BAT and its role in energy regulation in murine BAT, we hypothesized that twist1 might also be important for adipocyte function in human WAT. We therefore measured the expression of twist1 in human white adipocytes and WAT. We performed in vitro studies where twist1 was downregulated by RNA interference (RNAi) in primary cultures of human adipocytes and assessed the effects on gene expression and different aspects of fat cell function, including FA oxidation, basal lipolysis, and adipokine secretion. Finally, the interaction between twist1 and promoters was investigated by chromatin immunoprecipitation (ChIP) and reporter assays.
RESEARCH DESIGN AND METHODS
Patient samples.
This study was conducted in accordance with the guidelines in the Declaration of Helsinki and approved by the ethics committee at Karolinska University Hospital and at the Institutional Review Board of INSERM and Toulouse University Hospital. The study was explained in detail to each subject, and written informed consent was obtained.
In cohort 1, WAT, pancreas, liver, and skeletal muscle samples were obtained from nonmalignant tissues of subjects undergoing elective surgery. Cohort 2, described in detail elsewhere (15), comprised 14 healthy subjects (3 men and 11 women) (age 37.5 ± 10 years; BMI 29.2 ± 7 kg/m2) from whom mRNA of both intact subcutaneous WAT pieces and isolated adipocytes were available. Biopsies of the subcutaneous abdominal WAT (0.5–2 g) were obtained in the morning after an overnight fast by needle aspiration under local anesthesia. In cohort 3, subcutaneous WAT was obtained from healthy women undergoing elective surgical procedures of fat removal for esthetic purposes (age 42.3 ± 8.8 years; BMI 27.9 ± 5.1 kg/m2) (16). The cells from the stroma-vascular fraction (SVF) and mature adipocytes from human WAT in cohort 3 were obtained after collagenase digestion as previously described (16). The different cell populations obtained from the SVF were capillary endothelial cells, defined as CD34+/CD31+; resident macrophages, defined as CD34−/CD14+; and cells negative for CD34, CD31, and CD14. Subjects in cohorts 1 and 3 were not selected for age, sex, or BMI.
Cell culture.
Cell culture experiments were performed on in vitro–differentiated human adipocytes derived from preadipocytes isolated from WAT. Isolation, culture, and differentiation of preadipocytes were performed as previously described (17). For quantification of twist1 mRNA during adipocyte differentiation, preadipocytes were lysed at days 4, 8, and 12 of differentiation.
Small interfering RNA (siRNA) and ChIP experiments were performed at differentiation days 10–12. For reporter assays, 3T3-L1 cells were grown according to standard protocols from ATCC (Manassas, VA).
RNAi by siRNA.
Human in vitro–differentiated adipocytes were treated with 50 nmol/l ON-TARGETplus SMARTpool TWIST1 (l-006434-00) siRNA (Thermo Fisher Scientific, Lafayette, CO) and 9 μl HiPerFect Transfection Reagent (QIAGEN, Hilden, Germany) according to the manufacturers' protocols. The optimal transfection conditions were determined in separate titration experiments. To control for unspecific effects, control cells were treated with AllStars Negative Control siRNA (QIAGEN). These oligonucleotides have no known similarities to human sequences and have no effect on gene expression and metabolism of human in vitro–differentiated adipocytes (18). Cells were incubated for 48 h and conditioned media were analyzed for release of glycerol and adipokines and/or palmitate oxidation (described below). Cells were subsequently lysed for RNA or protein extraction as described below.
Quantitative real-time RT-PCR.
Cells were treated as described above and total RNA was extracted using the NucleoSpin RNA II (Macherey-Nagel, Düren, Germany). RNA concentration and purity were assessed spectrophotometrically with a NanoDrop ND-1000 (Thermo Scientific, Wilmington, DE) and with an Agilent Bioanalyzer. Half a microgram of total RNA was reverse-transcribed to cDNA using Omniscript reverse transcription kit (QIAGEN) and random hexamer primers (Invitrogen, Carlsbad, CA). For SYBR Green assays, 5 ng of cDNA was mixed with 2× iQ SYBR Green Supermix (Eurogentec SA, Ougrée, Belgium) and primers (Invitrogen) in a final volume of 25 μl. cDNA (10 ng) was mixed with 2× TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA) and TaqMan primers (Applied Biosystems) in a final volume of 20 μl. Primer sequences and catalog numbers are listed in supplementary Tables 1 and 2, available in an online appendix at http://diabetes.diabetesjournals.org/cgi/content/full/db09-0997/DC1. Quantitative RT-PCR was performed in an iCycler IQ (Bio-Rad Laboratories, Hercules, CA). mRNA levels were determined by a comparative threshold cycle (Ct) method. Ct values were normalized to the reference genes glyceraldehyde 3-phosphate dehydrogenase (GAPDH), LDL receptor–related protein 10 (LRP10), or 18S, according to the formula 2ΔCt−target gene/2ΔCt−reference gene = arbitrary units (AU). The PCR efficiency in all runs was close to 100%, and all samples were run in duplicate.
