14-3-3 proteins bind and regulate phosphorylated client proteins involved in diverse biological processes in eukaryotic organisms. This work reports that a tomato 14-3-3 protein regulates plant immunity by altering the abundance of a positive regulator of cell death.
Abstract
Programmed cell death (PCD) is triggered when Pto, a Ser-Thr protein kinase, recognizes either the AvrPto or AvrPtoB effector from Pseudomonas syringae pv tomato. This PCD requires mitogen-activated protein kinase kinase kinase (MAPKKK α ) as a positive regulator in tomato (Solanum lycopersicum) and Nicotiana benthamiana. To examine how PCD-eliciting activity of the tomato MAPKKK α protein is regulated, we screened for MAPKKK α -interacting proteins in tomato and identified a 14-3-3 protein, TFT7. Virus-induced gene silencing using the TFT7 gene in N. benthamiana compromised both Pto- and MAPKKK α -mediated PCD, and coexpression of TFT7 with tomato MAPKKK α enhanced MAPKKK α -mediated PCD. TFT7 was also required for PCD associated with several other disease resistance proteins and contributed to resistance against P. syringae pv tomato. Coexpression of TFT7 with MAPKKK α in vivo caused increased accumulation of the kinase and enhanced phosphorylation of two MAP kinases. TFT7 protein contains a phosphopeptide binding motif that is present in human 14-3-3 ϵ, and substitutions in this motif abolished interaction with MAPKKK α in vivo and also the PCD-enhancing activity of TFT7. A 14-3-3 binding motif, including a putative phosphorylated Ser-535, is present in the C-terminal region of MAPKKK α. An S535A substitution in MAPKKK α reduced interaction with TFT7 and both PCD-eliciting ability and stability of MAPKKK α. Our results provide new insights into a role for 14-3-3 proteins in regulating immunity-associated PCD pathways in plants.
INTRODUCTION
Plants often respond to attempted pathogen infection by undergoing a localized programmed cell death (PCD) referred to as the hypersensitive response (HR) (van Doorn and Woltering, 2005). In particular, the HR typically occurs upon recognition of a pathogen effector protein by a plant resistance (R) protein complex. Pto, a Ser/Thr protein kinase in tomato (Solanum lycopersicum) recognizes the effectors AvrPto and AvrPtoB from Pseudomonas syringae pv tomato, leading to host resistance to bacterial speck disease (Tang et al., 1996; Kim et al., 2002). This recognition event requires Prf, which belongs to the coiled coil–nucleotide binding site–leucine-rich repeat class of R proteins in order to activate downstream signaling pathways to trigger PCD (Salmeron et al., 1996; Mucyn et al., 2006). Eventually, this immune response results in reduced pathogen growth and restriction of pathogen movement from the sites of attempted infection.
Several components that are involved in Pto/Prf-mediated PCD in tomato and Nicotiana benthamiana have been identified and characterized, including protein kinases, an F box E3 ligase, and transcription factors (Zhou et al., 1997; Mysore et al., 2002; Ekengren et al., 2003; del Pozo et al., 2004; van den Burg et al., 2008). One of these components is mitogen-activated protein kinase kinase kinase α (MAPKKK α ), which functions as a positive regulator of Pto-mediated PCD (del Pozo et al., 2004). The gene encoding tomato MAPKKK α (Sl MAPKKK α ; previously Le MAPKKK α ) was identified from a large screen for genes involved in Pto/Prf-mediated PCD, using potato virus X–based virus-induced gene silencing (VIGS) in N. benthamiana. When MAPKKK α gene expression was attenuated by VIGS in tomato or N. benthamiana, PCD mediated by Pto/AvrPto, AvrPtoB1-387, and Cf9/Avr9 was compromised (del Pozo et al., 2004; Rosebrock et al., 2007). Expression of either the full-length tomato MAPKKK α or the kinase domain alone in leaves of N. benthamiana caused PCD (del Pozo et al., 2004). Components of two MAP kinase cascades were discovered to act downstream of MAPKKK α based on epistasis analysis. Moreover, expression of MAPKKK α in leaves of N. benthamiana activated two MAP kinases, MPK2 and MPK3, which are tomato orthologs of salicylic acid-induced protein kinase (SIPK) and wounding-induced protein kinase (WIPK) (Pedley and Martin, 2004). These observations suggest that MAPKKK α protein activity is likely to be tightly controlled to prevent unnecessary induction of MAPK cascades and the resulting PCD. However, the mechanism by which MAPKKK α is regulated has been unclear.
In mammalian systems, several proteins have been characterized as regulators of MAPKKK protein activities, particularly in their role as inducers of apoptosis. For example, among four MEK (mitogen-activated protein kinase/extracellular signal-regulated kinase kinase) kinases in humans, MEKK1 plays a key role in apoptosis induced by extracellular stimuli, such as growth factors, cytokines, and UV irradiation (Fanger et al., 1997). Its cleavage by caspase 3 releases the C-terminal kinase domain from the N-terminal regulatory region where a 14-3-3 protein binds, resulting in activation of downstream signaling to induce apoptosis (Cardone et al., 1997). Another human kinase, MEKK3, is phosphorylated on Ser-526 to promote its catalytic activity and it then recruits a 14-3-3 protein (Fritz et al., 2006). Binding of the 14-3-3 protein at this residue blocks dephosphorylation by the protein Ser/Thr phosphatase, PP2A, and probably preserves activated forms of MEKK3 (Fritz et al., 2006). Two isoforms of 14-3-3 proteins, 14-3-3 ζ and 14-3-3 ϵ, are known to interact with MEKK1, MEKK2, and MEKK3 (Fanger et al., 1998).
14-3-3 proteins are conserved phosphopeptide binding proteins in eukaryotic organisms (Ferl et al., 2002; Ferl, 2004; Bridges and Moorhead, 2005). Two 14-3-3 binding motifs have been identified in certain 14-3-3 client proteins: RSXpSXP and RXXXpSXP, where pS and X represent a phosphoserine and any amino acid, respectively. 14-3-3 proteins typically function as a homodimer or heterodimer and are involved in many biological processes, such as primary metabolism, signal transduction, cell cycle control, apoptosis, protein trafficking, transcription, and stress responses (Bridges and Moorhead, 2005; Darling et al., 2005; Morrison, 2009). These proteins have various roles including (1) stabilizing client proteins by preventing their accessibility to proteases and phosphatases, (2) triggering a conformational change to activate or inhibit a client proteins' activities, (3) acting as scaffolding proteins to recruit two different target proteins at the right time and location, and (4) shuttling target proteins between different organelles or locations in the cell (Darling et al., 2005). Each eukaryotic organism has multiple 14-3-3 protein isoforms, with seven occurring in human, two in yeast, and approximately 12 in plants, including Arabidopsis thaliana, tomato, and tobacco (Ferl et al., 2002; Bridges and Moorhead, 2005).
14-3-3 proteins are also regulators of important biological processes in plants, such as metabolism, transcription, organellar protein trafficking, and stress responses (Roberts et al., 2002; Sehnke et al., 2002; Roberts, 2003). There have been some indications that 14-3-3 proteins play a role in plant immune responses. For example, expression of several 14-3-3 genes was induced in the incompatible interaction between barley (Hordeum vulgare) and avirulent strains of powdery mildew fungus (Erysiphe graminis f sp hordei) and between soybean (Glycine max) and P. syringae pv glycinea (Seehaus and Tenhaken, 1998; Finnie et al., 2002). In the barley/powdery mildew interaction, a complex between 14-3-3 proteins and an H+-ATPase might be important for triggering PCD (Finnie et al., 2002). In addition, expression of three out of 10 14-3-3 genes in tomato was specifically induced during Cf9/Avr9-mediated host immunity (Roberts and Bowles, 1999). Most recently, an Arabidopsis 14-3-3 protein was reported to interact with the RPW8.2 protein and to play a role in certain host responses mediated by that R protein (Yang et al., 2009). However, the extent to which 14-3-3 proteins play a role in plant immunity and their underlying mechanisms are essentially unknown. In addition, although there are two cases where plant 14-3-3 proteins were shown to interact with plant MAPKs (Lalle et al., 2005; Chang et al., 2009), there are no reports related to a possible role of these proteins in controlling plant MAPK cascades as they do in mammalian systems.
In order to investigate how MAPKKK α protein activity is controlled in plants, we first identified tomato MAPKKK α -interacting proteins by screening a tomato yeast two-hybrid library. We discovered a tomato 14-3-3 protein, TFT7, as a MAPKKK α -interacting protein and report a role for TFT7 in regulation of PCD. TFT7 was found to increase the accumulation of MAPKKK α in plant cells and to be required for PCD caused by Pto- and MAPKKK α -mediated signaling. In addition, we discovered that TFT7 plays a role in several other R protein–mediated pathways, indicating it plays a broad and important role in plant immunity.
