This work uses reverse genetic and biochemical approaches to provide evidence supporting the hypothesis that an Hsp70 system plays a heretofore unrecognized role in chloroplast protein import.
Abstract
Heat shock protein 70s (Hsp70s) are encoded by a multigene family and are located in different cellular compartments. They have broad-ranging functions, including involvement in protein trafficking, prevention of protein aggregation, and assistance in protein folding. Hsp70s work together with their cochaperones, J domain proteins and nucleotide exchange factors (e.g., GrpEs), in a functional cycle of substrate binding and release accompanied by ATP hydrolysis. We have taken advantage of the gene targeting capability of the moss Physcomitrella patens to investigate the functions of chloroplast Hsp70s. We identified four Hsp70 genes and two GrpE cochaperone homolog genes (CGE) in moss that encode chloroplast proteins. Disruption of one of the Hsp70 genes, that for Hsp70-2, caused lethality, and protein import into heat-shocked chloroplasts isolated from temperature-sensitive hsp70-2 mutants was appreciably impaired. Whereas the double cge null mutant was not viable, we recovered a cge1 null/cge2 knock down mutant in which Hsp70-2 was upregulated. Chloroplasts isolated from this mutant demonstrated a defect in protein import. In addition, two different precursors staged as early import intermediates could be immunoprecipitated with an Hsp70-2–specific antibody. This immunoprecipitate also contained Hsp93 and Tic40, indicating that it represents a precursor still in the Toc/Tic translocon. Together, these data indicate that a stromal Hsp70 system plays a crucial role in protein import into chloroplasts.
INTRODUCTION
The majority of chloroplast-localized proteins are synthesized in the cytosol as precursors bearing N-terminal cleavable transit peptides and are posttranslationally imported into chloroplasts. The import process can be divided into three stages: recognition, translocation, and maturation (Kessler and Schnell, 2006). The translocons at the outer (Toc) and inner (Tic) membrane of chloroplasts play essential roles in recognition and translocation during protein import. Toc75, Toc34, and Toc159 are core components in the Toc complex. Precursor protein interacts with GTPase receptors Toc159 and Toc34 and is translocated across outer membrane through the channel thought to be formed by Toc75 in a GTP- and ATP-dependent manner. Formation of the Tic complex appears to be dynamic since a stable Tic complex containing stoichiometric amounts of its components has not been isolated (Kessler and Schnell, 2006). Tic subunits were identified using coimmunoprecipitation, cross-linking, and genetic approaches. Among them, Tic110, Tic40, Tic22, and Tic20 have been studied relatively extensively. Tic22 is localized in the intermembrane space and is peripherally associated with the outer face of the inner membrane. Its proposed function is to maintain contact between the Tic and Toc complexes during protein import so that precursor proteins can be translocated from the cytosol to the stroma directly (Kouranov et al., 1998). Tic110 appears to play a crucial role in the assembly of the Tic translocon and possibly contributes to the formation of the translocation channel (Heins et al., 2002; Inaba et al., 2003). It also provides a docking site for preproteins as they emerge into the stroma (Inaba et al., 2003). Tic20 has been proposed to be the inner membrane protein-conducting channel (Chen et al., 2002; Kikuchi et al., 2009). Tic40 is an inner envelope membrane protein that is found in close physical proximity to translocating precursor proteins and Tic110 (Wu et al., 1994; Ko et al., 1995; Stahl et al., 1999; Chou et al., 2006). It has a single transmembrane anchor and tetratricopeptide and stress inducible (Sti1) domains, characteristics of Hip/Hop-type cochaperones (Chou et al., 2003; Bedard et al., 2007). Tic110 interacts with two stromal chaperones, Hsp93 (Akita et al., 1997; Nielsen et al., 1997) and Cpn60 (Kessler and Blobel, 1996). Cpn60, together with the stromal processing peptidase (Richter and Lamppa, 1998), is involved in maturation of newly imported proteins.
Eukaryotic cells have evolved multiple transport pathways (Schatz and Dobberstein, 1996). One remarkable shared common feature in the mechanisms of transport across the endoplasmic reticulum (ER) and mitochondrial membranes is the involvement of heat shock protein 70s (Hsp70s). Eukaryotic Hsp70s are the homologs of the Escherichia coli chaperone DnaK and are encoded by a multigene family. They are localized in different cellular compartments and participate in numerous cellular processes, probably through a common mechanism (De Los Rios et al., 2006). Hsp70s possess two major domains, an ATPase domain at the N terminus followed by a substrate binding domain (Stevens et al., 2003). In prokaryotic Hsp70 systems (e.g., in bacteria and mitochondria), it is well established that the chaperone completes its functional cycle with assistance of at least two cochaperones: a member of the J-domain protein family and a nucleotide exchange factor, GrpE (Harrison et al., 1997). J-domain proteins are required for tight coupling of ATP hydrolysis to substrate association, while GrpE facilitates the release of ADP from the chaperone, allowing for the binding of new ATP in preparation for the next functional cycle.
Proteins can be imported into the ER either co- or posttranslationally. For proteins translocated posttranslationally, an Hsp70 located on the receiving (trans) side of the membrane (BiP) is recruited to drive the preprotein through the heteromeric Sec61 import channel into the ER lumen (Matlack et al., 1999; Alder et al., 2005). As with chloroplasts, most mitochondrial proteins are synthesized in the cytosol as precursors posttranslationally imported into the organelle and processed to their mature forms in the matrix. Mitochondrial Hsp70 (mtHsp70) is thought to bind the incoming precursor as it appears in the matrix, and through ATP hydrolysis, provide the driving force for the translocation reaction (Gambill et al., 1993). This protein is also postulated to be involved in substrate refolding after translocation (Horst et al., 1997).
It has long been known that pea (Pisum sativum) chloroplasts contain at least three Hsp70s, one associated with the outer envelope membrane and two residing in the stroma (Marshall et al., 1990). The functions of these Hsp70s are still not known. In green algae, it has been suggested that the stromal Hsp70B is involved in protection against photoinhibition (Schroda et al., 1999; Yokthongwattana et al., 2001; Schroda, 2004). Recently, this chaperone, along with two stromal cochaperones, have been implicated in the assembly and disassembly of large homomeric Vipp1 complexes, although the physiological significance of this function is yet unknown (Liu et al., 2005, 2007).
With respect to protein import, it was reported that newly imported ferredoxin-NADP+ reductase could be coimmunoprecipitated with a chloroplast Hsp70 antibody (Tsugeki and Nishimura, 1993). Another newly imported protein, the Rieske iron-sulfur protein, was recovered in a stromal Hsp70-containing protein complex (Madueno et al., 1993). More recently, stromal Hsp70 has been observed in contact with importing proteins belonging to a subset of Toc/Tic substrates (Vojta et al., 2007). These latter data suggest that stromal Hsp70s might be involved in protein import. In addition, an interaction between a pea stromal Hsp70 and a synthesized transit peptide of the small subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) has been reported (Ivey et al., 2000). In another study, Hsp70 from pea and tomato (Solanum lycopersicum) plastids was seen to interact with the transit peptide of ferredoxin-NADP+ reductase precursor (Rial et al., 2000). Zhang and Glaser (2002) analyzed 277 confirmed chloroplast transit peptides and determined that 82.5% contained at least one Hsp70 binding site.
In numerous studies designed to identify protein transport machinery components in the chloroplast envelope membranes, a stromal Hsp93 chaperone (a member of Hsp100 family), but not Hsp70, has consistently been observed in association with the translocation complex (Akita et al., 1997; Nielsen et al., 1997). Deletion of one stromal Hsp93 (ClpC1) causes a reduction in protein import capacity of isolated chloroplasts (Constan et al., 2004; Kovacheva et al., 2005; Kovacheva et al., 2007), while the double knockout mutant appears to be lethal. From these findings, Hsp93 has been hypothesized to replace in chloroplasts the function of the mtHsp70 in mitochondria and to be the ATPase that provides the driving force for chloroplast protein import.
We have examined the function of the chloroplast stromal Hsp70s by generating targeted Hsp70 knockout mutants in the moss Physcomitrella patens, an emerging model plant system in which exogenous DNA can be incorporated into the chromosomes by homologous recombination. We have previously shown that the chloroplast protein import machinery in this moss is functionally equivalent to that of flowering plants (Hofmann and Theg, 2003). Other developments in P. patens research, such as the release of the complete genomic sequence (Rensing et al., 2008) and the development of RNA interference techniques (Bezanilla et al., 2005), make the moss an attractive model system in which to continue to study chloroplast protein transport.
In this report, we show that the insertional inactivation of the P. patens gene for one of three stromal Hsp70s, designated herein as Hsp70-2, results in lethality. Plants in which the chromosomal copy of this gene was interrupted were viable only when they were cotransformed with a cDNA copy of the original gene. Impaired import was observed in chloroplasts in which Hsp70-2 function was conditionally diminished, as well as in those with reduced GrpE-homologous cochaperones (termed chloroplast GrpE homologs [CGEs]). Moreover, two precursor proteins were demonstrated to be associated with Hsp70-2 protein while they were in the translocon and in contact with other translocon components. These results lead us to conclude that a stromal Hsp70 system plays a previously unrecognized critical role in the import of proteins across the chloroplast envelope membranes.
RESULTS
Cloning and Characterization of Genes Encoding Chloroplast Hsp70s from P. patens
The first step in characterization of the moss plastidic Hsp70s was their identification. A search of the P. patens EST database using the Arabidopsis thaliana stromal Hsp70 protein sequences and prediction of subcellular locations using TargetP and ChloroP (Emanuelsson et al., 2007) identified two candidate genes, which we designated Hsp70-1 and Hsp70-2. Further analysis of the completed moss genome sequence revealed the presence of another candidate gene, designated Hsp70-3. We subsequently came to realize that the entire Hsp70-3 gene appears twice in the moss genome, reproduced with 100% fidelity through both exon and intron regions and into the 3 ′ untranslated region; these two identical genes are referred to as Hsp70-3a and Hsp70-3b.
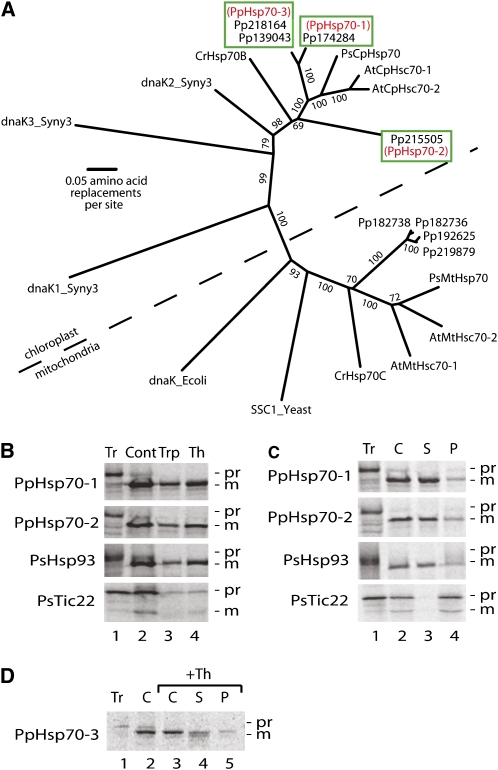
Figure 1A shows a cladistic analysis in which Hsp70-1 and Hsp70-3 reside on the same branch, clearly separated from Hsp70-2. With respect to the predicted mature forms, Hsp70-1 is 95% similar to Hsp70-3 at the amino acid level and 70% similar to Hsp70-2. The three P. patens proteins cluster with known plastid-localized Hsp70 chaperones in Arabidopsis, pea, and Chlamydomonas reinhardtii, as well as with three cyanobacterial Hsp70s; they are each in turn separated from chaperones known or predicted to reside in mitochondria (Figure 1A).
Figure 1.
Three Hsp70s of the Moss P. patens Are Localized in the Chloroplast Stroma.