For determination of twist1 mRNA in human SVF of WAT, RNA was extracted using RNeasy kit from QIAGEN. RNA concentrations were measured by fluorimetric assay (Ribogreen; Invitrogen) and reverse-transcribed using the Superscript II kit (Invitrogen). Reverse transcription was also performed without the superscript enzyme on RNA samples to ensure the absence of contaminating genomic DNA. The amplification reaction was performed in duplicate on 15 ng of the cDNA samples in a final volume of 20 μl in 96-well reaction plates (Applied Biosystems) in a GeneAmp 7500 detection system (Applied Biosystems).
Palmitate oxidation.
Cells were treated with siRNA for 48 h as described above. The medium was subsequently changed to a glucose-free medium supplemented with 2 mmol/l l-carnitine, 1% FA-free BSA, 14C-labeled palmitate, and unlabeled palmitate for 3 h. FA oxidation was measured as described (19). Liberated 14CO2 (free fraction) was collected on Carbosorb and 14C-labeled β-oxidation metabolites (acid-soluble fraction) were extracted using butanol. Both fractions were measured by scintillation counting. The cells were lysed for RNA and/or protein, and palmitate/FA oxidation (total counts from both fractions) was normalized to protein amount in each experimental group. All conditions were measured in duplicate or triplicate.
Lipolysis measurements.
Condition medium aliquots were taken from in vitro–differentiated adipocytes. Glycerol release into the medium (μmol/l) was used as a measurement of the lipolysis rate and was analyzed using a bioluminescence method (20).
Western blotting.
Cells were lysed in radioimmunoprecipitation assay buffer and protein concentrations were measured as previously described (18). Protein lysates were boiled in 1× SDS loading buffer, separated by SDS-PAGE, and blotted as described before (21). Antibodies were α-twist1 (catalog number T6451; Sigma, St. Louis, MO) and α-β-actin (catalog number A2066; Sigma). Antibody-antigen complexes were detected by chemiluminescence using the Chemidoc XRS system (Bio-Rad Laboratories), and images were analyzed using Quantity One Software (Bio-Rad Laboratories).
Adipokine measurements.
Human IL-6, monocyte chemoattractant protein-1 (MCP-1), and adiponectin were measured in medium aliquots using a Quantikine Immunoassay for human IL-6 (catalog number D6050), MCP-1 (catalog number DCP00), or adiponectin (catalog number DRP300), respectively (R&D Systems, Minneapolis, MN), according to the manufacturer's instructions.
Chromatin immunoprecipitation.
ChIP was performed using a kit (Acetyl-Histone H4 Immunoprecipitation [ChIP] Assay Kit, catalog number 17-229; Millipore, Temecula, CA) designed for detecting transcriptionally active chromatin from mammalian cells. ChIP was performed according to the manufacturer's instructions. Sonication conditions were optimized in separate experiments to produce 200- to 1,000-base pair (bp) DNA fragments. Immunoprecipitation was performed with α-twist1 (catalog number T6451; Sigma) or α-acetyl-histone H4 antibodies overnight at 4°C. Anti-GLUT4 (catalog number 4670-1704; AbD Serotec, Oxford, U.K.) or no antibody was used as negative control.
The extracted DNA was used for PCR analysis. PCRs were conducted using primers spanning regions in human MCP-1, TNF-α, and IL-6 promoters. Primer sequences and PCR product sizes are listed in supplementary Table 3. All primers were designed to amplify promoter regions spanning E-boxes predicted by MatInspector software from Genomatix BiblioSphere. PCRs were carried out at 25 μl in the following conditions: 94°C for 3 min, followed by 32 cycles of 94°C for 30 s, 55/58/60°C for 30 s (depending on primer pair), 72°C for 45 s, and a final step at 72°C for 10 min. PCR products were separated on 2% agarose gels and visualized by ethidium bromide staining.
Reporter assays.
The pIL6 promoter construct was a gift from Dr Chiara Constanzo at the University of Verona, Italy, and has been described in detail elsewhere (22). In brief, it was constructed from a part of the human IL-6 promoter, spanning from −541 to +68, relative to the transcriptional start site (NM_000600), inserted into the pGL2-basic vector (Promega, Madison, WI). This promoter fragment spans several E-boxes predicted by MatInspector software and overlaps with the region amplified with the ChIP primers IL-6 proximal (supplementary Table 3). For construction of the twist1 expression vector, the coding sequence for human twist1 was excised from Full-Length Mammalian Gene Collection, Clone 4125830 (reference sequence Hs.66744) (Invitrogen) and cloned into pcDNA3.1 (Invitrogen). Expression of twist1 was confirmed by in vitro translation using a TnT T7/SP6 Coupled Reticulocyte Lysate System (catalog number L5020; Promega). Gene reporter assays were performed in undifferentiated 3T3-L1 cells transfected with Lipofectamine and Plus reagent (both from Invitrogen) as described (23) using 0.2 μg of pGL2-basic or pIL6 together with either 0.2 μg of pcDNA3.1 carrying the human twist1 sequence (pTwist1) or the empty expression vector pcDNA3.1. A cytomegalovirus-β-galactosidase–containing vector (0.1 μg) was included in all transfections to control for transfection efficiency. Luciferase activity, was normalized for β-galactosidase activity, and all experiments were run in triplicate and repeated at least four times.