RESULTS
A Tomato 14-3-3 Protein Interacts with MAPKKK α
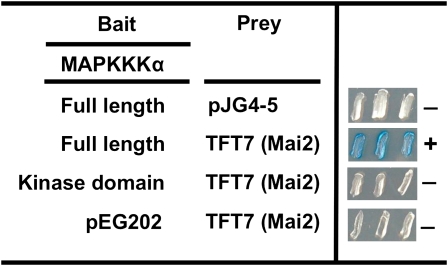
The full-length or kinase domain (KD) alone of tomato MAPKKK α triggers PCD when either one is overexpressed in leaves of tomato or N. benthamiana (del Pozo et al., 2004). To understand molecular events underlying this PCD, we used a yeast two-hybrid screen to identify MAPKKK α -interacting proteins from tomato. Both the full-length and KD of tomato MAPKKK α were used separately as baits to screen 2 × 106 initial transformants from a prey library generated from Rio Grande-PtoR tomato leaves inoculated with P. syringae pv tomato (Zhou et al., 1995). From the screening, a clone was identified whose protein interacted with the full-length MAPKKK α but not with the KD alone (Figure 1).
Figure 1.
A Tomato 14-3-3 Protein Interacts with Full-Length Tomato MAPKKK α in Yeast.
A tomato cDNA prey library in pJG4-5 vector was screened to search for proteins that interact with either the full-length or KD only of MAPKKK α in a bait vector, pEG202. Blue (or dark) patches indicate positive interaction. Photographs were taken 2 d after streaking onto plates with X-gal. +, positive interaction; −, no interaction.
[See online article for color version of this figure.]
This clone contained the full open reading frame of a protein that is identical to TFT7, a previously known tomato 14-3-3 protein (Roberts and Bowles, 1999). Tomato has 12 isoforms of 14-3-3 proteins and, based on phylogenetic analysis with these proteins from tomato and 10 available from tobacco, TFT7 is most similar to Isoform A (GenBank accession number S76737) from tobacco (see Supplemental Figures 1 and 2 online).
TFT7 Regulates PCD Mediated by Pto and MAPKKK α
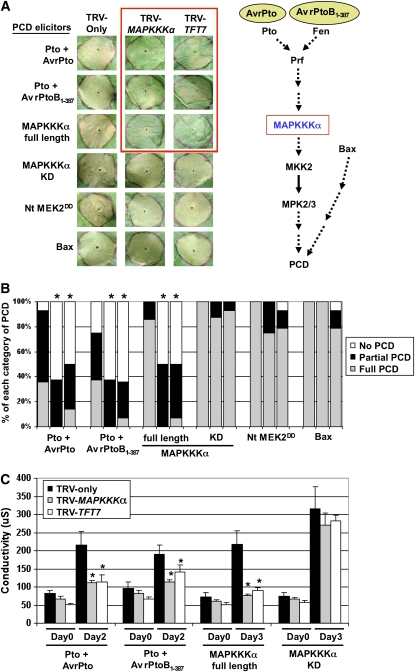
We first assessed whether TFT7 is involved in PCD mediated by Pto or MAPKKK α. A fragment containing nucleotides 83 to 369 of the TFT7 gene was cloned into a tobacco rattle virus (TRV) vector and used for VIGS in N. benthamiana (see Supplemental Figure 2 online). N. benthamiana was used instead of tomato due to higher efficiency of VIGS and better transient expression of PCD elicitors in this species. The abundance of transcripts homologous to TFT7 was greatly reduced in the silenced plants, compared with TRV-only infected plants (see Supplemental Figure 3 online). Six different PCD elicitors, as shown in Figure 2A, were transiently expressed, and PCD was monitored over the subsequent 5 d. To induce expression of full-length Sl MAPKKK α, Sl MAPKKK α -KD, or Nt MEK2DD (a constitutively active form of Nt MAP kinase kinase 2), 2 μ M estradiol was applied 48 h after agroinfiltration, and for Bax (a mouse pro-apoptotic protein), 10 μ M dexamethasone was applied 48 h after agroinfiltration. The degree of PCD was visually scored and categorized into three classes, based on how much of the Agrobacterium tumefaciens–infiltrated area developed cell death: 80 to 100% (+), 10 to 80% ( ± ), or <10% (−). Silencing with TFT7 in N. benthamiana greatly reduced PCD induced by Pto/AvrPto, Pto/AvrPtoB1-387, and full-length MAPKKK α but had little or no effect on PCD induced by MAPKKK α -KD, MEK2DD, or Bax, as shown in Figures 2A and 2B. The visual scores of PCD were substantiated by an electrolyte leakage assay, as shown in Figure 2C. This pattern of PCD suppression was similar to the effect of silencing MAPKKK α (Figures 2A to 2C). Based on these results and a previous signaling model (del Pozo et al., 2004), TFT7 appears to act at the same point in this PCD pathway as MAPKKK α.
Figure 2.
TFT7 Positively Regulates PCD Mediated by Pto and MAPKKK α.
(A) Silencing with TFT7 in N. benthamiana diminishes PCD mediated by Pto and MAPKKK α. TRV was used to silence MAPKKKα or TFT7 in N. benthamiana. TRV-only was used as a control. The PCD elicitors shown were expressed by infiltrating Agrobacterium carrying T-DNA with the genes indicated into the silenced leaves; the infiltrated area is outlined by a circle on each leaf. Treatments inside the red box showed diminished PCD. Photos were taken 4 d after agroinfiltration for Pto/AvrPto and Pto/AvrPtoB1-387 and 5 d after agroinfiltration for the others. The PCD pathway on the right is derived from del Pozo et al. (2004).
(B) The degree of elicitor-induced PCD as shown in the x axis was monitored visually and categorized into three classes: +, complete (80 to 100%); +/−, partial (10 to 80%); −, none (<10%). With the number of spots in each class, the degree of PCD was statistically analyzed, using Fisher's exact test (P < 0.05). Asterisks indicate significant reduction of PCD by silencing of either MAPKKKα (second lane for each PCD elicitor) or TFT7 (third lane for each PCD elicitor), compared with TRV-only plants (first lane for each PCD elicitor). The results are representative of at least three independent experiments, and in each experiment, four to six plants and two to three leaves were used (8 to18 infiltrations for each PCD elicitor).
(C) To measure elicitor-induced electrolyte leakage as shown in the x axis, two leaf discs (9 mm in diameter) were collected from the agroinfiltrated area at the indicated time points and incubated in 2 mL of water for 2 h at 25 ° C. Electrolyte leakage was measured, using an Acorn CON 5 ion conductivity meter. Asterisks indicate a significant reduction (t test; P < 0.05) in ion leakage by silencing of either MAPKKKα or TFT7, compared with TRV-only plants. Data presented are means of six infiltrated areas (one per plant), and error bars indicate standard errors.
TFT7 Contributes to Host Resistance to P. syringae pv tomato and Is Required for PCD Induction Mediated by Diverse R Proteins
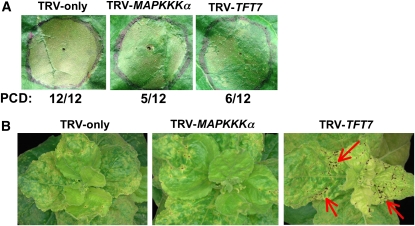
To further examine if TFT7 affects immunity-associated PCD, we assessed the host response to P. syringae pv tomato DC3000, a bacterium that expresses the HopQ1-1 effector, which is recognized by an R gene in N. benthamiana (Wei et al., 2007). Leaves of N. benthamiana silenced for either MAPKKK α or TFT7 were infiltrated with 5 × 106 colony-forming units (CFU)/mL of DC3000, and PCD formation was monitored over the subsequent 3 d. PCD was delayed and weaker in both MAPKKK α - and TFT7-silenced plants compared with TRV-only plants, further supporting a role for these proteins as positive regulators of PCD (Figure 3A).
Figure 3.
Silencing of TFT7 in N. benthamiana Reduces Immunity-Associated PCD and Resistance against the Bacterial Pathogen P. syringae pv tomato.
(A) Assay for immunity-associated PCD. A suspension of 5 × 106 CFU/mL of P. syringae pv tomato strain DC3000 was infiltrated into the circled area of leaves of gene-silenced N. benthamiana plants. The photos were taken 44 h after inoculation, and the numbers below the images indicate the number of regions that developed PCD over the total number of infiltrated regions. The results are representative of three independent experiments (12 different infiltrations per each experiment).