(A) Phylogenetic analysis of Hsp70 sequences. Seven Hsp70 protein sequences from the moss P. patens (labels begin with Pp) and 13 from other species (see Methods for accession numbers) were aligned and a phylogenetic tree was generated using the neighbor-joining method. The scale bar indicates 0.05 substitutions per site. Bootstrap values that are >70% from 1000 replications for each branch are shown. The dotted line separates the chloroplast clade from the mitochondrial one. In the Hsp70-3 box, there are two protein ID numbers that are derived from the Hsp70-3a and -3b genes, respectively. The alignment used to generate the tree is available as Supplemental Data Set 1 online.
(B) Import assay of Hsp70-1 and Hsp70-2. [35S]-labeled precursor proteins were incubated with pea chloroplasts followed by either trypsin or thermolysin treatment. All controls were treated the same manner except without adding proteases. Proteases activities were quenched and chloroplasts were subjected to SDS-PAGE followed by phosphor image analysis.
(C) Fractionation of chloroplasts and localization of the Hsp70-1 and Hsp70-2 Proteins. After incubation with precursor proteins, chloroplasts were reisolated, lysed, and separated into soluble and pellet fractions, which were analyzed using SDS-PAGE and phosphor imaging.
(D) Localization of Hsp70-3. [35S]-labeled Hsp70-3 precursor protein was incubated with pea chloroplasts followed by thermolysin treatment. Chloroplasts were reisolated, lysed, and separated into soluble and pellet fractions, which were analyzed using SDS-PAGE and phosphor imaging.
Tr, translation products; Th, thermolysin treatment; Trp, trypsin treatment; Cont, mock protease treatment; C, chloroplasts; S, soluble fraction; P, membrane fraction; Pr, precursor protein; M, mature protein; Hsp93 (from pea) is a marker for the stroma; Tic22 (from pea) is a marker for the inner envelope membrane and faces the intermembrane space.
[See online article for color version of this figure.]
The Three Identified P. patens Hsp70s Are Imported into Chloroplasts and Localized in the Stroma
To verify that the three Hsp70s we identified are indeed chloroplast proteins, we isolated their respective cDNA clones, transcribed and translated them in the presence of 35S-Met, and incubated the radiolabeled proteins with isolated pea chloroplasts. As shown in Figures 1B to 1D, the precursor proteins were imported into the plastids and cleaved to their mature size. The mature proteins were resistant to digestion with either trypsin or thermolysin, indicating that they had penetrated both envelope membranes (Cline et al., 1984). In addition, these proteins remained in the soluble phase upon chloroplast fractionation. Comparison of their behavior toward these treatments with those of the stromal marker Hsp93 (Moore and Keegstra, 1993; Zheng et al., 2002), and the membrane-associated marker for the intermembrane space Tic22 (Kouranov et al., 1998, 1999), indicated that all three Hsp70 proteins analyzed reside in the stroma, as predicted.
Hsp70-2 Is an Essential Gene
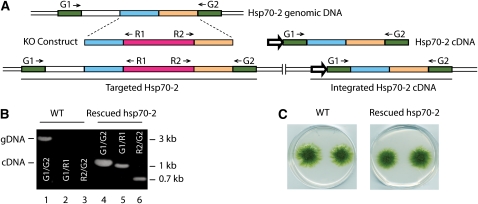
To gain insights into the chloroplast Hsp70 function, we took advantage of the homologous recombination activity of moss to create targeted knockout plants. We readily obtained targeted knockouts of Hsp70-1 and -3; a full analysis of these plants is in progress. In contrast with Hsp70-1, -3a, and -3b, we were unable to isolate an Hsp70-2 knockout plant using similar techniques despite 23 independent transformations and after screening 29 stable transformants. Only when a cDNA copy of the Hsp70-2 mRNA under control of the cauliflower mosaic virus 35S promoter was supplied during cotransformation with the knockout construct were we able to recover any plants. Subsequent PCR analysis identified a number of plants in which the chromosomal copy of Hsp70-2 had been interrupted by incorporation of the resistance gene and in which the cDNA copy persisted through two rounds of antibiotic selection, even though no further selection was supplied to maintain the cDNA (Figure 2). This indicates that Hsp70-2 is an essential gene required for plant viability. It is noteworthy that neither Hsp70-1 nor Hsp70-3 could substitute for the loss of Hsp70-2, even though they are almost 78 and 80% identical, respectively.
Figure 2.
Hsp70-2 cDNA Rescues the hsp70-2 Lethal Phenotype.
(A) Schematic representation of Hsp70-2 locus with a targeting construct (KO construct) and a rescuing cDNA. Positions of primers used for PCR are shown. The wider arrow represents the direction of 35S promoter.
(B) PCRs for Hsp70-2. Targeting products and integrated product using genomic DNA remplates from the wild type or an Hsp70-2 cDNA rescued knockout strain. Primers used in each PCR are indicated on gel images and in the scheme. gDNA, PCR product amplified from the genomic locus; cDNA, PCR product amplified from the cDNA construct that is incorporated in the genome of the rescued hsp70-2 strain.
(C) Photos of moss colonies of 1-month-old wild type and an Hsp70-2 rescued knockout strain.
Conditional hsp70-2 Mutants Show Reduced Capacity for Chloroplast Protein Import
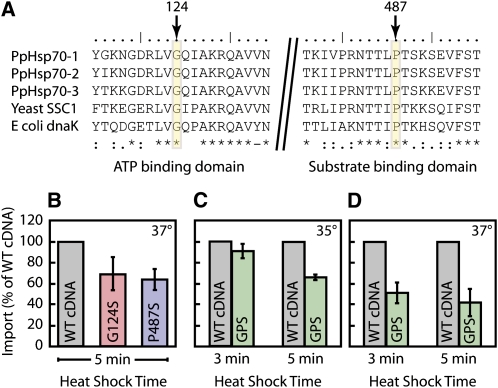
The hsp70-2 mutant could be obtained only when the moss was cotransformed with the knockout construct and rescuing cDNA, indicating that the Hsp70-2 gene is essential for viability. Since the null mutant is lethal, we needed additional tools to study the function of this gene. Two single amino acid substitutions have been reported to confer temperature sensitivity to the mitochondrial matrix Hsp70 in yeast (Ssc1p) (Kang et al., 1990; Gambill et al., 1993). One of these mutations is in the ATPase domain (Gly to Ser), and the other is in the substrate binding domain (Pro to Ser) of Ssc1p. The target amino acids for these mutations are highly conserved in Hsp70s and are present in the moss Hsp70-2 (Figure 3A). We introduced these mutations, either singly (G124S and P487S) or in combination (GPS), in the Hsp70-2 cDNA, which we then used as the rescuing copy during cotransformation with the knockout construct. The resulting plants were analyzed, and some of them were identified as potential temperature-sensitive mutants by PCR and by sequencing (see Supplemental Figure 1 online). G124S, P487S, and GPS plants showed no phenotype under normal growth conditions. We found that wild-type P. patens stops growing at ∼ 34 ° C and dies at 37 ° C, the nonpermissive temperature at which these substitutions were described in yeast (Kang et al., 1990; Gambill et al., 1993). Accordingly, we were unable to demonstrate a temperature-sensitive growth phenotype in these mutants, which is likely explained by our inability to probe these elevated temperatures.
Figure 3.
Protein Import into Chloroplasts Isolated from Temperature-Sensitive Mutants Is Inhibited by Heat Shock.
(A) Partial amino acid sequences of P. patens Hsp70-1, Hsp70-2, and Hsp70-3 are aligned with the yeast mitochondrial Hsp70 (SSC1) and E. coli DnaK. The conserved residues whose mutation resulted in a temperature-sensitive mutant in yeast are shown indicated by arrows and rectangles; the numbers correspond to the Hsp70-2 protein. Asterisks, amino acid residues in that column are identical in all sequences in the alignment; colons, conserved substitutions; period, semiconserved substitutions.
(B) to (D) Chloroplasts were isolated from protoplasts of the two single temperature-sensitive mutants (G124S and P487S), the double temperature-sensitive mutant (GPS), and a control strain cotransformed with a wild-type cDNA. Chloroplasts were heat shocked in 35 ° C (C) or 37 ° C ([B] and [D]) water baths for 3 or 5 min, as indicated. Radiolabeled precursor protein prCGE1 was then added to the heat-shocked chloroplasts to initiate the import reactions. Samples were subjected to SDS-PAGE and phosphor image analysis. Means + se are shown (n = 3).
[See online article for color version of this figure.]
Despite our inability to test for temperature-sensitive growth in moss, we reasoned that if Hsp70-2 was involved in protein import into chloroplasts, we should see an effect of these mutations when isolated plastids were subjected to a heat shock prior to initiation of an in vitro import reaction. To this end, chloroplasts were isolated from Hsp70-2 knockout plants rescued with wild-type cDNA (controls) and from plants rescued with G124S, P487S (Figure 3B), or GPS (Figures 3C and 3D) cDNAs. They were heat shocked at 35 or 37 ° C for 3 or 5 min and then incubated with CGE1 precursor under import conditions. Although the wild type import reaction is itself relatively heat sensitive, the chloroplasts from strains bearing temperature-sensitive mutated site(s) in the Hsp70-2 cDNA displayed reduced import capacity compared with those from the control plants (Figures 3B to 3D). We routinely observed that after heat shock the mutant plastids imported proteins at roughly 50% the rate of the controls. These data strongly suggest that Hsp70-2 is involved in protein import into chloroplasts.
Cloning and Characterization of Two CGE Genes from P. patens
As described in the Introduction, Hsp70 chaperones operate in a manner dependent on cochaperones. To further probe the involvement of the chloroplast Hsp70 system in protein import into this organelle, we sought to modify its activity by mutating its nucleotide exchange cochaperone, CGE. Without assuming specific cochaperone-chaperone partnerships, we reasoned that CGE knockout (or knockdown) mutants would be compromised in their ability to import proteins if Hsp70-2 was involved in this process.
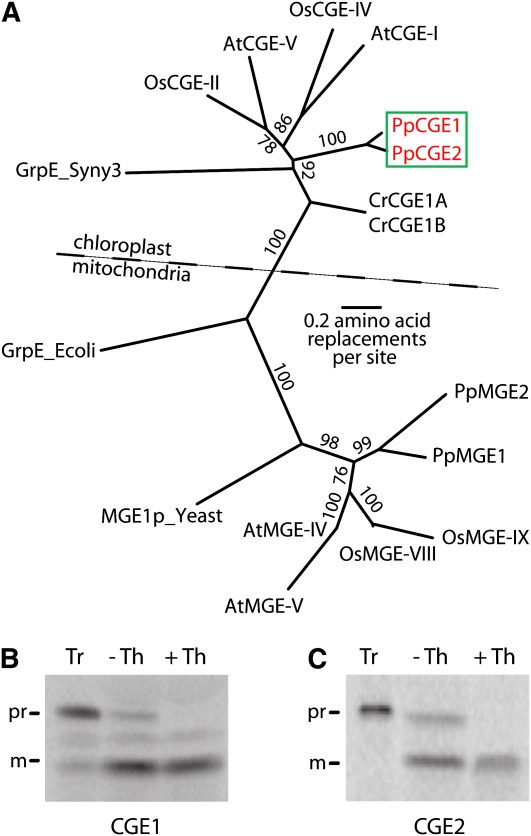
An analysis of the moss genome sequence revealed that the plastids contain two GrpE homologs, designated CGE1 and CGE2, with a second two homologs (MGE1 and MGE2) likely residing in the mitochondria. Cladistic analysis placed CGE1 and CGE2 proteins in the chloroplast clade, while the other GrpEs were in the mitochondrial clade, as expected from the localization predictions (Figure 4A). With the transit peptide excluded from the analysis, CGE1 and CGE2 display 83% sequence identity.
Figure 4.