Statistical analysis.
Values shown are mean ± SEM. Results were analyzed with suitable nonparametric statistical methods including Mann-Whitney, Wilcoxon, and Friedman tests.
RESULTS
Twist1 is expressed predominantly in the adipocytes of human WAT.
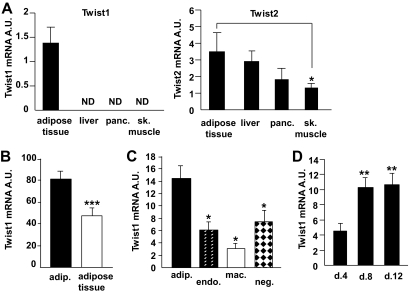
Twist1 and twist2 mRNA expression was investigated by quantitative RT-PCR in paired samples from human WAT, liver, pancreas, and skeletal muscle (Fig. 1A, cohort 1). Twist1 was detected only in WAT, whereas twist2 mRNA expression was readily detected in all examined tissues (Fig. 1A) and at similar levels in adipose tissue, liver, and pancreas. In paired samples of human WAT and isolated adipocyte fractions, twist1 appeared to be expressed predominantly in adipocytes because twist1 mRNA levels were higher in the adipocyte fraction compared with WAT (Fig. 1B, cohort 2). This was confirmed in a more detailed cell fraction analysis of adipose tissue demonstrating that twist1 expression was significantly higher in adipocytes compared with macrophages, endothelial cells, or cells expressing neither CD34, CD31, nor CD14 (Fig. 1C, cohort 3). Moreover, the expression of twist1 increased during in vitro differentiation of human preadipocytes, reaching a maximum level at differentiation day 8 (Fig. 1D).
FIG. 1.
Twist1 mRNA levels in different cell types from adipose tissue and during adipocyte differentiation. A: Twist1 and twist2 mRNA levels were measured in paired samples from human white adipose tissue (n = 3), liver (n = 2), pancreas (panc., n = 3), and skeletal muscle (sk. muscle, n = 3) (cohort 1). B: Twist1 expression was measured in 14 samples from human mature adipocytes (adip.) and the corresponding adipose tissue (cohort 2). C: Twist1 was also measured in samples from human mature adipocytes (n = 6) and the corresponding CD34+/CD31+ capillary endothelial cells (endo., n = 10), CD14+/CD34− resident macrophages (mac., n = 9), and cells negative for CD34, CD31, and CD14 (neg., n = 10) (cohort 3). D: Twist1 mRNA levels were measured during in vitro differentiation of human preadipocytes, n = 9. Cells were lysed at days 4, 8, and 12 of differentiation. RNA from earlier stages is not relevant to isolate because other types of cells from the stroma-vascular fraction (e.g., endothelial cells) may be present during the first few days of cell culture. Nonadipocyte cells are, however, not supported in the adipogenic differentiation medium and consequently die off at approximately day 2. All mRNA levels were normalized to the reference gene 18S and are shown as arbitrary units (A.U.). Bars indicate mean ± SEM. ND, not detected. ***P < 0.0001, **P < 0.01, and *P < 0.05.
Twist1 RNAi reduces FA oxidation but does not influence lipolysis in human cultured adipocytes.