(B) Assay for bacterial speck disease. A suspension of 2 × 104 CFU/mL of P. syringae pv tomato strain DC3000 ΔhopQ1-1/ΔavrPto/ΔavrPtoB was inoculated into gene-silenced N. benthamiana plants, and the photos were taken 5 d later. Necrotic lesions were more severe on plants silenced for TFT7 (red arrows) compared with the MAPKKKα and TRV-only plants. The photos are representative of three independent experiments (four plants for each experiment).
We next examined whether TFT7 or MAPKKK α affect host susceptibility to a virulent P. syringae pv tomato strain. For this assay, we used a DC3000 strain that lacks HopQ1-1, as it is a pathogen of N. benthamiana and is attenuated for virulence due to deletions in the avrPto/avrPtoB genes (Kvitko et al., 2009). A low titer (2 × 104 CFU/mL) of the DC3000 ΔhopQ1-1/ΔavrPto/ΔavrPtoB strain was infiltrated into leaves of either MAPKKK α- or TFT7-silenced plants. Disease symptoms and bacterial growth were monitored for 6 d after infiltration. Although there was no significant difference in bacterial growth (see Supplemental Figure 4 online), more severe disease symptoms developed on leaves of TFT7-silenced plants than on leaves of MAPKKK α-silenced or TRV-only plants (Figure 3B). TFT7 may therefore play an additional role compared with MAPKKK α in basal defense against this bacterial pathogen.
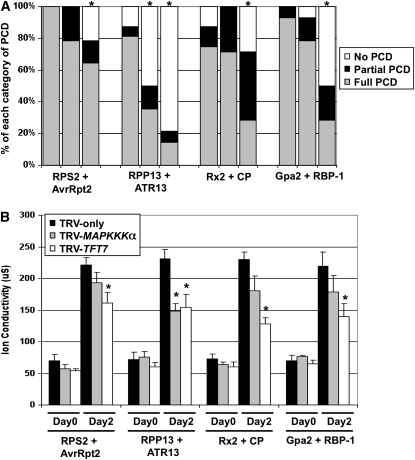
The potentially broader role of TFT7 in plant defense prompted us to examine if it also contributes to the host response against other elicitors of PCD. Four additional PCD-inducing R gene/effector gene pairs, originating from diverse organisms, were selected for this experiment: Arabidopsis RPS2/AvrRpt2 from P. syringae (Day et al., 2005), Arabidopsis RPP13/ATR13Emco5Δ 41aa from the oomycete Hyaloperonospora parasitica (Rentel et al., 2008), potato (Solanum tuberosum) Rx2/coat protein from PVX (Bendahmane et al., 2000), and potato Gpa2/RBP-1 from the potato cyst nematode (Sacco et al., 2009). PCD was monitored visually for 6 d after agroinfiltration of these gene pairs into leaves of TFT7-silenced plants and compared with MAPKKK α-silenced or TRV-only plants (Figure 4A). PCD normally elicited by these four gene pairs was significantly diminished in TFT7-silenced plants. However, three PCD responses induced by RPS2/AvrRpt2, Rx2/CP, and Gpa2/RBP-1 were notably unaffected in plants silenced for MAPKKK α. These results were substantiated using an electrolyte leakage assay (Figure 4B). Thus, TFT7 appears to play a role in diverse immunity-associated PCD responses in N. benthamiana.
Figure 4.
Silencing with TFT7 in N. benthamiana Compromises PCD Associated with Multiple R Gene/Effector Gene Interactions.
(A) The degree of PCD induced by four R gene/effector gene interactions was visually measured as described in Figure 2B and statistically analyzed using Fisher's exact test (P < 0.05). Asterisks indicate significant reduction of PCD by silencing of either MAPKKKα (second lane for each PCD elicitor) or TFT7 (third lane for each PCD elicitor) compared with PCD in TRV-only plants (first lane for each PCD elicitor). The results are from two independent experiments, and in each experiment, four to six plants and two to three leaves were used (8 to18 infiltrations for each PCD elicitor).
(B) Electrolyte leakage was measured as described in Figure 2C. Asterisks indicate significant reduction (t test; P < 0.05) in ion leakage by silencing of either MAPKKKα or TFT7 compared with TRV-only plants. Data presented are means of six infiltrated areas (one per plant), and error bars indicate standard errors.
The Phosphopeptide Binding Motif of TFT7 Is Required for in Vivo Interaction with MAPKKK α and for Enhancement of MAPKKK α -Mediated PCD
The three-dimensional structures of many 14-3-3 proteins are known, including the seven 14-3-3 proteins expressed by humans (Yang et al., 2006). Based on amino acid similarity ( ∼77%; E-value: 8.92e-91), 14-3-3 ϵ is the human 14-3-3 that is most similar to TFT7. The human 14-3-3 ϵ protein interacts with MEKKs and plays an important role in apoptosis induced by extracellular stimuli (Fanger et al., 1997). We used SWISS-MODEL (http://swissmodel.expasy.org) to compare these proteins and found that TFT7 is predicted to have nine helices and is structurally highly similar to human 14-3-3 ϵ (see Supplemental Figures 5 and 6 online). The 14-3-3 ϵ structure solved by x-ray crystallography is bound to a phosphoserine peptide, RRARpSAP (Protein Data Bank number 2br9A). Three amino acid residues (R57-R130-Y131) in 14-3-3 ϵ form a contact surface for binding to the phosphoserine peptide, and these residues are perfectly conserved in the TFT7 protein (R58-R131-Y132), as shown in Supplemental Figure 6 online.
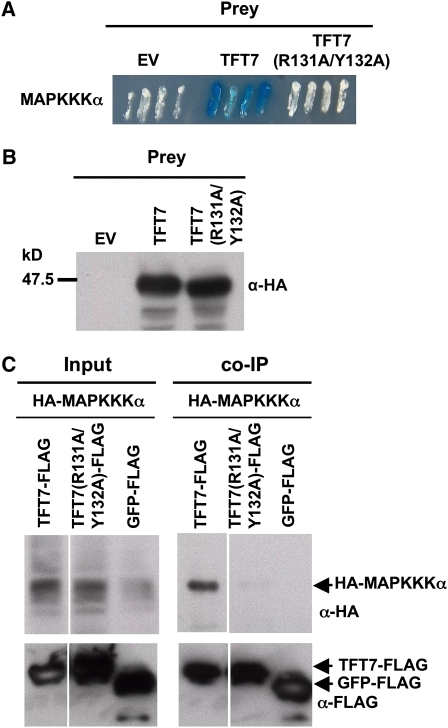
Based on previous results with 14-3-3 ϵ, we hypothesized that R131 and Y132 of TFT7 might be important for its binding to MAPKKK α. To test this hypothesis, Ala substitutions were made at the R131 and Y132 positions of TFT7 to generate TFT7(R131A/Y132A). Using the yeast two-hybrid system, we found that TFT7 interacted strongly with full-length MAPKKK α, whereas TFT7(R131A/Y132A) did not interact (Figure 5A). Both wild-type TFT7 and TFT7(R131A/Y132A) were expressed in yeast (Figure 5B).
Figure 5.
Substitutions in the Phosphopeptide Binding Motif of TFT7 Abolish Interaction of TFT7 with MAPKKK α in Yeast and Plant Cells.
(A) TFT7 or TFT7(R131A/Y132A) genes in the prey vector were analyzed in a yeast two-hybrid system with Sl MAPKKKα in the bait vector. Yeast transformants were streaked onto plates with X-gal, and photographs were taken 2 d later. Blue (or dark) patches indicate positive interaction.
(B) Expression of the TFT7 and TFT7(R131A/Y132A) proteins in the yeast two-hybrid system was verified using immunoblotting with an anti-HA antibody.
(C) The TFT7-FLAG or TFT7(R131A/Y132A) and HA-MAPKKK α proteins were coexpressed in N. benthamiana. Estradiol was applied to the leaves 48 h after agroinfiltration. Leaf discs were collected for protein extraction 24 h after estradiol treatment for co-IP. GFP-FLAG was used a negative control. Immunoblotting was performed with anti-HA for MAPKKK α and with anti-FLAG for TFT7 or GFP.
[See online article for color version of this figure.]