Two CGEs of the Moss P. patens Are Localized in the Chloroplast Stroma.
(A) Phylogenetic analysis of CGE sequences. Four GrpE protein sequences from the moss P. patens (PpCGE1, PpCGE2, PpMGE1, and PpMGE2) and 13 from other species (see Methods for accession numbers) were aligned and a phylogenetic tree was generated using the neighbor-joining method. The scale bar indicates 0.2 substitutions per site. Bootstrap values that are >70% from 1000 replications for each branch are shown. The dotted line separates the chloroplast clade from the mitochondrial one. The alignment used to generate the tree is available as Supplemental Data Set 2 online.
(B) and (C) Import assay of CGE1 and CGE2. [35S]-labeled precursor proteins (Tr) were incubated with chloroplasts, followed by thermolysin (Th) treatments. All controls (-Th) were treated in the same manner excluding protease addition. Protease activities were quenched and chloroplasts were subjected to SDS-PAGE followed by phosphor image analysis as in Figure 1.
[See online article for color version of this figure.]
We isolated cDNA clones of the identified CGE1 and CGE2 genes. After in vitro translation, the radiolabeled precursor proteins were imported into pea chloroplasts and were processed to mature forms that were protease protected (Figures 4B and 4C). In this manner, CGE1 and CGE2 were identified as bona fide chloroplast proteins.
Characterization of cge Mutants
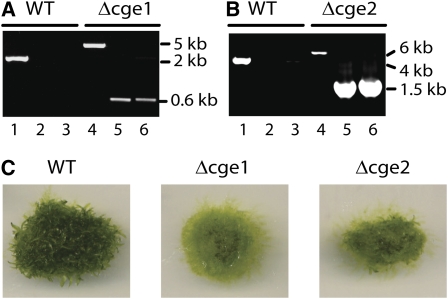
CGE1 and CGE2 genes were individually knocked out by means of polyethylene glycol (PEG)-mediated transformation. PCR (Figures 5A and 5B), RT-PCR (Figure 6A, lanes 2 and 3), and protein blotting (Figure 6C, lanes 2 and 3) showed that the two isolated transformants were null mutants; numerous independent lines were obtained for each type of mutant. The single cge1 and cge2 mutants are viable on phototrophic media but displayed similar slow growth phenotypes and were delayed in development of leafy shoots compared with the wild type (Figure 5C).
Figure 5.
Identification of CGE Single Mutants.
(A) PCR for CGE1 and targeting products using genomic DNA templates from wild-type or cge1 knockout plants. A scheme in Supplemental Figure 2A online shows the primer positions.
(B) PCR for CGE2 and targeting products using genomic DNA templates from wild-type or cge2 knockout plants. A scheme in Supplemental Figure 2B online shows the primer positions.
(C) Wild type, cge1, and cge2 mutants grown for 5 weeks on BCD medium.
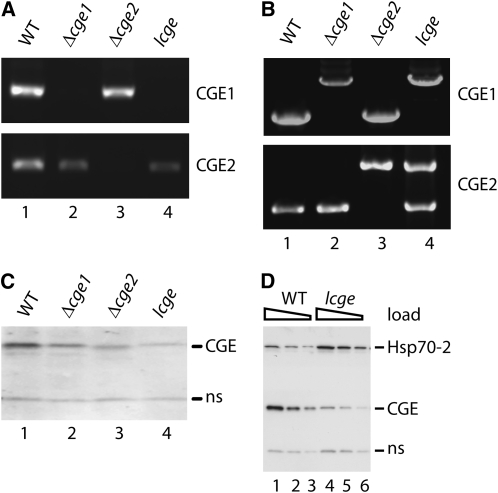
Figure 6.
Characterization of cge Single and the lcge Double Mutants.
(A) RT-PCR. DNA templates were cDNA prepared from wild-type and the mutant strains. One pair of primers complementary to either the CGE1 gene (top panel) or the CGE2 gene (bottom panel) was used.
(B) Diagnostic PCR. Genomic DNA was isolated from the wild type and from cge1, cge2, and lcge mutants as templates. One pair of gene-specific primers was used for each gene. The primers were complementary to the 5 ′ and 3 ′ homologous regions, respectively, of each knockout cassette.
(C) Immunoblotting. Chloroplasts were isolated from wild-type, cge1, cge2, and lcge plants. The antibody used was raised against the moss CGE2 protein and cross-reacted with the CGE1 protein.
(D) Immunoblotting using antibodies against Hsp70-2 and CGE. Chloroplasts were isolated from the wild type and the lcge mutant. ns, protein nonspecifically reacting with the CGE antibody. Samples were loaded on the basis of chlorophyll content: 2.0 μ g (lanes 1 and 4), 1.0 μ g (lanes 2 and 5), and 0.5 μ g (lanes 3 and 6).
To generate a cge double mutant, we used the cge1 mutant as background to knock out the CGE2 gene or, reciprocally, used the cge2 mutant as background to knock out the CGE1 gene. After screening 66 stable transformants, we identified one plant as a candidate double mutant. By employing the same strategies we used for identifying cge single mutants, we found that the patterns of PCR reactions using this mutant's genomic DNA as template were just like that of the singles (Figures 5A and 5B). This suggested that both wild-type CGE genes were disrupted by the knockout cassettes. Surprisingly, further analysis by RT-PCR showed that whereas the mutant plant did not express CGE1 mRNA (Figure 6A, top panel, lane 4), it did express some CGE2 mRNA (Figure 6A, bottom panel, lane 4). Protein blotting revealed that the mutant contained 10 to 20% of the CGE2 protein compared with the wild type (Figure 6C, lane 4). These results suggested that this plant is a null cge1 and knockdown cge2 mutant. We refer to this plant as the low CGE (lcge) mutant throughout the rest of this article. As with the hsp70-2 mutant, a true double cge knockout was only obtained when a rescue cDNA of the CGE2 gene was cotransformed with the knockout construct (see Supplemental Figure 2 online). Further PCR analysis suggested that, although the lcge mutant carried an insertion in both CGE loci, the CGE2 gene, along with part of its promoter, had become incorporated in an ectopic site in the genome (Figure 6B). These data are consistent with the postulate that a cge1 cge2 double knockout is lethal and that the lcge mutant we recovered survived on the basis of the low levels of CGE2 produced from the displaced gene.
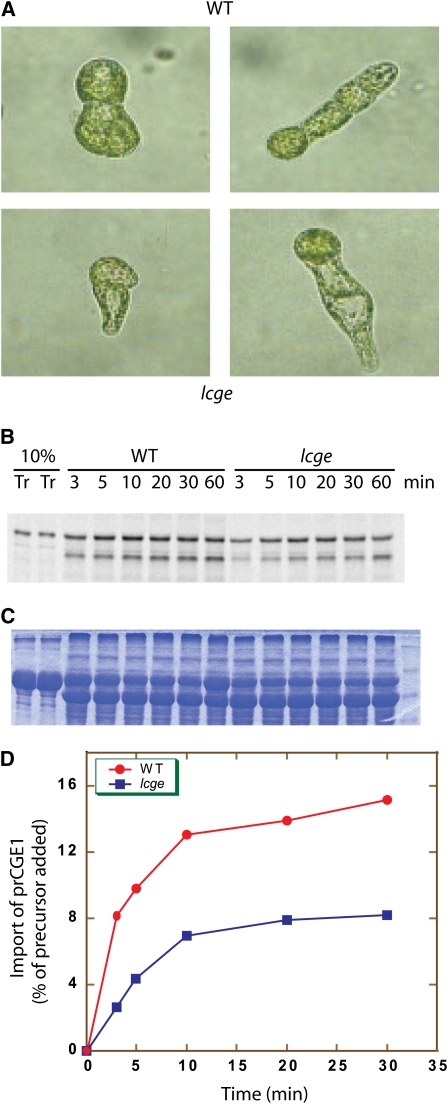
The low level of CGE2 expression in the lcge mutant background resulted in a phenotype more severe than that observed with either of the single cge knockout plants. We observed a 20 to 40% reduction in growth rate of the lcge mutant compared with the wild type, and a large portion of protonemal cells regenerated from lcge protoplasts had smaller and fewer chloroplasts, especially in the newly forming cells (Figure 7A). Strikingly, chloroplasts isolated from the lcge mutant often showed severe defects in protein import (Figures 7B to 7D). We conclude from these data that the stromal CGEs, and by extension, at least one of their chaperone partners, most likely Hsp70-2, play a crucial role in the chloroplast protein import reaction.
Figure 7.
Phenotyping and Protein Import Assay for the lcge Mutant.
(A) Colonies of two- and four-cell (left and right columns, respectively) stage wild-type (top) and lcge (bottom) plants were regenerated from protoplasts.
(B) Protein import assay. [35S]-labeled CGE1 precursor protein (Tr) was incubated with moss chloroplasts at room temperature. Samples were withdrawn at the indicated time points. A phosphor image is shown.
(C) Coomassie blue–stained gel showing the loading of samples used in (B).
(D) Quantification of the phosphor image in (B).One of three experiments is shown here.
The majority of protein import experiments performed with the lcge mutant revealed a defect in the ability of mutant chloroplasts to take up proteins, as in Figure 7. However, on occasion, chloroplasts from this plant were able to import proteins at the wild-type level. In an effort to understand this, we performed quantitative protein blotting of different cultures of the mutant for CGE and Hsp70-2. As expected, the level of CGE2 in the lcge mutant was consistently low. Interestingly, we observed a consistent concomitant increase in the level of Hsp70-2 (Figure 6D). We further found that the level of Hsp93 was not altered in the mutant (see Supplemental Figure 3 online). We hypothesize that Hsp70-2 was upregulated in the lcge mutant in an effort to compensate for the lower overall chaperone activity due to the low levels of CGE proteins and that this occasionally resulted in an activity of Hsp70-2 sufficient for protein import. This would require that Hsp70-2 manifests some chaperone activity in the absence of CGE. Such a weak Hsp70 activity in the absence of cochaperones has indeed been reported in other chaperone-dependent systems (Zylicz et al., 1989; Azem et al., 1997).
Immunoprecipitation of an Early Import Intermediate Residing in the Toc/Tic Complex with an Hsp70-2 Antibody
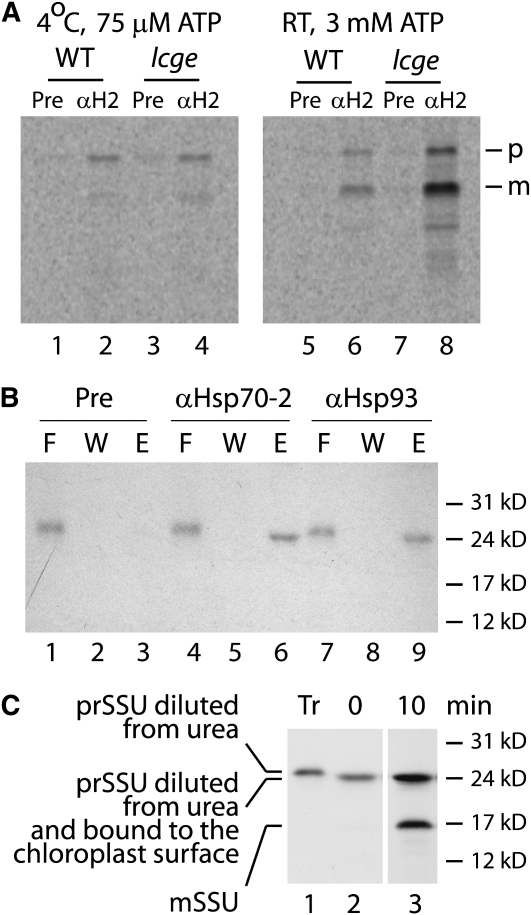
When precursor proteins are incubated with chloroplasts at low temperatures and with low concentrations of ATP, they form the so-called early import intermediate, in which precursor proteins are inserted into the Toc/Tic translocon but do not pass all the way into the stroma (Nielsen et al., 1997; Chen and Schnell, 1999). Precursors at this stage of the import process can be immunoprecipitated by antibodies directed at both Toc and Tic subunits. Under these conditions, Hsp93 has repeatedly been found to be associated with the translocon, and this stable association has been taken as evidence that Hsp93 provides the ATPase activity required for chloroplast protein import (Akita et al., 1997; Nielsen et al., 1997; Kovacheva et al., 2005, 2007; Chou et al., 2006). Our experiments with Hsp70-2 described above led us to reexamine whether this chaperone might also be associated with the translocon under conditions leading to the early import intermediate. In particular, we reasoned that this association might be more stable in the lcge mutant, as the chaperone is expected to be stranded in the high affinity substrate binding state after ATP hydrolysis but before CGE-mediated nucleotide exchange.