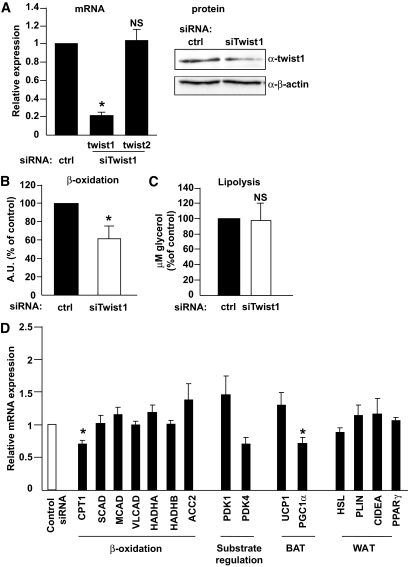
To assess biological functions of twist1 in human adipocytes, we performed in vitro gene silencing of twist1 in human in vitro–differentiated adipocytes using siRNA. The RNAi treatment was efficient and reduced the mRNA expression of twist1 up to 95% (Fig. 2A, left panel). Twist1 protein levels were also reduced, albeit to a lesser extent (∼35%, Fig. 2A, right panel). This effect was selective because the expression of twist2 mRNA (Fig. 2A) and several reference genes (GAPDH and LRP10, values not shown) was not affected by twist1 siRNA. In contrast to murine data, in vitro downregulation of twist1 significantly reduced FA oxidation by ∼40% (P < 0.05, n = 3), measured as release of CO2 and β-oxidation metabolites into conditioned media (Fig. 2B). This effect was specific and not secondary to off-target or dedifferentiating effects because lipolysis was unaltered by twist1 gene silencing in similar experiments (Fig. 2C). We also measured glucose transport in cultured adipocytes treated with siRNA but did not observe any differences between control- and twist1 siRNA—treated cells (graph not shown).
FIG. 2.
In vitro silencing of twist1 in human cultured adipocytes. Human in vitro–differentiated adipocytes were treated with 50 nmol/l control siRNA or siRNA directed against twist1. A: Twist1 and twist2 mRNA was measured; n = 5 (left panel). Twist1 protein levels were measured by Western blot and normalized for β-actin (right panel). The result shown is a representative experiment. The effect of siRNA at the protein level was less pronounced than that at the mRNA level, a finding that we have observed previously for several other genes (18,31). B: FA oxidation was measured in conditioned media from adipocytes treated with siRNA as above. C: Lipolysis was measured as release of glycerol into conditioned media from adipocytes treated with siRNA as above. D: mRNA levels of a set of genes were measured after siRNA treatment (n = 3–6). mRNA levels were normalized to the reference gene GAPDH. Results shown are mean ± SEM, and all mRNA levels are shown as relative expression (fold change to control siRNA). *P < 0.05.
Selective downregulation of gene expression after twist1 RNAi in human cultured adipocytes.
Given the selective effect of twist1 downregulation, we measured the mRNA levels of several genes involved in FA oxidation (acyl-CoA synthetase [ACC2], CPT-1, acyl-CoA dehydrogenases [SCAD, MCAD, and VLCAD], hydroxyacyl-CoA dehydrogenase [HADHA], and β-ketothiolase [HADHB]). We also determined the expression of genes involved in the regulation of substrate usage (pyruvate dehydrogenase kinases 1 and 4 [PDK1/4]), as well as genes important for brown (UCP-1, PGC-1α) and white (hormone-sensitive lipase [HSL], perilipin [PLIN], cell death–inducing DNA fragmentation factor-α–like effector A [CIDEA], and peroxisome proliferator–activated receptor γ [PPARγ]) adipocyte function. Only CPT-1 and PGC-1α were significantly reduced (∼25%) after siRNA treatment, but there were no significant effects on the expression of other genes involved in FA oxidation (Fig. 2D). In accordance with the unaltered lipolytic capacity in twist1 siRNA–treated cells, the expression of HSL, PLIN, CIDEA, and PPARγ was not changed, further suggesting that there were no off-target effects of siRNA treatment.
Twist1 RNAi suggests a role in the regulation of adipokine production.
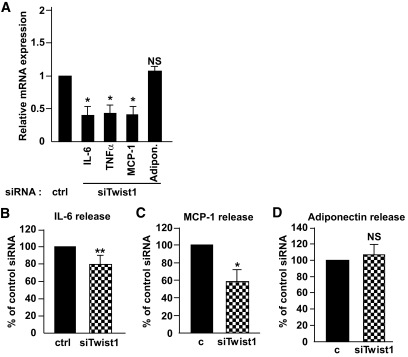
Because Twist1 appears to be involved in inflammatory pathways and in order to screen for additional functional roles of twist1, we assessed the effect of twist1 RNAi on a set of genes coding for adipokines. The expression of IL-6, TNF-α, and MCP-1 was markedly reduced in cells treated with twist1 siRNA compared with control-treated cells (Fig. 3A). This effect was selective because the expression of the fat-specific secreted factor adiponectin was not affected (Fig. 3A). To determine whether mRNA changes were reflected at the protein level, we measured the release of adipokines in conditioned media from the same experiments. Silencing of twist1 reduced the release of IL-6 by ∼20% (P < 0.01, n = 5) and MCP-1 by ∼40% (P < 0.05, n = 3) (Fig. 3B and C), whereas the release of adiponectin was unaltered (P = 0.59, n = 3) (Fig. 3D). The protein levels of TNF-α in the medium were below the detection limit of our assay.
FIG. 3.