To examine the possible effect of these mutations on the in vivo interaction between TFT7 and MPKKK α in plant cells, we conducted coimmunoprecipitation (co-IP) experiments. We first checked protein expression of the wild type and the variant form of TFT7 fused with a FLAG tag. Both TFT7 proteins were expressed at similar levels in N. benthamiana (see Supplemental Figure 7 online). Coexpression experiments revealed that MAPKKK α was more abundant when coexpressed with wild-type TFT7 than with TFT7(R131A/Y132A) (see next section). To minimize the effect of this difference on our co-IP experiments, we adjusted the input MAPKKK α protein to be the same for the two TFT7 interaction experiments. Abundant HA-MAPKKK α coimmunoprecipitated with TFT7-FLAG, whereas little HA-MAPKKK α was observed to co-IP with TFT7(R131A/Y132A) or green fluorescent protein (GFP)-FLAG control (Figure 5C). These results indicate that the R131 and Y132 residues, which lie in a phosphopeptide binding motif of TFT7, are important for interaction with MAPKKK α in vivo.
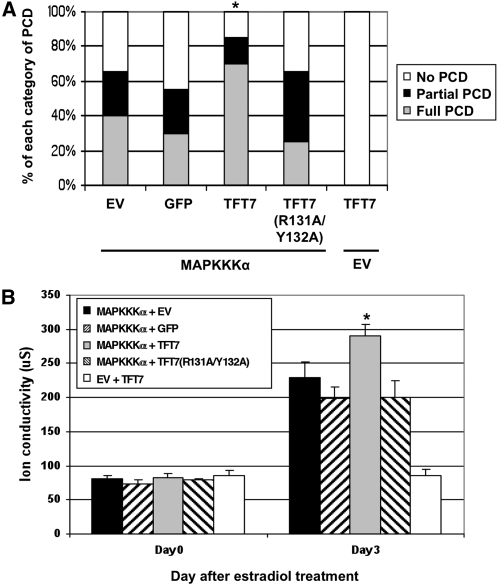
To determine if substitutions in the phosphopeptide binding motif affect TFT7 function as a positive regulator of PCD, we first examined whether TFT7 has a gain-of-function effect on PCD. For this assay, TFT7-FLAG protein was coexpressed with HA-MAPKKK α controlled by an estradiol-inducible system using an Agrobacterium-mediated transient assay. In this experiment, we adjusted the Agrobacterium concentration and the amount of estradiol to minimize the strength of MAPKKK α -induced PCD. PCD was induced and monitored over the subsequent 5 d, and the degree of PCD was visually scored and categorized, as shown in Figures 2A and 2B. Coexpression of TFT7-FLAG significantly enhanced PCD induced by HA-MAPKKK α by 30 to 40%, compared with the controls of an empty vector or coexpression of GFP-FLAG (Figure 6A). By contrast, coexpression of TFT7(R131A/Y132A)-FLAG with HA-MAPKKK α did not enhance PCD beyond the level seen with HA-MAPKKK α alone (Figure 6A). Expression of TFT7-FLAG alone did not induce PCD. This PCD enhancement by TFT7, but not by TFT7(R131A/Y132A), was substantiated by an electrolyte leakage assay (Figure 6B). These results indicate that physical interaction of TFT7 with MAPKKK α is critical for PCD-enhancing activity of TFT7.
Figure 6.
Substitutions in the Phosphopeptide Binding Motif of TFT7 Abolish Its Ability to Enhance PCD Induced by MAPKKK α.
(A) Coexpression of TFT7, but not TFT7(R131A/Y132A), enhances PCD induced by Sl MAPKKK α. TFT7-FLAG, TFT7(R131A/Y132A)-FLAG, or GFP-FLAG under control of the cauliflower mosaic virus 35S promoter and HA-MAPKKKα controlled by an estradiol-inducible system were coexpressed in N. benthamiana by Agrobacterium-mediated transient transformation. Estradiol was applied 48 h after agroinfiltration, and the degree of PCD was scored 5 d later and statistically analyzed as described in Figure 2B. Asterisks indicate significant enhancement of PCD based on Fisher's exact test (P < 0.05). Results are representative of three independent experiments (20 infiltrations for each experiment).
(B) Electrolyte leakage was measured as described in Figure 2C. Asterisks indicate significant increase (t test; P < 0.05) in ion leakage by coexpression of TFT7 with MAPKKK α. Data presented are means of six infiltrated areas (one per plant), and bars indicate standard errors.
TFT7 Increases Protein Abundance of MAPKKK α and Enhances in Vivo Phosphorylation of Two MAP Kinases Downstream of MAPKKK α
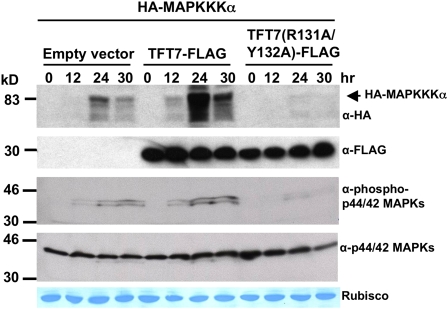
Although the above data support a role for TFT7 as a positive regulator of PCD induction by MAPKKK α, they do not address the molecular basis of this activity. One role of 14-3-3 proteins is to stabilize their target protein. We therefore followed up on our earlier observation that TFT7 did in fact appear to increase the abundance of MAPKKK α in leaves of N. benthamiana. We examined MAPKKK α abundance at several time points after induction of MAPKKK α expression in the presence of TFT7-FLAG or TFT7(R131A/Y132A)-FLAG. More HA-MAPKKK α was detected in leaves expressing TFT7-FLAG than in leaves expressing TFT7(R131A/Y132A)-FLAG or an empty vector control (Figure 7). For example, at 24 h after induction of MAPKKK α expression in the presence of TFT7, the kinase was more than twice as abundant as in leaves expressing TFT7(R131A/Y132A) or an empty vector control (Figure 7). Silencing of TFT7 greatly reduced protein abundance of MAPKKK α (see Supplemental Figure 8 online). These results suggest that TFT7 binding increases accumulation of MAPKKK α, possibly by preventing degradation of the kinase during PCD. The abundance of MAPKKK α gradually decreased after 24 h (Figure 7), as PCD progressed. We observed in two out of three experiments that coexpression of TFT7(R131A/Y132A) was associated with even less abundance of MAPKKK α protein than that in an empty vector control (as shown in Figure 7), indicating a possible dominant-negative effect of this variant protein on MAPKKK α protein abundance.
Figure 7.
TFT7 Increases Accumulation of MAPKKK α and Phosphorylation of Two MAP Kinases Downstream of MAPKKK α in Plant Cells.
TFT7-FLAG or TFT7(R131A/Y132A)-FLAG proteins were coexpressed with HA-Sl MAPKKK α, and total proteins were extracted at the designated time points after estradiol treatment. MAPKKK α, TFT7, phosphorylated MAP kinases, and MAP kinases were detected using anti-HA, anti-FLAG, anti-phospho-p44/42 MAPKs, and anti-p44/42 MAPKs, respectively. Coomassie staining of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) protein is shown as a loading control.
[See online article for color version of this figure.]
In all three treatments above, PCD first appeared at a similar time (i.e., around 20 h after induction of MAPKKK α expression), suggesting that the strength of PCD signaling and not its initiation by MAPKKK α is affected by TFT7. To investigate this issue, we looked at MAPKs that become phosphorylated (activated) upon expression of MAPKKK α. The antibody to detect phosphorylated p44/42 MAPKs in mammalian systems has been successfully used previously in N. benthamiana (Hann and Rathjen, 2007). In addition, MPK2 and MPK3, which are tomato orthologs of SIPK and WIPK in tobacco, are known to act downstream of MAPKKK α (Pedley and Martin, 2004). We therefore used the anti-phospho-p44/42 MAPKs to determine whether and when MAPKs might be phosphorylated during PCD induced by MAPKKK α expression. Phosphorylation of two MAPKs increased during MAPKKK α expression and was first detected 12 h after induction in the leaves expressing TFT7 and the empty vector control (Figure 7). However, the abundance of phosphorylated MAPKs was consistently higher in the presence of TFT7-FLAG than in leaves expressing the empty vector (Figure 7). In contrast with the decrease in MAPKKK α abundance at 30 h shown in Figure 6, the intensity of the phosphorylated MAPKs remained the same at 24 and 30 h after induction. Therefore, TFT7 appears to enhance MAPKKK α signaling but does not cause that signaling to initiate sooner. MAPK phosphorylation was markedly decreased in leaves coexpressing TFT7(R131A/Y132A), consistent with the lower abundance of MAPKKK α.