Figure 8A shows an experiment in which an isoform-specific antibody to Hsp70-2 immunoprecipitated the precursor to CGE1, either staged as the early import intermediate (left panel) or under import conditions (right panel), in both wild-type and lcge mutant chloroplasts. The presence of the precursor form in the immunoprecipitates indicates that the substrate protein had not reached the stroma and is consistent with it being captured while in the translocon. Control experiments demonstrated that relatively little precursor is pulled down if the antibody is added after chloroplast lysis. The antibody did not pull down a significant amount of mature protein unless import conditions were established. Clearly, more precursor and mature substrate protein were immunoprecipitated in the lcge mutant, consistent with the known role of the nucleotide exchange factor. The appearance of the mature protein in the pull downs could in principle be explained as the association of CGE with Hsp70-2 in its role as cochaperone. However, this cannot explain the pull downs containing the precursor as CGE functions as a cochaperone only as a dimer (Harrison et al., 1997). Instead, this experiment is consistent with the idea that Hsp70-2 associates with the translocon, does so more stably in the lcge mutant, and in so doing is able to immunoprecipitate precursors in transit through the Toc/Tic complex.
Figure 8.
Immunoprecipitation of Importing Precursors with Anti-Hsp70-2 and Anti-Hsp93 Antibodies.
(A) In vitro–translated prCGE1 was incubated with isolated chloroplasts from wild-type or lcge plants on ice at a low ATP concentration (to generate the early import intermediate; left panel) or at room temperature at a high ATP concentration (to allow complete import; right panel). Chloroplasts were reisolated, followed by immunoprecipitation using preimmune serum (Pre) or anti-Hsp70-2 antibody ( α H2) as described in Methods. Phosphor images are shown. The positions of the precursor (p) and mature (m) CGE1 protein are indicated to the right.
(B) Overexpressed precursor protein to the Rubisco small subunit (prSSU) was incubated with isolated chloroplasts from the lcge mutant for 20 min on ice at low ATP concentrations to generate the early import intermediate. Chloroplasts were reisolated and incubated with a cross-linker, DSP, on ice and then quenched with Tris buffer. After lysis, membranes were pelleted by centrifugation and solubilized with buffer containing Triton X-100. The supernatant obtained after further centrifugation was subjected to immunoprecipitation using preimmune serum or anti-Hsp70-2 or anti-Hsp93 antibodies. A fluorograph is shown. F, flow through; W, wash fraction; E, elution.
(C) Fluorograph of an import experiment showing that the slight change in mobility of prSSU observed in (B) is an artifact caused by the presence (lane 1) and absence (lanes 2 and 3) of urea. The import reaction times are indicated above lanes 2 and 3. prSSU, precursor; mSSU, mature form.
The precursor to the small subunit of Rubisco (prSSU) is probably the best studied chloroplast protein import substrate, although it is not imported as well in moss chloroplasts as the CGE1 precursor is. Under the conditions of the experiment shown in Figure 8A, we observed that little prSSU was immunoprecipitated with the Hsp70-2 antibody. In order to increase the amount of prSSU staged as the early import intermediate, we repeated this experiment with the lcge mutant using bacterially expressed precursor. Figure 8B shows that under these conditions, a substantial amount of prSSU was immunoprecipitated by the Hsp70-2 antibody. While the results of Figure 8A could possibly be interpreted as involving the cochaperone activity of CGE, this cannot be the explanation for Figure 8B since prSSU has no intrinsic affinity for Hsp70-2. This experiment then demonstrates that Hsp70-2 must be part of the Toc/Tic complex in which prSSU is trapped as the early import intermediate in the cold at low ATP concentrations in the lcge mutant.
The last three lanes of the gel in Figure 8B demonstrate that prSSU is also immunoprecipitated by an antibody to Hsp93. This chaperone is a known and accepted component of the active envelope translocon (Akita et al., 1997; Nielsen et al., 1997; Constan et al., 2004; Kovacheva et al., 2005, 2007; Chou et al., 2006). While our experiment was not strictly quantitative, Hsp70-2 and Hsp93 appear to pull down the same amount of prSSU, suggesting that they are both present in the same complex. Figure 8C shows that the small change in mobility of prSSU between the flow through and eluted samples observed in panel B is an artifact caused by dilution from urea.
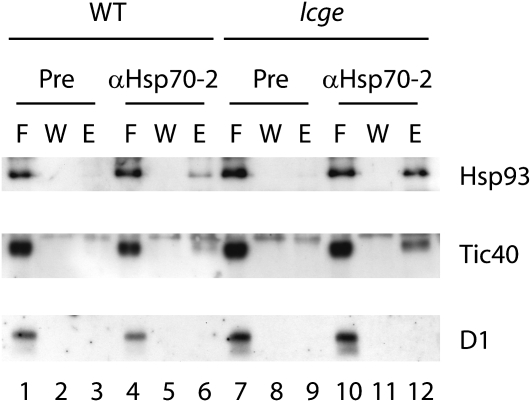
In order to demonstrate that Hsp70-2 is present in the Toc/Tic complex under conditions in which it coimmunoprecipitated the early import intermediate in lcge chloroplasts, we probed the immunoprecipitate with antibodies to other translocon subunits. Figure 9 shows that the complex pulled down with Hsp70-2 antibodies contained Hsp93 and Tic40. The specificity of the detection is demonstrated by the lack of reaction with an irrelevant antibody against the photosystem II core D1 protein. These experiments demonstrate that Hsp70-2 is present in the actively translocating Toc/Tic complex in chloroplasts and that this association is stabilized under conditions in which CGE is limiting.
Figure 9.
Immunoblotting of Immunoprecipitate Obtained with Anti-Hsp70-2 Antibody.
Blots with antibodies against two translocon components, Hsp93 and Tic40, and an irrelevant antibody, D1, are shown. The import and coimmunoprecipitation assays were performed as described in Figure 8B. F, flow through; W, wash fraction; E, elution.
DISCUSSION
In this report, we have provided three lines of evidence indicating that a stromal Hsp70 system is involved in protein import into chloroplasts. First, combining reverse genetic and biochemical approaches, we showed that Hsp70-2 is essential for viability of the moss P. patens. We generated conditional hsp70-2 mutants by knocking out this gene via gene targeting while providing cDNA copies that contained temperature-sensitive mutations to rescue the plants. Heat-shocked chloroplasts isolated from these temperature-sensitive mutants displayed lower competence for protein import into chloroplasts when compared with the wild type. Second, we generated a mutant with significantly lower than wild-type levels of the stromal cochaperone CGE, and chloroplasts isolated from this lcge mutant plant were generally compromised in their ability to import proteins. Finally, we showed that precursors in the process of traversing the Toc/Tic translocon could be immunoprecipitated by an antibody to Hsp70-2, as could other translocon components, indicating that this chaperone is also in the translocon complex. These data, combined with previous reports in which incoming precursors or newly imported proteins have been observed in contact with stromal Hsp70 (Madueno et al., 1993; Tsugeki and Nishimura, 1993; Nielsen et al., 1997; Vojta et al., 2007), make a strong case for a role of the moss Hsp70-2 in chloroplast protein import. In addition, a recent article by Ratnayake et al. (2008) suggests that the Hsp70 that was previously reported to reside in the envelope intermembrane space (Hsp70-IAP) and which is accepted to be in contact with translocating precursors (Schnell et al., 1994; Jackson-Constan et al., 2001) is in fact a stromal chaperone, perhaps analogous to Hsp70-2. Given the high degree of similarity among the chloroplast stromal Hsp70s, 83% between Hsp70-1 and Hsp70-2 for instance, it is surprising that the individual hsp70-2 null mutation is lethal and cannot be complemented by the remaining chaperones, even though they are each found in the moss EST database, indicating that they are each expressed. Recently, some of the disparate activities of Hsp70 chaperones have been unified mechanistically in the entropic pulling hypothesis (De Los Rios et al., 2006). Nevertheless, it is known from other systems that different individual chaperones can have individual functions that are not complemented by their close homologs (Nimura et al., 1996, 2001; Blanco-Rivero et al., 2005; Su and Li, 2008). It is not clear at present which region(s) of the protein confers the remarkable specificity of moss Hsp70-2.
We found only two CGE homologs in the moss chloroplast stroma. Since there are three stromal Hsp70s, this suggests that the CGE cochaperones do not exhibit specificity for their partner chaperones. Consistent with this, we found that both cochaperones were immunoprecipitated by Hsp70-2 from the stroma after their import (see Supplemental Figure 4 online). We also note that both individual cge1 and cge2 null mutants displayed similar slow growth and delayed development phenotypes, which again suggests that these cochaperones do not serve specific Hsp70s. This is not unexpected, as it is usually through the J-domain protein cochaperones that specificity for chaperone substrates is conferred (Schroda and Vallon, 2008; Sharma et al., 2009). Thus, we can anticipate that the low amount of CGE2 still present in the lcge mutant plastids would support only a low activity of all chloroplast Hsp70s, which would in turn be expected to have pleiotropic consequences for chloroplast functions. It is interesting that the level of Hsp70-2 increased in response to the CGE decrease (Figure 6D), in an apparent effort to maintain chaperone activity. It is noteworthy that the amount Hsp93 was not altered in lcge mutant compared with the wild type (see Supplemental Figure 3 online).
There are two potential interpretations of our data that do not include participation of Hsp70-2 in chloroplast protein import, neither of which we think likely, but which bear some discussion. In the first, the protein import defect observed in the hsp70-2ts and lcge mutants is seen as a consequence of a component required for this process misfolding after synthesis in the absence of the chaperone. While this interpretation could explain the effects of Hsp70-2 and CGE mutations on import in vivo, we believe it is a less tenable explanation for our in vitro heat shock experiments. In these latter experiments, the 3- to 5-min heat shock was delivered to isolated chloroplasts, and the subsequent import reaction was initiated immediately thereafter, leaving no possibility for the de novo synthesis of new proteins that might require the chaperone for folding. Were this interpretation to apply to the in vitro experiments, we would have to postulate that the chaperone system was needed to affect a rapid repair of a heat denatured (membrane) protein within a few minutes, and generally these repair cycles occur on somewhat longer time scales (Glover and Lindquist, 1998; Goloubinoff et al., 1999; Krzewska et al., 2001). In addition, this interpretation offers no explanation of our observation that the Hsp70-2 is present in the translocon during import (Figures 8 and 9).
A second possible interpretation of some of our data that does not include a role for Hsp70 in protein import is that Hsp93 is in fact the sole chaperone required for import but that it is titrated away from the translocon to fill in for a poorly functioning Hsp70 system to perform other housekeeping functions. While this interpretation could conceivably apply to our results obtained with the mutants we have generated, it offers no explanation for why we would recover Hsp70-2 in the translocon with the early import intermediate and why this recovery would be enhanced in the lcge mutant. As already stated, our combined data can best be interpreted as demonstrating the participation of Hsp70-2 in the import process.