Expression and release of IL-6, MCP-1, and adiponectin into conditioned media after siRNA treatment of cultured adipocytes. Human in vitro–differentiated adipocytes were treated with 50 nmol/l control siRNA or siRNA directed against twist1. Expression of the inflammatory genes IL-6, TNF-α, and MCP-1 as well as adiponectin was measured after siRNA treatment (A). mRNA levels were normalized to the reference gene GAPDH. Release of IL-6 (B), MCP-1 (C), and adiponectin (D) was measured in conditioned media; n = 4 (IL-6) and n = 3 (MCP-1/adiponectin). Results shown are mean ± SEM and mRNA levels are shown as relative expression (fold change to control siRNA). **P < 0.01, *P < 0.05.
Twist1 binds to human MCP-1, IL-6, and TNF-α promoters.
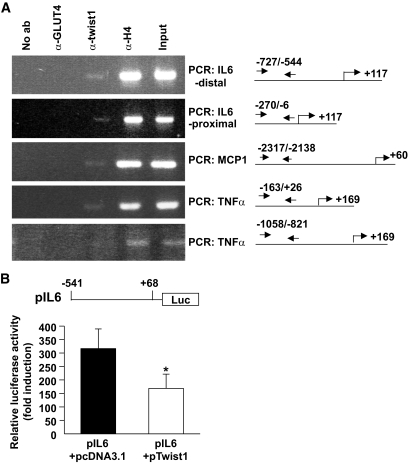
Because twist1 is a transcription factor, it can directly regulate the transcriptional expression of MCP-1, IL-6, and TNF-α by binding to E-boxes in the promoter regions of these genes. Using MatInspector software, we identified putative E-boxes in the promoter regions of these genes and designed PCR primers, which would generate PCR products overlapping them. We then performed ChIP analysis on human in vitro–differentiated adipocytes and were able to amplify E-box–containing regions from MCP-1, TNF-α, as well as IL-6 (Fig. 4A), suggesting that twist1 directly binds to these promoter regions in vivo. The binding of twist1 was specific because we could not detect any PCR products using another set of primers designed to generate a PCR fragment overlapping an E-box more upstream in the TNF-α promoter.
FIG. 4.
ChIP in human in vitro–differentiated adipocytes. A: ChIP was performed on in vitro–differentiated adipocytes. ChIP assays were performed using antibodies specifically recognizing twist1, acetyl-histone H4 (positive control), and GLUT4 (negative control). Input DNA and immunoprecipitated DNA were quantified by PCR using specific primer sets recognizing human MCP-1, IL-6, and TNF-α promoter regions overlapping E-boxes. Regions complementary to the primers, transcriptional start sites (indicated by an arrow), and translational start sites in the promoters are shown to the right. Primer sequences and expected PCR products are listed in supplementary Table 3. B: Undifferentiated 3T3-L1 cells were transiently transfected with pIL6 together with either empty pcDNA3.1 or pTwist1. An internal control (cytomegalovirus-β-galactosidase–containing vector) was included in all transfections. Relative luciferase activity was normalized for β-galactosidase expression and expressed as fold over empty vector (pGL2-basic) + empty pcDNA3.1 or pGL2-basic + pTwist1. Samples were run in triplicate, and the results display mean ± SEM of four independent experiments. *P < 0.05.
To verify the interaction between twist1 and cytokine promoters, we performed transient transfections using a luciferase reporter construct coupled to 609 bp of the human IL-6 promoter (which overlaps the proximal region amplified in the ChIP experiments) together with an expression vector containing the cDNA of human twist1. Assays were performed in undifferentiated 3T3-L1 cells. Coexpression of twist1 significantly reduced the activity of the IL-6 promoter compared with control cells cotransfected with an empty vector (Fig. 4B), suggesting that twist1 indeed influences the transcriptional activity of this promoter.
DISCUSSION
Although studies in mice suggest that twist1 is important in the regulation of BAT energy metabolism and inflammatory pathways, the role of this factor in WAT function has thus far been unknown. This is the first study of twist1 in white fat cells.
Among four different human organs of importance for metabolism, twist1 was expressed only in human WAT. This selective expression pattern is similar to the expression pattern observed in adult mice (14). In contrast, twist2 mRNA could readily be detected in liver, muscle, pancreas, and WAT. In mice, twist2 mRNA is detected in both fat tissue and liver, and ob/ob and db/db mice were reported to display elevated levels of twist protein in fat tissue (24). It is not clear whether these proteins represent twist2 or a mix of both twist proteins. Nevertheless, this suggests that twist2 has a different role than twist1. With regard to intra-organ expression of twist1, our cellular expression analysis, using two independent methods, demonstrates that twist1 expression is considerably higher in adipocytes than in other cell types of human WAT. First, the expression levels in isolated adipocytes were higher than in the corresponding adipose tissue from different donors. Second, when analyzing the expression in different cell fractions, the levels in adipocytes were significantly higher than in macrophages, endothelial cells, or a CD34-, CD31-, and CD14-negative cell population isolated from the same adipose tissue. Twist1 expression also increased during in vitro differentiation of preadipocytes, further supporting a functional role in mature adipocytes. Interestingly, twist1 expression decreases during myoblast and osteoblast differentiation (6–9), supporting the notion that twist1 has tissue-specific functions.