A Putative Phosphorylation Site of MAPKKK α Is Important for Interaction with TFT7 and for Both Stability and PCD-Eliciting Ability of MAPKKK α
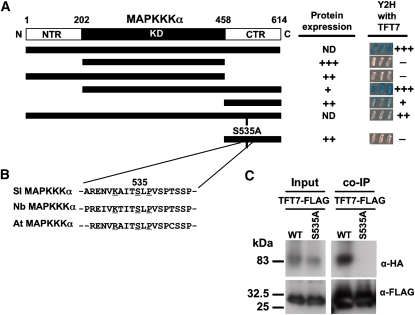
MAPKKK α can be viewed as having three regions: an N-terminal region (NTR), a KD, and a C-terminal region (CTR), as shown in Figure 8A. To determine which of these regions is important for interaction with TFT7, we tested the KD only, NTR-KD, KD-CTR, and CTR only of tomato MAPKKK α in a yeast two-hybrid system. These experiments revealed that the CTR plays the major role in interaction with TFT7. To identify a possible 14-3-3 binding motif in the CTR of MAPKKK α, amino acid sequences of Sl MAPKKK α and its two close orthologs, Nb MAPKKK α and At MAPKKK α, were aligned. From this alignment, one site (KXXXpSXP) in Sl MAPKKK α similar to the RXXXpSXP consensus motif, which is present perfectly in At MAPKKK α, was found (Figure 8B). Ser-535 in this motif of Sl MAPKKK α was substituted to Ala to see if this residue is important for interaction with TFT7. This substitution in the full length of MAPKKK α greatly reduced interaction with TFT7, and the same mutation in the CTR fragment completely abolished interaction with TFT7 (Figure 8A; see Supplemental Figure 9 online). Immunoblotting showed that all of the MAPKKK α proteins were detectable in yeast except for the full-length versions (see Supplemental Figure 10 online). Both full-length proteins were, however, shown to interact similarly with another MAPKKK α -interacting protein, Mai8, in yeast and so the S535A mutation is unlikely to have affected the overall conformation of MAPKKK α (see Supplemental Figure 11 online).
Figure 8.
Ser-535 Is a Putative Phosphorylation Site of MAPKKK α That Is Required for Its Interaction with TFT7 in Yeast and Plant Cells.
(A) Schematic showing the three regions of MAPKKK α : NTR, KD, and CTR. Four truncated forms and two mutated forms of Sl MAPKKK α were tested for expression and interaction with TFT7 in a yeast two-hybrid system, as described in Figure 1. Protein expression was determined using anti-LexA, as shown in Supplemental Figure 10 online, and the strength of interaction was confirmed by β -galactosidase assay, as shown in Supplemental Figure 9 online. +++, high expression or strong interaction; ++, moderate expression or interaction; +, low expression or interaction; −, no interaction; ND, not detected.
(B) Alignment of CTRs from Sl MAPKKK α and two homologs, Nb MAPKKK α and At MAPKKK α, showing a 14-3-3 protein binding motif (K/RXXXpSXP). The putative phosphorylated Ser (Ser-535) was substituted with Ala.
(C) Co-IP was performed as described in Figure 5C. WT, HA-Sl MAPKKK α ; S535A, HA-Sl MAPKKK α (S535A).
[See online article for color version of this figure.]
To examine the in vivo interaction between HA-MAPKKK α (S535A) and TFT7-FLAG, both proteins were coexpressed in N. benthamiana for co-IP. Although the wild-type MAPKKK α interacted with TFT7, we were unable to detect an interaction of the variant, MAPKKK α (S535A) with TFT7 (Figure 8C). We consistently observed slightly less MAPKKK α (S535A) protein compared with the wild-type MAPKKK α, and it is possible this difference affected these co-IP experiments (see input lanes in Figure 8C). However, based on the ratio of input and co-IP proteins, it still appears likely that the S535A mutation greatly reduced or eliminated the in vivo interaction with TFT7.
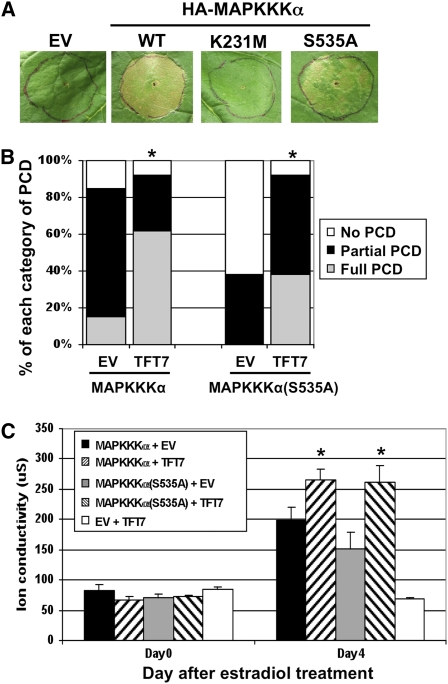
Our earlier results showed a correlation between TFT7 binding to MAPKKK α and its ability to enhance PCD-eliciting ability. To determine if the S535A mutation affects PCD-eliciting ability of MAPKKK α, HA-MAPKKK α (S535A) was expressed to induce PCD, and this ability was compared with that of the wild-type MAPKKK α. An empty vector and a kinase-inactive mutant of MAPKKK α (K231M) were used as negative controls. PCD induced by MAPKKK α (S535A) was weaker and delayed, compared with that by the wild-type protein (Figures 9A and 9B). To examine if TFT7 can still enhance PCD by MAPKKK α (S535A), both proteins were coexpressed, and PCD was monitored over 5 d after estradiol treatment. Unexpectedly, TFT7 also enhanced PCD by MAPKKK α (S535A) (Figures 9B and 9C). This enhancement may be due to the presence of a cryptic second TFT7 binding site in MAPKKK α ; the existence of such a second site is indicated by the interaction between TFT7 and MAPKKK α (S535A) in the yeast two-hybrid system (Figure 8; see Supplemental Figure 9 online).
Figure 9.
An S535A Substitution in MAPKKK α Reduces Its Ability to Elicit Cell Death but Still Allows PCD Enhancement by TFT7 Protein.
(A) HA-MAPKKK α (S535A) was expressed in N. benthamiana, and PCD was compared with the wild-type MAPKKK α and a kinase-inactive form (K231M). Transient assay was done as described in Figure 6. EV, empty vector.
(B) HA-MAPKKK α or HA-MAPKKK α (S535A) was coexpressed with TFT7-FLAG in N. benthamiana. The degree of PCD was scored 5 d after estradiol treatment and statistically analyzed as described in Figure 2B. Asterisks indicate significant enhancement of PCD based on statistical analysis. Results are representative of three independent experiments (13 infiltrations for each experiment).
(C) Electrolyte leakage was measured as described in Figure 2C. Asterisks indicate significant increase in ion leakage by coexpression of TFT7, based on a t test (P < 0.05). Data presented are means of six infiltrated areas (one per plant), and bars indicate standard errors.
DISCUSSION
We have found that TFT7 interacts with MAPKKK α and positively regulates PCD mediated by both Pto and MAPKKK α. The interaction of TFT7 is associated with an increase in the accumulation of MAPKKK α and enhancement of phosphorylation of two MAPKs activated by MAPKKK α. One mode of action of human 14-3-3 proteins is to block dephosphorylation of their client proteins and thereby block degradation by proteases (Darling et al., 2005). Our finding that Ser-535 of MAPKKK α is important for binding of TFT7 suggests that this 14-3-3 protein may also block dephosphorylation of this residue to delay degradation of MAPKKK α. Such a delay would allow sustained signal transfer to downstream MAPK cascades, resulting in enhancement of PCD. Binding of TFT7 to MAPKKK α is a critical step controlling PCD-enhancing activity because mutations in the phosphopeptide binding motif of TFT7 disrupted the interaction and abolished ability of the 14-3-3 protein to enhance MAPKKK α -mediated PCD. These results provide new insights into a role for 14-3-3 proteins in regulating PCD in plants and also point to potential similarities between plants and animals regarding their use of 14-3-3 proteins to regulate MAPK cascades.
Two MAPKs were phosphorylated upon estradiol-induced expression of MAPKKK α. Interestingly, the mechanism to regulate these two MAPKs by phosphorylation seems to be different. The larger MAPK is present under noninduced conditions (Figure 7, fourth panel), and only some of this protein appears to be phosphorylated upon MAPKKK α expression. By contrast, the smaller MAPK was not detected under noninduced conditions but was expressed after MAPKKK α induction. This timing matches the detection of the phosphorylated small MAPK and suggests that both expression and phosphorylation of this kinase may be coordinated. Although we don't know the identity of these MAPKs, these observations are reminiscent of the well-studied MAPKs, SIPK and WIPK (corresponding to tomato MPK2 and MPK3) (Zhang and Klessig, 2001; Pedley and Martin, 2005). Both of these MAPKs are activated during pathogen attack, and WIPK protein expression is itself induced during the immune response. Finally, it is notable that TFT7 affected only the intensity of MAPK phosphorylation, but not the timing at which it first occurred. This indicates that TFT7 enhances MAPKKK α -mediated PCD by increasing the strength of signal transfer and not by shortening the time for signal transfer to downstream signaling pathways.