Obviously, we were not the first to consider the possibility that Hsp70 is involved in protein import into chloroplasts. The idea that Hsp93 and not Hsp70 acts as the plastid protein import motor arose from experiments such as those shown in Figures 8 and 9, wherein Hsp93 was observed by coimmunoprecipitation experiments to be in contact with an incoming precursor and with the other translocon components. One can justifiably ask why we have seen Hsp70 in this location when others have not. To answer this, we note first that the vast majority of the work done in this area has been performed with prSSU. It has been suggested in the literature that different precursors appear to interact with different chaperones (Vojta et al., 2007), so the use of prSSU may have biased previous experiments toward Hsp93. Indeed, in our initial experiments, we did not observe prSSU in the Hsp70-2 immunoprecipitate, and it was not until we increased its concentration at the early import intermediate stage using bacterially overexpressed protein that this precursor was efficiently immunoprecipitated by Hsp70-2 antibody. However, it is conceivable that this is an artifact of the moss system because, in contrast with its behavior in pea chloroplasts, prSSU is not one of the better importing substrates in moss chloroplasts. In fact, a different substrate that is imported well by moss chloroplasts, the CGE1 precursor protein, was readily immunoprecipitated as an early import intermediate when produced in low concentrations by in vitro transcription/translation (Figure 8).
Perhaps a more relevant reason that we have observed Hsp70-2 in the translocon when others have not is due to our use of the lcge mutant, in which the levels of the CGE are lower than in the wild-type plants. The expected consequence of lowering the CGE activity is that the chaperone would become stabilized in the ADP-bound form, a state in which it has an elevated affinity for the substrate. We do not know at this point whether this increased affinity applies to a site on the translocon or on the transporting protein or both. In either case, we would expect to recover more Hsp70-2 in contact with the early import intermediate and/or the translocon in the lcge mutant compared with the wild type, which is what we observed (Figures 8 and 9).
It is noteworthy that immunoprecipitation of prSSU by anti-Hsp70 antibody was observed previously under binding and import conditions in pea chloroplasts (Akita et al., 1997; Nielsen et al., 1997; Jackson-Constan et al., 2001). The first publication to report this (Nielsen et al., 1997) also described for the first time the presence of Hsp93 in the translocon; indeed, the interaction of the latter chaperone with precursors and translocon components was stronger than that with Hsp70. Because of this apparent higher affinity of Hsp93 for the translocation machinery, the role of Hsp70 in protein import was deemed equivocal (Akita et al., 1997; Nielsen et al., 1997; Jackson-Constan et al., 2001) and was ultimately dropped from consideration. However, others have pointed out that interactions between chaperones and their substrates might be intrinsically dynamic in nature and difficult to detect, but important nonetheless (Glover and Lindquist, 1998), and we suggest that this is the case with the stromal Hsp70s.
Protein import into chloroplasts proceeds through three stages: recognition, translocation, and maturation (Kessler and Schnell, 2006). Considering the stromal localization of the chaperones and cochaperones addressed by this study, it seems most likely that the Hsp70 system is involved in the translocation or/and maturation stage. Although the driving force for protein translocation across chloroplast envelopes has been proposed to be provided by the stromal Hsp93, whose participation in protein import has been amply demonstrated, this does not preclude the additional involvement of the stromal Hsp70 system. This may explain our puzzling observation that protein import into chloroplasts containing putative temperature-sensitive Hsp70 proteins as the only functional chaperone was inhibited by only ∼ 50% after heat shock. This was observed when the temperature-sensitive conferring amino substitutions occurred in either of two locations in different functional domains of the protein or when both mutations were combined in a single protein. We can envision three possible explanations for this result. First, the two mutant substitutions each coincidentally destabilized the chaperone to the same extent, and one domain dominates the heat-induced denaturation pathway. Second, one of the other Hsp70s present in the stroma can function in protein import and complements this activity of Hsp70-2. In this case, one would have to postulate that the lethality of the hsp70-2 null mutant is caused by some chaperone function other than chloroplast protein import. Finally, the 50% inhibition limit could be the result of a shared role of Hsp70-2 with another ATPase to provide the driving force for import. In this model, Hsp93 would appear to be the most likely candidate for the other ATPase.
Interactions between members of the Hsp70 and Hsp100 families of chaperone are known from other systems. For instance, in the presence of Hsp70, but not in its absence, both bacterial ClpB and yeast Hsp104 are able to mediate the disentanglement of protein aggregates and their subsequent refolding (Glover and Lindquist, 1998; Goloubinoff et al., 1999). Even more to the point, some Hsp100 family members have been implicated in protein transport across membranes. mtHsp78 (an Hsp100 family member) participates in protein import when the mtHsp70 is compromised (Schmitt et al., 1995; Krzewska et al., 2001). Specifically, Δ hsp78 mutants show no phenotype in wild-type cells but do not grow in a temperature-sensitive mtHsp70 mutant, even at permissive temperatures. The import deficiency observed in this Hsp70ts mutant is suppressed by overexpression of Hsp78, and the latter protein has been shown under these conditions to associate with incoming precursors. In addition, an Hsp100 present in the ER (p97/Cdc48) is thought to participate in the retrotranslocation of misfolded proteins from the lumen to the cytoplasm (Nakatsukasa and Brodsky, 2008). Many of these proteins likely interact with Hsp70 en route to the dislocation channel (Nishikawa et al., 2005). A number of parallels between the action of chloroplast Hsp93 and that of p97/Cdc48 have been noted previously (Chou et al., 2006). A possible interaction between Hsp70-2 and Hsp93 is suggested by our observation that they are both present in the same translocon with the anti-Hsp70-2 immunoprecipitated early import intermediate (Figures 8 and 9). Future experiments will be directed toward determining whether these chaperones cooperate with each other to affect chloroplast protein import, as they do to untangle aggregated proteins (Glover and Lindquist, 1998; Goloubinoff et al., 1999) and in ER dislocation (Lilley and Ploegh, 2004) or whether they substitute for one another to drive protein import, as observed in mitochondria (Schmitt et al., 1995; Krzewska et al., 2001).
The hypothesis that an Hsp70 functions at the trans-side of the chloroplast envelope membranes during protein import will not be viewed as surprising by those who study protein import into the mitochondria and the ER (Schatz and Dobberstein, 1996). In those systems, the indispensible action of mtHsp70 and BiP are well known and accepted. What does appear to be unique to plastids is that two chaperone systems, Hsp70s and Hsp100s, participate obligatorily in this process.
METHODS
Growth of the Moss and Pea
Physcomitrella patens (subspecies patens, Gransden) was maintained as previously described (Hofmann and Theg, 2003). Cultures were grown on BCD medium supplemented with 5 mM ammonium tartrate and 1 mM CaCl2 (Knight et al., 2002) overlaid with cellophane at 23 ° C under continuous light (60 to 80 μ mol photons m−2 s−1).
Pea (Pisum sativum, var Little Marvel) was grown at 20 ° C in moist vermiculite. The growth chamber was set to a 16-h photoperiod with light intensities of 50 to 60 μ mol photons m−2 s−1. Pea seedlings were harvested 10 to 12 d after germinating.
Isolation and Cloning cDNAs of Stromal Hsp70s and CGEs from P. patens
The strategies of database searching for homologs of chloroplast proteins in P. patens were described previously (Hofmann and Theg, 2003). The protein sequences of chloroplast-located Hsp70s from Arabidopsis thaliana (At4g24280 and At5g49910) were used for homology searching (tBLASTn; Altschul et al., 1997) in the GenBank P. patens EST and NIBB contig (http://moss.nibb.ac.jp/) databases. Homologs with E-values lower than 10−4 were assembled using the software Vector NTI Advance 9, ContigExpress, and the transit peptides in the resulting sequences were predicted with the programs ChloroP and TargetP (Emanuelsson et al., 2007). Two candidates were designated Hsp70-1 and Hsp70-2. Full-length cDNAs were isolated using the GeneRacer kit (Invitrogen) and then cloned into pCR4-TOPO vectors (Invitrogen). Each cDNA was sequenced at ∼ 2.5 × coverage. Primers used for cloning are Hsp70CTFw and Hsp70CTRv (for Hsp70-1); Hsp70BJFw and Hsp70BJRv (for Hsp70-2); and GeneRacer 5 ′ and GeneRacer 3 ′ (Invitrogen, for both genes); for sequencing, the primers used are Hsp70CT-5A, Hsp70CT-3A, Hsp70CT-5B, Hsp70CT-3B, Hsp70CT-5C, Hsp70CT-3C, Hsp70BJ-5A, Hsp70BJ-3A, Hsp70BJ-5B, Hsp70BJ-3B, Hsp70BJ-5C, Hsp70BJ-3C, T7 and T3. Specific primers to Hsp70-1 and Hsp70-2 are listed in Supplemental Table 1 online.
The Arabidopsis CGE protein sequence (CAB40381) was used for homology searching in P. patens. Three ESTs (AW476917, AW699924, and AW739069) and one contig were identified, and these all described one gene. Full-length cDNA of this CGE gene was isolated using GeneRacer (Invitrogen) and cloned into a pCR-Blunt II TOPO vector (Invitrogen). The cDNA was sequenced and designated CGE1. Primers used for cloning (GrpE-GR-Fw, GrpE-GR-Rv, GrpE-5end, and GrpE-3end) of CGE1 cDNA are summarized in Supplemental Table 1 online.
After the full sequence of the P. patens genome was released (Rensing et al., 2008), we identified two more Hsp70 and one CGE genes with products predicted to be located in chloroplasts. The two new Hsp70 genes were 100% identical in their intron/exon structure and sequence, and through 250 untranslated base pairs 3 ′ to the stop codon; they differed only in their respective 5 ′ untranslated regions. These three newly discovered genes were designated Hsp70-3a, Hsp70-3b, and CGE2. The cDNAs of the Hsp70-3 (identical for Hsp703a and Hsp70-3b) and CGE2 genes were isolated and cloned into pCR Blunt II TOPO vector. Primers, Hsp70-3Fw, Hsp70-3Rv, PpGrpE2-Fw, and PpGrpE2-4, used for this purpose are compiled in Supplemental Table 1 online.
Protein Sequence Alignment and Phylogenetic Analysis
Moss Hsp70 and CGE sequences were obtained from the moss genome project (http://genome.jgi-psf.org/Phypa1_1/Phypa1_1. home.html) at the Joint Genome Initiative using BLAST and Searching servers. Protein sequences from species other than P. patens were retrieved from GenBank and SwissProt. Multiple alignments of protein sequences were constructed using ClustalX version 1.87 (Thompson et al., 1997) with default gap penalties (see Supplemental Data Sets 1 and 2 online). The unrooted tree was generated using programs in PHYLIP 3.67 (http://evolution.gs.washington.edu/phylip.html). Protein distance matrixes were computed from the sequences using Jones-Taylor-Thornton model in the PROTDIST program. These matrixes were then analyzed using NEIGHBOR and CONSENSE functions to obtain a bootstrapped neighbor-joining tree. A bootstrap analysis of 1000 replicates was performed.
Construction of Knockout and Rescue Cassettes
Knockout cassettes containing antibiotic resistance genes encoding either hygromycin-phosphotransferase (APH4, conferring resistance to hygromycin) or neomycin-phosphotransferase (NPTII, conferring resistance to G418 sulfate) flanked by 600 to 1500 bp moss DNA encoding Hsp70-2 or CGE1 genes were constructed using three-step sequential PCR.
To make the Hsp70-2 knockout construct, moss genomic DNA and pBI426 plasmid (a gift from C. Champagne), which is a pUC 9 based vector and contains the nptII gene behind the 35S promoter, were used as templates in PCR reactions. It is noteworthy that the 35S promoter is not a strong promoter in moss (Horstmann et al., 2004; see also the NIBB PHYSCObase website at http://moss.nibb.ac.jp/). Two pieces of Hsp70-2 gene-specific DNA that cover approximately the first half of the gene were amplified and bisected by the nptII gene via two more rounds of PCR reactions. Six primers [Hsp70BJ-1N, Hsp70BJ-2N, Hsp70BJ-3N, Hsp70BJ-4N, (HspBJ2N)R1, and (HspBJ3N)R2] were used in this series of PCRs (see Supplemental Table 1 online). The knockout cassette was then cloned into a pCR4 TOPO vector, and the resulting plasmid was used as the DNA template to produce knockout transformation substrates via PCR.