In humans, FA oxidation in WAT is normally of minor significance and accounts for only a small percentage of the total FA oxidation in the body (25). However, in certain specific conditions (e.g., cancer cachexia), lipid oxidation in human white adipocytes may increase significantly (26). PGC-1α is a pivotal regulator in brown fat cells, and overexpression in human white adipocytes results in a more brown phenotype that is characterized by increased FA oxidation and oxygen consumption (19). In murine brown fat cells, twist1 knockdown leads to increased expression of PGC-1α target genes and increased mitochondrial biogenesis (14). Furthermore, high-fat feeding of transgenic mice overexpressing twist1 under the fat-specific adipocyte fatty acid–binding protein promoter results in decreased expression of UCP-1 and FA oxidation genes in BAT. FA oxidation was not directly measured in brown fat cells but was shown to be reduced in C2C12 myotubes (14). These transgenic mice are more prone to diet-induced obesity with increased lipid accumulation in brown adipocytes. Interestingly, even though the adipocyte fatty acid–binding protein promoter is active in WAT as well, there was no effect on gene expression in this tissue, supporting the notion that twist1 selectively and negatively regulates BAT metabolism in mice (14). In contrast to these results, our RNAi experiments in human in vitro–differentiated adipocytes demonstrate that twist1 downregulation reduced FA oxidation as well as the mRNA expression of PGC-1α and the rate-limiting enzyme for FA oxidation, CPT-1. Moreover, the effect on lipid metabolism seems restricted to this process because basal lipolysis was unaltered. Therefore, under the present study conditions, twist1 appears to be a positive regulator of FA oxidation in human white adipocytes, possibly via effects on CPT-1 and PGC-1α. It should be stressed that although twist1 was reported to be expressed at similar levels in murine WAT compared with BAT (14), the functional role of twist1 in murine WAT has thus far not been assessed. At present, it is therefore not possible to establish whether there are qualitative differences in the function of twist1 in human versus murine fat cells or if the contrasting data between the present and previous study (14) merely reflect different roles of twist1 in brown and white adipocytes. Nevertheless, because FA oxidation in white adipocytes is normally of minor physiological importance, it is difficult to speculate on the impact of these findings at the in vivo level. Although recent studies demonstrate the existence of brown adipocytes in adult humans (27–29), it is not possible for ethical reasons to obtain sufficient amounts of human brown fat cells for mechanistic studies in vitro. Therefore, we did not continue with direct mechanistic studies of FA oxidation but searched for additional and possibly more important roles for twist1 in white adipocytes.
Results from twist knockout mice suggest that twist1 and twist2 regulate cytokine signaling by repressing nuclear factor-κB–dependent cytokine activation (5). Our adipocyte gene silencing data also imply a role for twist1 in the regulation of inflammatory cytokines, although somewhat different from that in mice, because mRNA expression of TNF-α, IL-6, and MCP-1 was downregulated and the secretion of the latter two was attenuated in twist1-silenced cells. This effect was specific, because adiponectin mRNA expression and secretion were unaltered. The discrepancy between the findings in knockout mice and our adipocyte gene silencing experiments may be due to species-specific differences, although one cannot exclude the possibility that a partial or complete lack of twist in all cell types (as in the knockout models) may result in different effects compared with cell-specific gene knockdowns in vitro.
Our ChIP analysis demonstrated that twist1 directly interacts with specific promoter regions of all three inflammatory factors, thereby suggesting a direct regulatory role of twist1. Furthermore, IL-6 promoter activity was significantly reduced when twist1 was coexpressed in undifferentiated murine 3T3-L1 cells. This further strengthens the notion that twist1 affects the activity of the human IL-6 promoter via a direct interaction. Admittedly, the effects in the gene reporter assays may at first appear to be in contrast with the siRNA data where transient twist1 silencing reduced IL-6 mRNA expression and secretion. However, it is well known that promoter activity in plasmid DNA can differ significantly from that in an intact chromatin environment (30). In addition, it should be stressed that the reporter assays were performed in undifferentiated murine fibroblasts using human twist1 and a short part of the human IL-6 promoter sequence. Therefore, the suppressive effect of twist1 on the IL-6 promoter could be secondary to the cell type used in these experiments and/or absence of appropriate endogenous regulatory cofactors. The most relevant interpretation of the reporter data is that twist1 binds and regulates the IL-6 promoter. Taken together, our results from siRNA, ChIP, and reporter assays reflect the same mechanism: a regulatory role of twist1 in inflammatory pathways.