TFT7 enhances PCD induced not only by the wild-type MAPKKK α, but also by the MAPKKK α (S535A) variant. However, MAPKKK α (S535A) does not interact with TFT7 in vivo. There are two possible explanations for this discrepancy. First, there may be more than one TFT7 binding site in MAPKKK α. In this regard, despite the fact Ser-535 is the major TFT7 binding site in MAPKKK α, the MAPKKK α (S535A) variant still interacts with TFT7 in yeast (Figure 8A; see Supplemental Figure 9 online). Although this interaction was not detectable in plant cells, it is possible that a weak in vivo interaction occurs, allowing enhancement of MAPKKK α -mediated PCD. Secondly, it is possible that TFT7 interacts with another downstream key component of MAPKKK α. This is consistent with the observation that TFT7 silencing compromised PCD induced by a broader range of R/Avr interactions than MAPKKK α. Good candidates for a downstream component are MAPKKs or MAPKs because these kinases participate in diverse signaling pathways, including PCD (Nakagami et al., 2005), and also are known to bind to 14-3-3 proteins (see Introduction).
Both TFT7 and MAPKKK α positively affect PCD induced by P. syringae pv tomato, but only TFT7 appears to be involved in suppressing disease symptoms caused by this bacterium. Previously, we found that silencing of MAPKKK α decreased PCD associated with Pto-mediated resistance and also that associated with disease (del Pozo et al., 2004). However, in this study, silencing with TFT7 enhanced disease symptoms, indicating that this gene contributes positively to disease resistance but not to disease-associated cell death. This discrepancy may be explained by the observation that TFT7 affects not only the Pto- and MAPKKK α -mediated pathway, but also other defense pathways as indicated by its effects on diverse R/effector interactions. In the future, it will be interesting to search for a possible general component of plant immunity pathways that may be regulated by TFT7 and possibly other 14-3-3 proteins.
The predicted overall structure of TFT7 is very similar to human 14-3-3 ϵ and, in addition, both proteins have a conserved phosphopeptide binding site. The human14-3-3 ϵ positively regulates apoptosis mediated by MEKKs in human, and its ortholog in Drosophila controls eye development via a Ras-mediated signaling pathway (Chang and Rubin, 1997). GF14h, which is the closest homolog of human14-3-3 ϵ in rice (Oryza sativa), binds to BZR1, which regulates brassinosteroid signaling. Another rice 14-3-3 protein, GF14c, binds to BZR1, negatively regulating brassinosteroid signaling by reducing nuclear localization of BZR1 (Bai et al., 2007) and also negatively regulates flowering in rice (Asih et al., 2009). Thus, 14-3-3 ϵ homologs appear to be involved in diverse signaling pathways in plants. In our work to date with silencing of TFT7 in N. benthamiana, we have observed no obvious detrimental phenotypes on plant development. However, TRV-TFT7 plants are consistently larger than TRV-only infected plants, indicating that TFT7 may be involved in other signaling pathways, particularly those involved in growth.
14-3-3 proteins typically function as dimmers, and each of them has a distinct dimerization preference to make either a homodimer or a heterodimer (Bridges and Moorhead, 2005). For example, by in vivo co-IP experiments with the human 14-3-3 ϵ, it was shown that it predominantly makes heterodimers (Chaudhri et al., 2003). Recently, target proteins of GF14omega in Arabidopsis were identified by combining TAP-tag affinity protein purification and tandem mass spectrometry (Chang et al., 2009). In this screening, it was shown that, in Arabidopsis, GF14omega interacts with 10 other 14-3-3 proteins, including itself and 14-3-3 ϵ. These findings may indicate that 14-3-3 ϵ has a preference to form heterodimers. In the future, it will be interesting to examine the dimerization preference of TFT7 with other tomato 14-3-3 proteins.
Phosphorylation plays an important role in regulating components of signaling pathways involved in both PCD and host immunity. Because of the central role played by 14-3-3 proteins in physically interacting with phosphorylated client proteins and mediating signaling transduction, it appears likely that there will be many target proteins for these proteins in defense signaling. Earlier reports presaged an important role in defense for 14-3-3 proteins as TFT1, TFT4, and TFT6 gene expression were found to be induced during the Cf9/Avr9-mediated PCD (Roberts and Bowles, 1999). This study and another recent article (Yang et al., 2009) now provide insights into the role of specific 14-3-3 proteins in plant immunity. In the future, it will be important to expand our knowledge of the repertoire of 14-3-3 proteins that contribute to plant defense, identify their client proteins, and understand the molecular basis by which they promote plant immunity. Finally, there is some indication that pathogen effectors may target plant 14-3-3 proteins to disturb plant immunity because XopN, a Xanthomonas effector protein, that suppresses plant immunity triggered by PAMPs was found to interact with four different 14-3-3 proteins (TFT1, TFT3, TFT5, and TFT6) in tomato (Kim et al., 2009). It will therefore be important to determine if pathogen effectors undermine plant immunity by interfering with 14-3-3 protein functions.
METHODS
Bacteria, Yeast, and Plant Materials
Agrobacterium tumefaciens strain GV2260 and Pseudomonas syringae pv tomato strains were grown at 30 ° C in Luria-Bertani and King's B medium with appropriate antibiotics, respectively. Yeast strain EGY48(pSH18-34) was grown at 30 ° C in synthetic dropout medium (SD) with glucose as a carbon source, but lacking uracil. Nicotiana benthamiana was grown in the greenhouse with 16 h of light and temperature at 24 to 26 ° C day and 20 to 22 ° C night. For VIGS, plants were kept in a growth chamber with 16 h of light and a temperature of 24 ° C day and 21 ° C night.
Yeast Two-Hybrid Assay
Yeast strain EGY48(pSH18-34) was used for the two-hybrid assays. The Sl MAPKKK α gene and its derivatives were cloned into pEG202 as bait, and the TFT7 gene was cloned into pJG4-5 as prey. Yeast transformation was performed by the LiAc/PEG method as described in the Yeast Protocols Handbook (Clontech). Yeast transformants were screened on SD with galactose and X-gal, but lacking uracil, histidine, and tryptophan. Blue colonies indicate a positive interaction between two proteins in yeast. In addition, β -galactosidease assay was performed as described in the Yeast Protocols Handbook.
Agrobacterium-Mediated Transient Assay
The Sl MAPKKK α gene and its derivatives were cloned into pER8 with a double hemagglutinin (HA) tag at the N terminus, and its expression was controlled by an estradiol-inducible system. The TFT7 gene was cloned into pBTEX with a FLAG tag at the C terminus, and it was expressed using the cauliflower mosaic virus 35S promoter. Each construct was transformed into A. tumefaciens strain GV2260. Pto/AvrPto, Pto/AvrPtoB1-387, RPS2/AvrRpt2, RPP13/ATR13Emco5Δ 41aa, Rx2/CP, and Gpa2/RBP-1 under the 35S promoter, Sl MAPKKK α full length or KD only and Nt MEK2DD under the estradiol-inducible system, and Bax under the dexamethasone-inducible system were expressed transiently to induce PCD in N. benthamiana, as described previously (Abramovitch et al., 2003). Two micromolar of estradiol or 10 μ M dexamethasone mixed with 0.02% Tween 20 were used to induce expression of the Sl MAPKKK α gene, Nt MEK2DD, and Bax, respectively, 48 h after agroinfiltration.
Electrolyte Leakage Assays
For the electrolyte leakage assays, two leaf discs (9 mm in diameter) were collected from the Agrobacterium-infiltrated area and incubated in 2 mL water for 2 h at 25 ° C with shaking at 165 rpm. Leaf discs were then removed, and ion conductivity was measured using an Acorn CON 5 ion conductivity meter (OAKTON). Each time, six areas per treatment (one area per plant) were used for the assay.