The CGE1 knockout construct was made similarly: the knockout cassette containing the nptII gene flanked by ∼ 500 bp of CGE1 specific cDNA was cloned into a pCR4 TOPO vector. The resulting plasmid was used as template to produce linear DNA via PCR, with which moss protoplasts were transformed. Primers [GrpE-1, (R1)GrpE-2, (R2)GrpE-3, GrpE-4, (GrpE-2)R1, and (GrpE-3)R2] for making this construct are listed in Supplemental Table 1 online.
To make the CGE2 knockout construct, two pieces of CGE2-specific genomic DNA were amplified by PCR using primrs KpnI-GrpE2-1, ClaI-GrpE2-2, NotI-GrpE2-3, and SacI-GrpE2-4 (see Supplemental Table 1 online) and were cloned on either side of the aph4 gene in the plasmid pTN86 (AB267705, a kind gift from M. Hasebe). The entire knockout cassette was amplified by PCR and used for moss transformation.
To make rescue plasmids, Hsp70-2 cDNA was amplified using primers Hsp70BJFw and Hsp70BJ3en (see Supplemental Table 1 online) and cloned into pCR-Blunt II TOPO vector (Invitrogen). The cDNA was then subcloned into the BamHI/XhoI sites of a pART7 vector (Gleave, 1992; a gift from J. Bowman) that contains the 35S promoter and the OCS terminator. CGE1 cDNA was cloned into the XmaI/ClaI sites of the pART7 vector using primers XmaGrpE5en and ClaGrpE-4 (see Supplemental Table 1 online). The CGE2 rescue plasmid was made by cutting the cDNA from the plasmid mentioned in the section above into the pART7 plasmid via KpnI and XbaI sites.
Since we had the Hsp70-2 cDNA rescue plasmid in hand and had successfully used it to prove that Hsp70-2 was an essential gene, in transformations, we reasoned that it would be a straightforward matter to use it to generate Hsp70-2 rescue plasmids with temperature-sensitive substitutions. To this end, we generated the described point mutations from the original Hsp70-2 cDNA using the QuikChange kit (Stratagene). Primers used for mutation of G to S in the ATPase domain are BJ-ATP-Fw and BJ-ATP-Rv; for mutation of P to S in the substrate binding domain, the primers are BJ-Subs-Fw and BJ-Subs-Rv (see Supplemental Table 1 online).
Constructs for Protein Import Assays
For in vitro chloroplast import reactions, Hsp70-1, Hsp70-2, Hsp70-3, and CGE1 cDNAs were amplified and cloned into pCR-Blunt II TOPO vector. Primers used in PCRs for each gene are listed in Supplemental Table 1 online: Hsp70CTFw and GeneRacer 3 ′ (for Hsp70-1); Hsp70BJFw and GeneRacer 3 ′ (for Hsp70-2); Hsp70-3Fw and Hsp70-3Rv (for Hsp70-3); GrpE-5end and GeneRacer 3 ′ (for CGE1), respectively. The resulting plasmids were linearized with restriction enzyme SpeI and transcribed (Promega) in vitro under the control of T7 promoter. The radiolabeled precursor proteins were then translated in rabbit reticulocyte lysate (Promega) in the presence of 35S-Met and a mixture of amino acids excluding Met.
The plasmid containing CGE2 cDNA in pCR-Blunt II TOPO vector was the same as mentioned in the section above and was linearized with NotI, and the mRNA was transcribed under control of SP6 promoter. The radioactive CGE2 precursor protein was translated in wheat germ in the presence of 35S-Met and other nonradioactive amino acids.
The clone for translation of pea Tic22 is as described by Kouranov et al. (1999). The clone for translation of the pea Hsp93 precursor protein (L09547) was a kind gift from K Inoue. The two plasmids were linearized with XhoI and transcribed with the T7 polymerase, and the 35S-precusor proteins were prepared in rabbit reticulocyte lysate.
The cDNA of the pea precursor protein to small subunit of Rubisco (prSSU) was amplified using primers pRBCD161S and pRBCD161A (see Supplemental Table 1 online) and cloned into pET161/GW/D-TOPO vector bearing a C-terminal 6XHis tag (Invitrogen). The resulting plasmids were introduced into Escherichia coli strain BL21-CodonPlus (Stratagen) via electroporation. The prSSU was overexpressed in presence of 3H-Leu. The cells were lysed with M9 medium containing CelLytic Express powder (Sigma-Aldrich). The 3H radiolabeled prSSU was affinity purified against Ni-NTA Agarose as described in the manual (Qiagen). prSSU was then concentrated by centrifugation using an Amicon Ultra Centrifugal Filter Device (Millipore) with a 10-kD cutoff. The protein was stored in 8 M urea, 8 mM DTT, 10 mM Tris, and 100 mM phosphate buffer, pH 7.0, at −80 ° C.
Import of Hsp70 Proteins into Pea Chloroplasts and Fractionation of Chloroplasts
Intact chloroplasts were isolated from 10- to 12-d-old pea shoots as described by Theg et al. (1989). Radiolabeled precursors were incubated with pea chloroplasts for 40 min under light in presence of 3 mM ATP. The chloroplasts were reisolated through a 35% Percoll pad, recovered by centrifugation at 2600g for 7 min, washed once with import buffer (IB; 330 mM sorbitol, 50 mM Tricine, and 3 mM MgCl2, pH 8.0), and resuspended in IB. Half of the reisolated chloroplasts were solubilized in Laemmli sample buffer (Laemmli, 1970) as a control. The other half was lysed with buffer containing 10 mM MES and 5 mM MgCl2, pH 6.5, and were centrifuged at 100,000g for 8 min. The supernatant was precipitated with 15% trichloroacetic acid and resuspended in Laemmli sample buffer; the pellets were resuspended in the same sample buffer. The intact chloroplast sample and both soluble and membrane fractions were subjected to SDS-PAGE and phosphor image analysis.
Protease Treatments of Chloroplasts
Following incubation of chloroplasts with radioactive precursor proteins, the chloroplasts were centrifuged and resuspended in 600 μ L ice-cold import buffer supplemented with either 200 μ g trypsin/mL or 200 μ g thermolysin/mL and 5 mM CaCl2. Protease treatments were incubated on ice for 30 min, after which the chloroplasts were recovered by centrifugation at 1000g for 1.5 min. The protease activities were quenched by adding cold IB containing twofold to fivefold excess of soybean (Glycine max) trypsin inhibitor or EDTA. All controls were treated the same manner except without adding proteases.
Generation of Knockouts and Rescued and Temperature-Sensitive Mutants
PEG-mediated transformation of P. patens protoplasts was performed essentially as described by Schaefer et al. (1991). The concentration of protoplasts isolated from 6- to 7-d-old subcultured protonemal tissues was adjusted to 1.6 × 106/mL. Protoplasts (300 μ L) were mixed with 300 μ L PEG solution and 10 μ g linear knockout DNA. In case of rescuing cotransformation, 10 μ g of rescue plasmids were also added. The total volume of DNA was <100 μ L. The protoplast mixture was heat shocked at 42 ° C for 5 min and cooled at room temperature for 10 min. The protoplasts were then diluted with 1, 2, and 7 mL of 8% mannitol at 10-min intervals. Protoplasts were pelleted, resuspended in 0.5 mL of 8% mannitol, embedded in 10 mL agar, and were spread onto three plates of protoplast regeneration medium overlaid with cellophane. Selection of stable transformants was performed according to Knight et al. (2002). After 7 d of regeneration, colonies were transferred to and grown on selection media containing 30 mg/L of either G418 or hygromycin for ∼ 14 d. Colonies were then grown on antibiotic-free medium for 1 week, followed by a second 2-week selection period. The surviving stable transformants were then subjected to genotyping.
Identification of the Genotypes of Stable Transformants
Genomic DNA of moss strains was isolated as described by Knight et al. (2002). Three or four PCR reactions were performed for genotyping of a single mutant (Figure 2). The first reaction contained gene-specific primers (marked as G1 and G2 in Figure 2; for instance, Hsp70BJFw and BJ-3B for Hsp70-2; IdGrpE-Fw and IdGrpE-Rv for CGE1; IdGrpE2-Fw and IdGrpE2-Rv for CGE2) that annealed to the regions outside the knockout cassette. In the second and third reactions, one gene-specific (G1 or G2) primer together with a primer annealing to the resistant gene (marked as R1 or R2 in Figure 2; for example R1LongRv or R2LongFw for identification of Hsp70-2 and CGE1 KO; Hyg-Fw or Hyg-Rv for identification of CGE2 KO) were used to determine if the 5 ′ and 3 ′ ends of the knockout cassette were incorporated at the gene locus. In case of the cge2 mutant, an additional PCR using KpnI-GrpE2-1 and SacI-GrpE2-4 as primers was performed to determine whether the replaced genomic DNA was inserted at another location in the genome. All sequences of primers mentioned here are summarized in Supplemental Table 1 online.
Isolation of RNA and cDNA for RT-PCR
Total RNA was isolated from moss tissues using the RNeasy plant mini kit (Qiagen). cDNAs were synthesized according to the manual of the SuperScript first-strand synthesis system for RT-PCR (Invitrogen).
Isolation of Intact Chloroplasts from P. patens and Import of Proteins into Heat-Shocked Moss Chloroplasts
Chloroplasts were isolated from protoplasts of 6- to 8-d-old P. patens protonemal tissues as described previously (Hofmann and Theg, 2003). Protein import assays were conducted as described above for pea. For heat shock treatment, 30 μ L of intact chloroplasts (15 μ g chlorophyll) were incubated in a 35 or 37 ° C water bath for the indicated time and then placed on ice. These heat-shocked chloroplasts were subsequently used for protein import reactions, as described above.
Production of Antibodies and Immunoblotting
For overexpression of CGE2-glutathione S-transferase (GST) fusion protein, cDNA encoding CGE2 mature protein according to prediction of ChloroP was amplified using primers pet-MaGE2-Fw and pet-GE2-Rv, which possess SmaI and BamHI sites (see Supplemental Table 1 online), respectively. The PCR product was cloned into pET-14b-GST plasmid (a kind gift from Xiao-Ping Zhang and Wolf Heyer), which contains a T7 promoter followed by GST insert and T7 terminator. CGE2 cDNA was inserted downstream of GST. Between the cDNA and GST sequences, there was a PreScission protease cleavage site for protein purification purposes. BL21(DE3) cells (Invitreogen) were transformed with the plasmid pET-MaCGE2, and CGE2-GST fusion protein was overexpressed using a standard method.
For purification of CGE2, BL21(DE3) cells were grown in Luria-Bertani medium supplemented with carbenicillin. After induction of the fusion protein CGE2-GST with 1 mM isopropyl- β -d-thiogalactopyranoside, cells were resuspended in PBS plus protease inhibitors (0.1 mM benzamidine, 2 μ M leupeptin, and 2 μ M pepstatin) and were broken by sonication. The lysate was centrifuged for 15 min at 12,000g. The supernatant was loaded onto a glutathione Sepharose 4B (GE Healthcare Life Sciences) column, which was then washed with 10 × bed volume of PBS. Cleavage of the fusion protein was performed in column with PreScission protease, which itself is a GST-tagged protein. The cleavage buffer containing 50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 0.5 mg/mL PreScission protease, pH 7.5, was applied to the column and incubated at 4 ° C overnight. CGE2 was eluted with cleavage buffer (minus PreScission protease), while the GST portion of the fusion protein and the protease stayed in the column at this step and were eluted with 10 mM glutathione later.