In summary, the transcription factor twist1 is expressed predominantly in human white adipocytes. In vitro gene silencing results in attenuated expression and secretion of several inflammatory adipokines but also in reduced FA oxidation, which is in contrast to findings in mice. We therefore propose a novel function for twist1 as a possible regulator of inflammatory adipokines. Given the rather low quantitative role of FA oxidation in human WAT, the effects of twist1 on this process may be less important.
ACKNOWLEDGMENTS
This work was supported by several grants from the Swedish Heart and Lung Foundation, Swedish Diabetes Association, Storstockholms Diabetesförening, Torsten och Ragnar Söderberg Foundation, Novo Nordisk Foundation, the European Union (HEPADIP, LSHM-CT-2005-018734), ADAPT (HEALTH-F2-2008-201100), COST action (BM0602), NordForsk (SYSDIET-070014), the Swedish Medical Association, and the Swedish Diabetes Foundation.
No potential conflicts of interest relevant to this article were reported.
We thank Elisabeth Dungner, Eva Sjölin, Kerstin Wåhlén, Gaby Åström, Britt-Marie Leijonhufvud, and Katarina Hertel for excellent technical assistance. We thank Chiara Costanzo, University of Verona, Italy, for the pIL6 construct.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Wang SM, Coljee VW, Pignolo RJ, Rotenberg MO, Cristofalo VJ, Sierra F: Cloning of the human twist gene: its expression is retained in adult mesodermally-derived tissues. Gene 1997; 187: 83– 92 [PubMed] [Google Scholar]
- 2.Castanon I, Baylies MK: A Twist in fate: evolutionary comparison of Twist structure and function. Gene 2002; 287: 11– 22 [DOI] [PubMed] [Google Scholar]
- 3.El Ghouzzi V, Legeai-Mallet L, Benoist-Lasselin C, Lajeunie E, Renier D, Munnich A, Bonaventure J: Mutations in the basic domain and the loop-helix II junction of TWIST abolish DNA binding in Saethre-Chotzen syndrome. FEBS Lett 2001; 492: 112– 118 [DOI] [PubMed] [Google Scholar]
- 4.Chen ZF, Behringer RR: Twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev 1995; 9: 686– 699 [DOI] [PubMed] [Google Scholar]
- 5.Sosić D, Richardson JA, Yu K, Ornitz DM, Olson EN: Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell 2003; 112: 169– 180 [DOI] [PubMed] [Google Scholar]
- 6.Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, Justice MJ, Karsenty G: A twist code determines the onset of osteoblast differentiation. Dev Cell 2004; 6: 423– 435 [DOI] [PubMed] [Google Scholar]
- 7.Isenmann S, Arthur A, Zannettino AC, Turner JL, Shi S, Glackin CA, Gronthos S: TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment. Stem Cells 2009; 27: 2457– 2468 [DOI] [PubMed] [Google Scholar]
- 8.Spicer DB, Rhee J, Cheung WL, Lassar AB: Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science 1996; 272: 1476– 1480 [DOI] [PubMed] [Google Scholar]
- 9.Gong XQ, Li L: Dermo-1, a multifunctional basic helix-loop-helix protein, represses MyoD transactivation via the HLH domain, MEF2 interaction, and chromatin deacetylation. J Biol Chem 2002; 277: 12310– 12317 [DOI] [PubMed] [Google Scholar]
- 10.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA: Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004; 117: 927– 939 [DOI] [PubMed] [Google Scholar]
- 11.Kahn SE, Hull RL, Utzschneider KM: Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 2006; 444: 840– 846 [DOI] [PubMed] [Google Scholar]
- 12.Niesner U, Albrecht I, Janke M, Doebis C, Loddenkemper C, Lexberg MH, Eulenburg K, Kreher S, Koeck J, Baumgrass R, Bonhagen K, Kamradt T, Enghard P, Humrich JY, Rutz S, Schulze-Topphoff U, Aktas O, Bartfeld S, Radbruch H, Hegazy AN, Löhning M, Baumgart DC, Duchmann R, Rudwaleit M, Häupl T, Gitelman I, Krenn V, Gruen J, Sieper J, Zeitz M, Wiedenmann B, Zipp F, Hamann A, Janitz M, Scheffold A, Burmester GR, Chang HD, Radbruch A: Autoregulation of Th1-mediated inflammation by twist1. J Exp Med 2008; 205: 1889– 1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharif MN, Sosic D, Rothlin CV, Kelly E, Lemke G, Olson EN, Ivashkiv LB: Twist mediates suppression of inflammation by type I IFNs and Axl. J Exp Med 2006; 203: 1891– 1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan D, Fujimoto M, Lopes A, Wang YX: Twist-1 is a PPARdelta-inducible, negative-feedback regulator of PGC-1alpha in brown fat metabolism. Cell 2009; 137: 73– 86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arner P, Stenson BM, Dungner E, Näslund E, Hoffstedt J, Ryden M, Dahlman I: Expression of six transmembrane protein of prostate 2 in human adipose tissue associates with adiposity and insulin resistance. J Clin Endocrinol Metab 2008; 93: 2249– 2254 [DOI] [PubMed] [Google Scholar]