VIGS
A 287-bp DNA fragment from TFT7 gene shown in Supplemental Figure 2 online was cloned into the pTRV2 vector for TRV-based VIGS in N. benthamiana, as described previously (Liu et al., 2002). pTRV2-Nb MAPKKK α and pTRV2 vector only were used as controls (del Pozo et al., 2004). Eighteen-day-old N. benthamiana seedlings were used for TRV infection, and gene-silenced N. benthamiana plants were used for the assays 4 to 5 weeks later. Four to six independent silenced plants with two to three leaves infiltrated each were used for the PCD assays each time.
RT-PCR and SYBR Green I Gel Staining
Two leaf discs (9 mm in diameter) were collected to extract total RNA using the RNeasy Plant Mini Kit (Qiagen). One microgram of total RNA was used to make cDNA with SuperScript III reverse transcriptase. Two microliters of cDNA was used for PCR. The forward and reverse primers, as follows, were used for the TFT7 gene homolog (Isoform A) in N. benthamiana (see Supplemental Figure 2 online): forward primer, 5 ′ -GGTTTACTTGGCTAGGCTGG-3 ′ and reverse primer, 5 ′ -GTGGCAGTAGCAGCCTCATAAG-3 ′. The EF1 α gene was used as an internal control with the following primers: the forward primer, 5 ′ -CATGGGCTTGGTGGGAATC-3 ′, and the reverse primer, 5 ′ -AGCCTGGTATGGTTGTGACTTTTG-3 ′ (del Pozo et al., 2004). The PCR products from 28 cycles for the TFT7 gene homolog and from 25 cycles for EF1 α were compared after staining the gel with SYBR Green I stain (Molecular Probes).
In Silico Sequence and Phylogenetic Analyses
DNA or protein sequence analysis was performed using Lasergene (DNASTAR). Amino acid alignments to generate a 14-3-3 protein phylogenetic tree were performed with ClustalX software. For this, the gap opening penalty and the gap extension penalty were used, as follows: 10 and 0.1 for pairwise alignment and 15 and 0.3 for multiple alignment. The midpoint-rooted tree was generated by the neighbor joining method, using PAUP* 4.0 Beta version (Sinauer Associates), and bootstrapping analysis was performed with 2000 trials. The tertiary structure of TFT7 protein was predicted using the SWISS-MODEL server. The predicted protein structure was analyzed with the Protein Data Bank viewer.
Site-Directed Mutagenesis
PCR-based site-directed mutagenesis (Ho et al., 1989) was performed to generate the substitutions S535A in Sl MAPKKK α and R131A/Y132A in TFT7.
Co-IP
HA-Sl MAPKKK α or HA-Sl MAPKKK α (S535A) and TFT7-FLAG or TFT7(R131A/Y132A)-FLAG were coexpressed in N. benthamiana by Agrobacterium-mediated transient expression. GFP-FLAG was used as a negative control. Eighteen leaf discs (9 mm in diameter) per treatment were collected 24 h after estradiol treatment for protein extraction, using 500 μ L of extraction buffer (GTEN buffer, 0.1% Triton X-100, 10 mM DTT, PVPP, 1 × plant protein protease inhibitor, and 1 × protein phosphatase inhibitor). During desalting, buffer was exchanged to IP buffer (GTEN buffer, 0.15% Nonidet P-40, and 1 mM DTT). Co-IP was performed as described previously with some modifications (Moffett et al., 2002). Briefly, protein extract was incubated for 30 min with goat IgG agarose beads to remove proteins binding to agarose beads. The extract was then incubated at 4 ° C for 18 h with anti-FLAG agarose beads. After washing with IP buffer six times, beads were resuspended with 100 μ L of 1 × SDS loading buffer. Input in Figures 5 and 8 indicates protein extract after incubating with goat IgG agarose beads.
Immunoblotting
Proteins were separated on 8% or 12% SDS-PAGE gel, transferred onto a polyvinylidene fluoride membrane using an electroblotter (Bio-Rad), and detected with antibodies as indicated in each figure. Anti-FLAG (1:2500), anti-HA (1:5000), anti-LexA (1:2000), anti-phospho-p44/42 MAPKs (1:1000), and anti-p44/42 MAPKs (1:1000) were purchased from Sigma-Aldrich and Cell Signaling Technologies, respectively. Signal was detected using the Amersham ECL-Plus Western blotting detection system (GE Healthcare).
Pathogen Assays
About 4 to 5 weeks after infection with the TRV vectors, gene-silenced N. benthamiana plants were used for pathogen assays. For observing the HR, 1.25 × 107 CFU/mL (OD600 = 0.005) or 5 × 106 CFU/mL (OD600 = 0.002) of P. syringae pv tomato wild-type strain DC3000 were infiltrated into leaves with a needleless syringe, and the response was monitored for 3 d. For the disease assays, 2 × 104 CFU/mL of P. syringae pv tomato strain DC3000 ΔhopQ1-1/ΔavrPto/ΔavrPtoB, which is pathogenic on N. benthamiana (Kvitko et al., 2009), was infiltrated by vacuum. Disease symptoms were photographed 5 d after infiltration. For measuring bacterial populations, two leaf discs (9 mm in diameter) were collected 0, 2, and 4 d after infiltration, and the bacterial cells were counted on King's B medium containing appropriate antibiotics.
Accession Numbers
Sequence data from this article can be found in the GenBank database under the following accession numbers: Sl MAPKKK α, AY500155; TFT7, X95905; and tobacco Isoform A, S76737.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. TFT7 Is Homologous to Isoform A among Tobacco 14-3-3 Proteins.
Supplemental Figure 2. Alignment of the DNA Sequences of TFT7 and Tobacco 14-3-3 Isoform A.
Supplemental Figure 3. TFT7 Gene Homolog Was Silenced in TRV-TFT7 Plants.
Supplemental Figure 4. Bacterial Growth in Gene-Silenced N. benthamiana Plants.
Supplemental Figure 5. TFT7 and the Human 14-3-3 ϵ Are Structurally Similar.
Supplemental Figure 6. TFT7 Has a Conserved Phosphopeptide Binding Motif in Common with Human 14-3-3 ϵ.
Supplemental Figure 7. TFT7 and TFT7(R131A/Y132A) Are Both Expressed Well in N. benthamiana.
Supplemental Figure 8. Silencing of TFT7 Reduces Protein Abundance of MAPKKK α.
Supplemental Figure 9. MAPKKK α (S535A) Still Interacts with TFT7 in Yeast.
Supplemental Figure 10. The Truncated and Mutated Forms of MAPKKK α Proteins Are Expressed in Yeast.
Supplemental Figure 11. The S535A Mutation Did Not Affect Interaction of MAPKKK α with Another Interacting Protein, Mai8 in Yeast.
Supplemental Data Set 1. Text File of Alignment Used to Generate Phylogenetic Tree in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Simona Despa at the Cornell University Statistical Consulting Office for help with data analysis, Peter Moffett for providing Rx2/CP and Gpa2/RBP-1 constructs and the co-IP protocol, and Hong Gu Kang for helping with the co-IP experiments and providing the GFP-FLAG construct for a control. We thank Brian Staskawicz for providing the RPS2/AvrRpt2 and RPP13/ATR13Emco5Δ 41aa constructs for the cell death assay. We thank Inhwa Yeam and Joe Matthieu for critical review of our manuscript. This work was supported by National Science Foundation Integrative Organismal Systems Grants 0444600 and 0841807 (G.B.M.).