CGE1 protein was overexpressed and purified using similar methods except gene-specific primers pet-MaGE1-Fw and pet-GE1-Rv (see Supplemental Table 1 online) were applied.
The purified CGE2 was injected to rabbits to produce CGE antibody. This antibody recognizes both CGE1 and CGE2, with higher affinity to the latter (see Supplemental Figure 5 online).
Peptides YNQPGGAPEGSAPGAQ and QPGSGAASSPPGDDAE from the C terminus of Hsp70-1 and Hsp70-2, respectively, were synthesized and conjugated to the carrier protein KLH. The conjugates were used for immunization in rabbits. The resultant antibodies are isoform specific and can distinguish Hsp70-1 from Hsp70-2 (see Supplemental Figure 6 online). Antibodies were purified through a column of Protein A Sepharose 4 Fast Flow (GE Healthcare Life Sciences).
Immunoblotting was performed according to Hofmann and Theg (2005). Other antibodies used in this work are as follows: Hsp93 antibody raised against pea ClpC (Shanklin et al., 1995), D1 antibody raised against Chlamydomonas reinhardtii C-terminal peptide of D1, a photosystem II reaction center protein (a gift from Richard Sayre), and Tic40 antibody raised against the Arabidopsis protein (Chou et al., 2006; a gift from Hsou-min Li). Goat-anti-rabbit IgG conjugated to horseradish peroxidase (Bio-Rad) diluted to 1:10,000 was used for immunodecorating for all proteins except Tic40, which was detected using goat-anti-mouse IgG-HRP (Dako) at concentration 1:2000.
Immunoprecipitation of CGEs with anti-Hsp70-2 Antibody
[35S]-labeled CGE1 and CGE2 were incubated with moss chloroplasts (60 μ g chlorophyll) at room temperature in presence of 3 mM ATP for 10 min under light. The reactions were stopped by adding 1.2 mL of cold IB containing 6.7 μ M nigericin. After washing once with the same buffer, chloroplasts were spun down and solubilized with IP buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% Triton X-100, 1 mM MgCl2, 1 mM CaCl2, 1 mM benzamidine, 2 μ M leupeptin, and 1 μ M pepstain) on ice for 15 min. In the ATP depleting treatments, 2.5 units/mL apyrase were included. Insoluble materials were removed by centrifugation at 16,000g for 10 min. To the supernatant, 20 μ L of affinity purified Hsp70-2 antibody and 30 μ L of Protein A Sepharose (50% slurry) were added. Preimmune serum was used as a control. The mixture was rotated end-over-end at 4 ° C overnight. The beads were pelleted at 3000g for 5 min at 4 ° C. The supernatants were saved as flow-through, and the pellets were washed six times with wash buffer (50 mM Tris, pH 8.0, 150 mM NaCl, and 0.1% Triton X-100). Supernatants of the last wash were subjected to trichloroacetic acid precipitation and resuspended with SDS-PAGE sample buffer. The washed beads were also resuspended with the sample buffer. All fractions were analyzed using SDS-PAGE and phosphor imaging.
Immunoprecipitation of Precursors Staged as Early Import Intermediates
[35S]-labeled CGE1 precursor protein (prCGE) or overexpressed [3H]-prSSU was incubated with isolated moss chloroplasts from either wild-type or lcge mutant plants on ice for 20 min at low ATP concentration (75 μ M) in darkness. prSSU was overexpressed in bacteria for this experiment to maximize the number of precursor molecules that might be in the translocon during our immunoprecipitations. Reactions were stopped by adding cold IB and centrifuging in a microfuge. Pellets were washed once with the same buffer. Chloroplasts were incubated in IB that contained the membrane-penetrating DSP (Pierce) cross-linker at 2.5 mM on ice for 15 min and then quenched with 50 mM Tris buffer, pH 8.0, on ice for 15 min. Chloroplasts were then lysed with buffer containing 10 mM MES and 5 mM MgCl2, and membranes were pelleted by centrifugation at 100,000g for 30 min at 4 ° C and solubilized with IP buffer supplemented with 1% Triton X-100 and 2.5 units/mL of apyrase. The supernatant obtained after 3000g centrifugation was subjected to immunoprecipitation as described above using preimmune serum, Hsp70-2 antibody, or Hsp93 antibody. Phosphor image analysis and fluorography were conducted to visualize the coimmunoprecipitating products.
In order to identify proteins immunoprecipitated with the early import intermediate by the anti-Hsp70-2 antibody, the IgGs from Hsp70-2 antiserum or preimmune serum were cross-linked using DMP (Abcam) to the Protein A Sepharose beads (GE Healthcare Life Sciences) prior to the immunoprecipitation experiment. Antibodies raised against Tic40 and Hsp93 were used to probe the corresponding immunoprecipitates associated with Hsp70-2. Antibody to the photosystem II reaction center protein D1 was used as a negative control.
Accession Numbers
Accession numbers used for homology searching, At4g24280 and At5g49910 from Arabidopsis, were used for searching the moss Hsp70s. CAB40381 (At5g17710) was used for searching moss CGEs. Accession numbers of protein sequences used in the tree in Figure 1A are as follows: Arabidopsis, AtCpHsc70-1, NP_194159.1; AtCpHsc70-2, NP_199802.1; AtMtHsc-1, NP_195504.2; AtMtHsc-2, NP_196521.1; Synechocystis sp strain PCC 6803, DNAK1_SYNY3, Q55154.1; DNAK2_SYNY3, P22358.1; SYNY3_DNK3, P73098.2; E. coli, DNAK_ECOLI, P0A6Y8; Saccharomyces cerevisiae, SSC1_YEAST, P12398.1; C. reinhardtii, CrHsp70B, XP_001696432.1; CrHsp70C, XP_001694468.1; P. sativum, PsCpHsp70, Q02028 and PsMtHsp70, P37900. Protein ID numbers of P. patens (http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html) are as follows: chloroplast Hsp70-1, 174284; Hsp70-2, 215505; Hsp70-3, 139043 and 218164; mitochondral Hsp70s, 182736, 182738, 192625, and 219879. Accession numbers of protein sequences used in the tree in Figure 4A are as follows: Arabidopsis, AtCGE-V, NP_850840.1; AtCGE-I, NP_849751.1; AtMGE-V, NP_200331.1; AtMGE-IV, NP_567757.1; Oryza sativa, OsMGE-VIII, NP_001061586.1; OsMGE-IX, NP_001062776.1; OsCGE-II, NP_001047409.1; Os0CGE-IV, NP_001052822.1; C. reinhardtii, Cr_CGE1A, Q945T2; Cr_CGE1B, Q945T1; E. coli, GrpE_ECOLI, NP_417104.1; Synechocystis sp strain PCC 6803, GrpE_SYNY3, NP_442221.1; and S. cerevisiae, MGE1p_YEAST, NP_014875.1. Protein ID numbers of P. patens (http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html) are as follows: PpCGE1, 172680; PpCGE2, 223954; PpMGE1, 168948; and PpMGE2, 110204. Accession numbers of mRNAs are as follows: PpHsp70-1, GU223149; PpHsp70-2, GU223150; pPCGE1, GU223151; and PpCGE2, GU223152.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Identification of Temperature-Sensitive Mutants.
Supplemental Figure 2. Identification of a cge Double Mutant Rescued by CGE2 cDNA.
Supplemental Figure 3. The Protein Levels of Hsp93 in the Wild Type and the lcge Mutant Are Comparable.
Supplemental Figure 4. Interaction of Hsp70-2 with CGEs.
Supplemental Figure 5. The Antibody Raised against the CGE2 Protein Reacts with Both CGE1 and CGE2 Proteins That Were Overexpressed in E. coli.
Supplemental Figure 6. Antibodies Raised against the Moss Hsp70-1 and Hsp70-2 Are Isoform Specific.
Supplemental Table 1. Primers Used in This Work.
Supplemental Data Set 1. Text File of the Hsp70 Alignment Used to Generate the Phylogenetic Tree in Figure 1A.
Supplemental Data Set 2. Text File of the GrpE Alignment Used to Generate the Phylogenetic Tree in Figure 4A.
Acknowledgments
We thank Kentaro Inoue, Connie Champagne, John Bowman, and Mitsuyasu Hasebe for providing clones, Hsou-min Li and Danny Schnell for antibodies, and Bo Liu, Julie Lee, and Byung-Kook Ham for sharing their expertise in microscopy. We also thank Nancy Hofmann, Connie Champagne, Shari Lo, and Karen Yip for valuable discussions, and we thank Thang Nguyen, Hannah Hsu, Christina Reginaldo, Fefe Wu, and Ani Tejirian for their excellent technical assistance. This work was supported by grants from the National Science Foundation and Department of Energy to S.M.T.