- 16.Bourlier V, Zakaroff-Girard A, Miranville A, De Barros S, Maumus M, Sengenes C, Galitzky J, Lafontan M, Karpe F, Frayn KN, Bouloumié A: Remodeling phenotype of human subcutaneous adipose tissue macrophages. Circulation 2008; 117: 806– 815 [DOI] [PubMed] [Google Scholar]
- 17.van Harmelen V, Skurk T, Hauner H: Primary culture and differentiation of human adipocyte precursor cells. Methods Mol Med 2005; 107: 125– 135 [DOI] [PubMed] [Google Scholar]
- 18.Stenson BM, Rydén M, Steffensen KR, Wåhlén K, Pettersson AT, Jocken JW, Arner P, Laurencikiene J: Activation of liver X receptor (LXR) regulates substrate oxidation in white adipocytes. Endocrinology 2009; 150: 4104– 4113 [DOI] [PubMed] [Google Scholar]
- 19.Tiraby C, Tavernier G, Lefort C, Larrouy D, Bouillaud F, Ricquier D, Langin D: Acquirement of brown fat cell features by human white adipocytes. J Biol Chem 2003; 278: 33370– 33376 [DOI] [PubMed] [Google Scholar]
- 20.Hellmér J, Arner P, Lundin A: Automatic luminometric kinetic assay of glycerol for lipolysis studies. Anal Biochem 1989; 177: 132– 137 [DOI] [PubMed] [Google Scholar]
- 21.Ryden M, Dicker A, van Harmelen V, Hauner H, Brunnberg M, Perbeck L, Lonnqvist F, Arner P: Mapping of early signaling events in tumor necrosis factor-alpha-mediated lipolysis in human fat cells. J Biol Chem 2002; 277: 1085– 1091 [DOI] [PubMed] [Google Scholar]
- 22.Faggioli L, Costanzo C, Donadelli M, Palmieri M: Activation of the Interleukin-6 promoter by a dominant negative mutant of c-Jun. Biochim Biophys Acta 2004; 1692: 17– 24 [DOI] [PubMed] [Google Scholar]
- 23.Pettersson AT, Laurencikiene J, Nordström EA, Stenson BM, van Harmelen V, Murphy C, Dahlman I, Rydén M: Characterization of the human CIDEA promoter in fat cells. Int J Obes (Lond) 2008; 32: 1380– 1387 [DOI] [PubMed] [Google Scholar]
- 24.Lee YS, Lee HH, Park J, Yoo EJ, Glackin CA, Choi YI, Jeon SH, Seong RH, Park SD, Kim JB: Twist2, a novel ADD1/SREBP1c interacting protein, represses the transcriptional activity of ADD1/SREBP1c. Nucleic Acid Res 2003; 31: 7165– 7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frayn KN, Langin D, Karpe F: Fatty acid-induced mitochondrial uncoupling in adipocytes is not a promising target for treatment of insulin resistance unless adipocyte oxidative capacity is increased. Diabetologia 2008; 51: 394– 397 [DOI] [PubMed] [Google Scholar]
- 26.Laurencikiene J, Stenson BM, Arvidsson Nordström E, Agustsson T, Langin D, Isaksson B, Permert J, Rydén M, Arner P: Evidence for an important role of CIDEA in human cancer cachexia. Cancer Res 2008; 68: 9247– 9254 [DOI] [PubMed] [Google Scholar]
- 27.Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, Kolodny GM, Kahn CR: Identification and importance of brown adipose tissue in adult humans. N Engl J Med 2009; 360: 1509– 1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ: Cold-activated brown adipose tissue in healthy men. N Engl J Med 2009; 360: 1500– 1508 [DOI] [PubMed] [Google Scholar]
- 29.Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerbäck S, Nuutila P: Functional brown adipose tissue in healthy adults. N Engl J Med 2009; 360: 1518– 1525 [DOI] [PubMed] [Google Scholar]
- 30.Smith CL, Hager GL: Transcriptional regulation of mammalian genes in vivo: a tale of two templates. J Biol Chem 1997; 272: 27493– 27496 [DOI] [PubMed] [Google Scholar]
- 31.Rydén M, Jocken J, van Harmelen V, Dicker A, Hoffstedt J, Wirén M, Blomqvist L, Mairal A, Langin D, Blaak E, Arner P: Comparative studies of the role of hormone-sensitive lipase and adipose triglyceride lipase in human fat cell lipolysis. Am J Physiol Endocrinol Metab 2007; 292: E1847– E1855 [DOI] [PubMed] [Google Scholar]