References
- Abramovitch R.B., Kim Y.-J., Chen S., Dickman M.B., Martin G.B. (2003). Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 22: 60–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asih P.Y., Yuka O., Shojiro T., Hiroyuki T., Ko S. (2009). The 14-3-3 protein GF14c acts as a negative regulator of flowering in rice by interacting with the florigen Hd3a. Plant Cell Physiol. 50: 429–438 [DOI] [PubMed] [Google Scholar]
- Bai M.Y., Zhang L.Y., Gampala S.S., Zhu S.W., Song W.Y., Chong K., Wang Z.Y. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 104: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A., Querci M., Kanyuka K., Baulcombe D.C. (2000). Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: Application to the Rx2 locus in potato. Plant J. 21: 73–81 [DOI] [PubMed] [Google Scholar]
- Bridges D., Moorhead G.B.G. (2005). 14-3-3 proteins: A number of functions for a numbered protein. Sci. STKE 2005: 2–10 [DOI] [PubMed] [Google Scholar]
- Cardone M.H., Salvesen G.S., Widmann C., Johnson G.L., Frisch S.M. (1997). The regulation of Anoikis: MEKK-1 activation requires cleavage by caspases. Cell 90: 315–323 [DOI] [PubMed] [Google Scholar]
- Chang H.C., Rubin G.M. (1997). 14-3-3e positively regulates Ras-mediated signaling in Drosophila.. Genes Dev. 11: 1132–1139 [DOI] [PubMed] [Google Scholar]
- Chang I.F., Curran A., Woolsey R., Quilici D., Cushman J.C., Mittler R., Harmon A., Harper J.F. (2009). Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana.. Proteomics 9: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhri M., Scarabel M., Aitken A. (2003). Mammalian and yeast 14-3-3 isoforms form distinct patterns of dimers in vivo.. Biochem. Biophys. Res. Commun. 300: 679–685 [DOI] [PubMed] [Google Scholar]
- Darling D.L., Yingling J., Wynshaw-Boris A. (2005). Role of 14-3-3 proteins in eukaryotic signaling and development. Curr. Top. Dev. Biol. 68: 281–315 [DOI] [PubMed] [Google Scholar]
- Day B., Dahlbeck D., Huang J., Chisholm S.T., Li D., Staskawicz B.J. (2005). Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell 17: 1292–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo O., Pedley K.F., Martin G.B. (2004). MAPKKK α is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 23: 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekengren S.K., Liu Y., Schiff M., Dinesh-Kumar S.P., Martin G.B. (2003). Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J. 36: 905–917 [DOI] [PubMed] [Google Scholar]
- Fanger G.R., Gerwins P., Widmann C., Jarpe M.B., Johnson G.L. (1997). MEKKs, GCKs, MLKs, PAKs, TAKs, and Tpls: upstream regulators of the c-Jun amino-terminal kinases?. Curr. Opin. Genet. Dev. 7: 67–74 [DOI] [PubMed] [Google Scholar]
- Fanger G.R., Widmann C., Porter A.C., Sather S., Johnson G.L., Vaillancourt R.R. (1998). 14-3-3 proteins interact with specific MEK kinases. J. Biol. Chem. 273: 3476–3483 [DOI] [PubMed] [Google Scholar]
- Ferl R.J. (2004). 14-3-3 proteins: Regulation of signal-induced events. Physiol. Plant. 120: 173–178 [DOI] [PubMed] [Google Scholar]
- Ferl R.J., Manak M.S., Reyes M.F. (2002). The 14-3-3s. Genome Biol. 3: reviews3010.3011–3010.3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnie C., Anderson C.H., Borch J., Gjetting S., Christensen A.B., de Boer A.H., Thordal-Christensen H., Collinge D.B. (2002). Do 14-3-3 proteins and plasma membrane H+-ATPases interact in the barley epidermis in response to the barley powdery mildew fungus?. Plant Mol. Biol. 49: 137–147 [DOI] [PubMed] [Google Scholar]
- Fritz A., Brayer K.J., McCormick N., Adams D.G., Wadzinski B.E., Vaillancourt R.R. (2006). Phosphorylation of serine 526 is required for MEKK3 activity, and association with 14-3-3 blocks dephosphorylation. J. Biol. Chem. 281: 6236–6245 [DOI] [PubMed] [Google Scholar]
- Hann D.R., Rathjen J.P. (2007). Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana.. Plant J. 49: 607–618 [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt H.D., Horton R.M., Pullen R.M., Pease L.R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- Kim J.G., et al. (2009). Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell 21: 1305–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Lin N.-C., Martin G.B. (2002). Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109: 589–598 [DOI] [PubMed] [Google Scholar]
- Kvitko B.H., Park D.H., Velasquez A.C., Wei C.F., Russell A.B., Martin G.B., Schneider D.J., Collmer A. (2009). Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5: e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalle M., Visconti S., Marra M., Camoni L., Velasco R., Aducci P. (2005). ZmMPK6, a novel maize MAP kinase that interacts with 14–3-3 proteins. Plant Mol. Biol. 59: 713–722 [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S.P. (2002). Virus-induced gene silencing in tomato. Plant J. 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Moffett P., Farnham G., Peart J., Baulcombe D.C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21: 4511–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D.K. (2009). The 14-3-3 proteins: Integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 19: 16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucyn T.S., Clemente A., Andriotis V.M., Balmuth A.L., Oldroyd G.E., Staskawicz B.J., Rathjen J.P. (2006). The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell 18: 2792–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore K.S., Crasta O.R., Tuori R.P., Folkerts O., Swirsky P.B., Martin G.B. (2002). Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato.. Plant J. 32: 299–315 [DOI] [PubMed] [Google Scholar]
- Nakagami H., Pitzschke A., Hirt H. (2005). Emerging MAP kinase pathways in plant stress signaling. Trends Plant Sci. 10: 339–346 [DOI] [PubMed] [Google Scholar]
- Pedley K.F., Martin G.B. (2004). Identification of MAPKs and their possible MAPK kinase activators involved in the Pto-mediated defense response of tomato. J. Biol. Chem. 279: 49229–49235 [DOI] [PubMed] [Google Scholar]
- Pedley K.F., Martin G.B. (2005). Role of mitogen-activated protein kinases in plant immunity. Curr. Opin. Plant Biol. 8: 541–547 [DOI] [PubMed] [Google Scholar]
- Rentel M.C., Leonelli L., Dahlbeck D., Zhao B., Staskawicz B.J. (2008). Recognition of the Hyaloperonospora parasitica effector ATR13 triggers resistance against oomycete, bacterial, and viral pathogens. Proc. Natl. Acad. Sci. USA 105: 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M.R. (2003). 14-3-3 proteins find new partners in plant cell signaling. Trends Plant Sci. 8: 218–223 [DOI] [PubMed] [Google Scholar]
- Roberts M.R., Bowles D.J. (1999). Fusicoccin, 14-3-3 proteins, and defense responses in tomato plants. Plant Physiol. 119: 1243–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M.R., Salinas J., Collinge D.B. (2002). 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol. Biol. 50: 1031–1039 [DOI] [PubMed] [Google Scholar]
- Rosebrock T.R., Zeng L., Brady J.J., Abramovitch R.B., Xiao F., Martin G.B. (2007). A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448: 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco M.A., Koropacka K., Grenier E., Jaubert M.J., Blanchard A., Goverse A., Smant G., Moffett P. (2009). The cyst nematode SPRYSEC protein RBP-1 elicits Gpa2- and RanGAP2-dependent plant cell death. PLoS Pathog. 5: e1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron J.M., Oldroyd G.E.D., Rommens C.M.T., Scofield S.R., Kim H.-S., Lavelle D.T., Dahlbeck D., Staskawicz B.J. (1996). Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 86: 123–133 [DOI] [PubMed] [Google Scholar]
- Seehaus K., Tenhaken R. (1998). Cloning of genes by mRNA differential display induced during the hypersensitive reaction of soybean after inoculation with Pseudomonas syringae pv. glycinea. Plant Mol. Biol. 38: 1225–1234 [DOI] [PubMed] [Google Scholar]
- Sehnke P.C., DeLille J.M., Ferl R.J. (2002). Consummating signal transduction: the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. Plant Cell 14: 339–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Frederick R.D., Zhou J., Halterman D.A., Jia Y., Martin G.B. (1996). Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274: 2060–2063 [DOI] [PubMed] [Google Scholar]
- van den Burg H.A., Tsitsigiannis D.I., Rowland O., Lo J., Rallapalli G., MacLean D., Takken F.L.W., Jones J.D.G. (2008). The F-box protein ACRE189/ACIF1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. Plant Cell 20: 697–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn W.G., Woltering E.J. (2005). Many ways to exit? Cell death categories in plants. Trends Plant Sci. 10: 117–122 [DOI] [PubMed] [Google Scholar]
- Wei C.F., Kvitko B.H., Shimizu R., Crabill E., Alfano J.R., Lin N.C., Martin G.B., Huang H.C., Collmer A. (2007). A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana.. Plant J. 51: 32–46 [DOI] [PubMed] [Google Scholar]
- Yang X., Wang W., Coleman M., Orgil U., Feng J., Ma X., Ferl R.J., Turner J.G., Xiao S. (2009). Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. Plant J. 60: 539–550 [DOI] [PubMed] [Google Scholar]
- Yang X.Y., Lee W.H., Sobott F., Papagrigoriou E., Robinson C.V., Grossmann G., Sundstrom M., Doyle D.A., Elkins J.M. (2006). Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc. Natl. Acad. Sci. USA 103: 17237–17242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Klessig D.F. (2001). MAPK cascades in plant defense signaling. Trends Plant Sci. 6: 520–527 [DOI] [PubMed] [Google Scholar]
- Zhou J., Tang X., Martin G.B. (1997). The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 16: 3207–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.-M., Loh Y.-T., Bressan R.A., Martin G.B. (1995). The tomato gene Pti1 encodes a serine-threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 83: 925–935 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.