References
- Akita M., Nielsen E., Keegstra K. (1997). Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J. Cell Biol. 136: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder N.N., Shen Y., Brodsky J.L., Hendershot L.M., Johnson A.E. (2005). The molecular mechanisms underlying BiP-mediated gating of the Sec61 translocon of the endoplasmic reticulum. J. Cell Biol. 168: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J.H., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azem A., Oppliger W., Lustig A., Jeno P., Feifel B., Schatz G., Horst M. (1997). The mitochondrial Hsp70 chaperone system: Effect of adenine nucleotides, peptide substrate, and mGrpE on the oligomeric state of mHsp70. J. Biol. Chem. 272: 20901–20906 [DOI] [PubMed] [Google Scholar]
- Bedard J., Kubis S., Bimanadham S., Jarvis P. (2007). Functional similarity between the chloroplast translocon component, Tic40, and the human co-chaperone, Hsp70-interacting protein (Hip). J. Biol. Chem. 282: 21404–21414 [DOI] [PubMed] [Google Scholar]
- Bezanilla M., Perroud P.F., Pan A., Klueh P., Quatrano R.S. (2005). An RNAi system in Physcomitrella patens with an internal marker for silencing allows for rapid identification of loss of function phenotypes. Plant Biol. (Stuttg.) 7: 251–257 [DOI] [PubMed] [Google Scholar]
- Blanco-Rivero M.C., Takabe T., Viale A.M. (2005). Functional differences between cyanobacterial DnaK1 chaperones from the halophyte Aphanothece halophytica and the freshwater species Synechococcus elongatus expressed in Escherichia coli. Curr. Microbiol. 51: 164–170 [DOI] [PubMed] [Google Scholar]
- Chen X., Schnell D.J. (1999). Protein import into chloroplasts. Trends Cell Biol. 9: 222–227 [DOI] [PubMed] [Google Scholar]
- Chen X., Smith M.D., Fitzpatrick L., Schnell D.J. (2002). In vivo analysis of the role of atTic20 in protein import into chloroplasts. Plant Cell 14: 641–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou M.L., Chu C.C., Chen L.J., Akita M., Li H.M. (2006). Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts. J. Cell Biol. 175: 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou M.L., Fitzpatrick L.M., Tu S.L., Budziszewski G., Potter-Lewis S., Akita M., Levin J.Z., Keegstra K., Li H.M. (2003). Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J. 22: 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K., Werner-Washburne M., Andrews J., Keegstra K. (1984). Thermolysin is a suitable protease for probing the surface of intact pea chloroplasts. Plant Physiol. 75: 675–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constan D., Froehlich J.E., Rangarajan S., Keegstra K. (2004). A stromal hsp100 protein is required for normal chloroplast development and function in Arabidopsis. Plant Physiol. 136: 3605–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Los Rios P., Ben-Zvi A., Slutsky O., Azem A., Goloubinoff P. (2006). Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proc. Natl. Acad. Sci. USA 103: 6166–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Gambill B.D., Voos W., Kang P.J., Miao B., Langer T., Craig E.A., Pfanner N. (1993). A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J. Cell Biol. 123: 109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Glover J.R., Lindquist S. (1998). Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82 [DOI] [PubMed] [Google Scholar]
- Goloubinoff P., Mogk A., Zvi A.P., Tomoyasu T., Bukau B. (1999). Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. USA 96: 13732–13737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison C.J., Hayer-Hartl M., Di Liberto M., Hartl F., Kuriyan J. (1997). Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science 276: 431–435 [DOI] [PubMed] [Google Scholar]
- Heins L., Mehrle A., Hemmler R., Wagner R., Kuchler M., Hormann F., Sveshnikov D., Soll J. (2002). The preprotein conducting channel at the inner envelope membrane of plastids. EMBO J. 21: 2616–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann N.R., Theg S.M. (2003). Physcomitrella patens as a model for the study of chloroplast protein transport: Conserved machineries between vascular and non-vascular plants. Plant Mol. Biol. 53: 643–654 [DOI] [PubMed] [Google Scholar]
- Hofmann N.R., Theg S.M. (2005). Toc64 is not required for import of proteins into chloroplasts in the moss Physcomitrella patens. Plant J. 43: 675–687 [DOI] [PubMed] [Google Scholar]
- Horst M., Oppliger W., Rospert S., Schonfeld H.J., Schatz G., Azem A. (1997). Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J. 16: 1842–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstmann V., Huether C.M., Jost W., Reski R., Decker E.L. (2004). Quantitative promoter analysis in Physcomitrella patens: A set of plant vectors activating gene expression within three orders of magnitude. BMC Biotechnol. 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T., Li M., Alvarez-Huerta M., Kessler F., Schnell D.J. (2003). atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J. Biol. Chem. 278: 38617–38627 [DOI] [PubMed] [Google Scholar]
- Ivey R.A., III, Subramanian C., Bruce B.D. (2000). Identification of a Hsp70 recognition domain within the rubisco small subunit transit peptide. Plant Physiol. 122: 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Constan D., Akita M., Keegstra K. (2001). Molecular chaperones involved in chloroplast protein import. Biochim. Biophys. Acta 1541: 102–113 [DOI] [PubMed] [Google Scholar]
- Kang P.J., Ostermann J., Shilling J., Neupert W., Craig E.A., Pfanner N. (1990). Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature 348: 137–143 [DOI] [PubMed] [Google Scholar]
- Kessler F., Blobel G. (1996). Interaction of the protein import and folding machineries in the chloroplast. Proc. Natl. Acad. Sci. USA 93: 7684–7689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler F., Schnell D.J. (2006). The function and diversity of plastid protein import pathways: A multilane GTPase highway into plastids. Traffic 7: 248–257 [DOI] [PubMed] [Google Scholar]
- Kikuchi S., Oishi M., Hirabayashi Y., Lee D.W., Hwang I., Nakai M. (2009). A 1-megadalton translocation complex containing Tic20 and Tic21 mediates chloroplast protein import at the inner envelope membrane. Plant Cell 21: 1781–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight C.D., Cove D.J., Cuming A.C., Quatrano R.S. (2002). Moss gene technology. Molecular Plant Biology, Gilmartin P.M., Bowler C., (Oxford, UK: Oxford University Press; ), pp. 285–301 [Google Scholar]
- Ko K., Budd D., Wu C., Seibert F., Kourtz L., Ko Z.W. (1995). Isolation and characterization of a cDNA clone encoding a member of the Com44/Cim44 envelope components of the chloroplast protein import apparatus. J. Biol. Chem. 270: 28601–28608 [DOI] [PubMed] [Google Scholar]
- Kouranov A., Chen X., Fuks B., Schnell D.J. (1998). Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol. 143: 991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A., Wang H., Schnell D.J. (1999). Tic22 is targeted to the intermembrane space of chloroplasts by a novel pathway. J. Biol. Chem. 274: 25181–25186 [DOI] [PubMed] [Google Scholar]
- Kovacheva S., Bedard J., Patel R., Dudley P., Twell D., Rios G., Koncz C., Jarvis P. (2005). In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J. 41: 412–428 [DOI] [PubMed] [Google Scholar]
- Kovacheva S., Bedard J., Wardle A., Patel R., Jarvis P. (2007). Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. Plant J. 50: 364–379 [DOI] [PubMed] [Google Scholar]
- Krzewska J., Langer T., Liberek K. (2001). Mitochondrial Hsp78, a member of the Clp/Hsp100 family in Saccharomyces cerevisiae, cooperates with Hsp70 in protein refolding. FEBS Lett. 489: 92–96 [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lilley B.N., Ploegh H.L. (2004). A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429: 834–840 [DOI] [PubMed] [Google Scholar]
- Liu C., Willmund F., Golecki J.R., Cacace S., Hess B., Markert C., Schroda M. (2007). The chloroplast HSP70B–CDJ2-CGE1 chaperones catalyse assembly and disassembly of VIPP1 oligomers in Chlamydomonas. Plant J. 50: 265–277 [DOI] [PubMed] [Google Scholar]
- Liu C., Willmund F., Whitelegge J.P., Hawat S., Knapp B., Lodha M., Schroda M. (2005). J-domain protein CDJ2 and HSP70B are a plastidic chaperone pair that interacts with vesicle inducing protein in plastids 1. Mol. Biol. Cell 16: 1165–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madueno F., Napier J.A., Gray J.C. (1993). Newly imported Rieske iron-sulfur protein associates with both Cpn60 and Hsp70 in the chloroplast stroma. Plant Cell 5: 1865–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J.S., DeRocher A.E., Keegstra K., Vierling E. (1990). Identification of heat shock protein hsp70 homologues in chloroplasts. Proc. Natl. Acad. Sci. USA 87: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack K.E., Misselwitz B., Plath K., Rapoport T.A. (1999). BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell 97: 553–564 [DOI] [PubMed] [Google Scholar]
- Moore T., Keegstra K. (1993). Characterization of a cDNA clone encoding a chloroplast-targeted Clp homologue. Plant Mol. Biol. 21: 525–537 [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K., Brodsky J.L. (2008). The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic 9: 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E., Akita M., Davila-Aponte J., Keegstra K. (1997). Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 16: 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimura K., Takahashi H., Yoshikawa H. (2001). Characterization of the dnaK multigene family in the cyanobacterium Synechococcus sp. strain PCC7942. J. Bacteriol. 183: 1320–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimura K., Yoshikawa H., Takahashi H. (1996). DnaK3, one of the three DnaK proteins of cyanobacterium Synechococcus sp. PCC7942, is quantitatively detected in the thylakoid membrane. Biochem. Biophys. Res. Commun. 229: 334–340 [DOI] [PubMed] [Google Scholar]
- Nishikawa S., Brodsky J.L., Nakatsukasa K. (2005). Roles of molecular chaperones in endoplasmic reticulum (ER) quality control and ER-associated degradation (ERAD). J. Biochem. 137: 551–555 [DOI] [PubMed] [Google Scholar]
- Ratnayake R.M., Inoue H., Nonami H., Akita M. (2008). Alternative processing of Arabidopsis Hsp70 precursors during protein import into chloroplasts. Biosci. Biotechnol. Biochem. 72: 2926–2935 [DOI] [PubMed] [Google Scholar]
- Rensing S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Rial D.V., Arakaki A.K., Ceccarelli E.A. (2000). Interaction of the targeting sequence of chloroplast precursors with Hsp70 molecular chaperones. Eur. J. Biochem. 267: 6239–6248 [DOI] [PubMed] [Google Scholar]
- Richter S., Lamppa G.K. (1998). A chloroplast processing enzyme functions as the general stromal processing peptidase. Proc. Natl. Acad. Sci. USA 95: 7463–7468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer D., Zryd J.P., Knight C.D., Cove D.J. (1991). Stable transformation of the moss Physcomitrella patens. Mol. Gen. Genet. 226: 418–424 [DOI] [PubMed] [Google Scholar]
- Schatz G., Dobberstein B. (1996). Common principles of protein translocation across membranes. Science 271: 1519–1525 [DOI] [PubMed] [Google Scholar]
- Schmitt M., Neupert W., Langer T. (1995). Hsp78, a Clp homologue within mitochondria, can substitute for chaperone functions of mt-hsp70. EMBO J. 14: 3434–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell D.J., Kessler F., Blobel G. (1994). Isolation of components of the chloroplast protein import machinery. Science 266: 1007–1012 [DOI] [PubMed] [Google Scholar]
- Schroda M. (2004). The Chlamydomonas genome reveals its secrets: Chaperone genes and the potential roles of their gene products in the chloroplast. Photosynth. Res. 82: 221–240 [DOI] [PubMed] [Google Scholar]
- Schroda M., Vallon O. (2008). Chaperones and Proteases. In The Chlamydomonas Sourcebook, Stern D., (Oxford, UK: Academic Press; ), pp. 671–730 [Google Scholar]
- Schroda M., Vallon O., Wollman F.A., Beck C.F. (1999). A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11: 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J., DeWitt N.D., Flanagan J.M. (1995). The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: An archetypal two-component ATP-dependent protease. Plant Cell 7: 1713–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S.K., Christen P., Goloubinoff P. (2009). Disaggregating chaperones: An unfolding story. Curr. Protein Pept. Sci. 10: 432–446 [DOI] [PubMed] [Google Scholar]
- Stahl T., Glockmann C., Soll J., Heins L. (1999). Tic40, a new “old” subunit of the chloroplast protein import translocon. J. Biol. Chem. 274: 37467–37472 [DOI] [PubMed] [Google Scholar]
- Stevens S.Y., Cai S., Pellecchia M., Zuiderweg E.R. (2003). The solution structure of the bacterial HSP70 chaperone protein domain DnaK(393–507) in complex with the peptide NRLLLTG. Protein Sci. 12: 2588–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P.H., Li H.M. (2008). Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol. 146: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theg S.M., Bauerle C., Olsen L.J., Selman B.R., Keegstra K. (1989). Internal ATP is the only energy requirement for the translocation of precursor proteins across chloroplastic membranes. J. Biol. Chem. 264: 6730–6736 [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugeki R., Nishimura M. (1993). Interaction of homologues of Hsp70 and Cpn60 with ferredoxin-NADP+ reductase upon its import into chloroplasts. FEBS Lett. 320: 198–202 [DOI] [PubMed] [Google Scholar]
- Vojta L., Soll J., Bolter B. (2007). Requirements for a conservative protein translocation pathway in chloroplasts. FEBS Lett. 581: 2621–2624 [DOI] [PubMed] [Google Scholar]
- Wu C., Seibert F.S., Ko K. (1994). Identification of chloroplast envelope proteins in close physical proximity to a partially translocated chimeric precursor protein. J. Biol. Chem. 269: 32264–32271 [PubMed] [Google Scholar]
- Yokthongwattana K., Chrost B., Behrman S., Casper-Lindley C., Melis A. (2001). Photosystem II damage and repair cycle in the green alga Dunaliella salina: Involvement of a chloroplast-localized HSP70. Plant Cell Physiol. 42: 1389–1397 [DOI] [PubMed] [Google Scholar]
- Zhang X.P., Glaser E. (2002). Interaction of plant mitochondrial and chloroplast signal peptides with the Hsp70 molecular chaperone. Trends Plant Sci. 7: 14–21 [DOI] [PubMed] [Google Scholar]
- Zheng B., Halperin T., Hruskova-Heidingsfeldova O., Adam Z., Clarke A.K. (2002). Characterization of chloroplast Clp proteins in Arabidopsis: Localization, tissue specificity and stress responses. Physiol. Plant. 114: 92–101 [DOI] [PubMed] [Google Scholar]
- Zylicz M., Ang D., Liberek K., Georgopoulos C. (1989). Initiation of λ DNA replication with purified host- and bacteriophage-encoded proteins: The role of the dnaK, dnaJ and grpE heat shock proteins. EMBO J. 8: 1601–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]