This work examines the molecular basis of heterosis by comprehensively describing the epigenetic modifications and transcriptional output, including both mRNA and small RNAs, of two rice subspecies and their reciprocal hybrids.
Abstract
The behavior of transcriptomes and epigenomes in hybrids of heterotic parents is of fundamental interest. Here, we report highly integrated maps of the epigenome, mRNA, and small RNA transcriptomes of two rice (Oryza sativa) subspecies and their reciprocal hybrids. We found that gene activity was correlated with DNA methylation and both active and repressive histone modifications in transcribed regions. Differential epigenetic modifications correlated with changes in transcript levels among hybrids and parental lines. Distinct patterns in gene expression and epigenetic modifications in reciprocal hybrids were observed. Through analyses of single nucleotide polymorphisms from our sequence data, we observed a high correlation of allelic bias of epigenetic modifications or gene expression in reciprocal hybrids with their differences in the parental lines. The abundance of distinct small RNA size classes differed between the parents, and more small RNAs were downregulated than upregulated in the reciprocal hybrids. Together, our data reveal a comprehensive overview of transcriptional and epigenetic trends in heterotic rice crosses and provide a useful resource for the rice community.
INTRODUCTION
In eukaryotic organisms, the regulation of gene activity involves both genetic and epigenetic mechanisms. Epigenetic regulation is mediated by differences in DNA methylation at cytosine residues and by posttranslational histone modifications (Henderson and Jacobsen, 2007). DNA methylation is primarily regarded as an epigenetic silencing mechanism, which maintains genomic stability by inactivating transposons and other repetitive sequences (Bird, 2002; Chan et al., 2005; Goll and Bestor, 2005). In addition to extensive distribution in heterochromatic regions, DNA methylation is also found in euchromatic regions, including transcribed regions of genes (Zhang et al., 2006; Zilberman et al., 2007). Histone modifications provide a dynamic and reversible mechanism to regulate gene expression through changes in the chromatin state and the recruitment of protein complexes that regulate transcription. Numerous modifications of histones have been discovered, most of which occur near their N terminus. To date, acetylation and methylation of Lys residues are the most intensely studied modifications (Kouzarides, 2007). Whereas Lys acetylation is generally linked to chromatin accessibility and gene activation, Lys methylation can be associated with either transcriptional activation or repression, depending on the position of the Lys residue and the nature of methylation (Berger, 2007; Li et al., 2007). Recent studies discovered that both activating and repressive histone modifications correlated with gene activity (Bernstein et al., 2006; Roh et al., 2006; Barski et al., 2007; Mikkelsen et al., 2007; Wang et al., 2009a), indicating a dynamic regulation of gene expression through a combinatorial interplay between opposing modifications. In addition, small RNAs, including microRNAs (miRNAs) and small interfering RNAs (siRNAs), provide RNA-mediated regulation of genome stability (Ghildiyal and Zamore, 2009).
Natural allelic variation in nucleotide sequence and transcript abundance between different genotypes is an important genetic component of phenotypic diversity (Benfey and Mitchell-Olds, 2008). In hybrids, novel patterns of gene action result from the combination of allelic variants and are thought to be involved in heterosis, which is defined as an increased performance of hybrid offspring relative to its inbred parents (Birchler et al., 2003; Swanson-Wagner et al., 2006; Springer and Stupar, 2007). Several studies have revealed that epigenetic variations can contribute to the molecular mechanisms of complex traits (Cubas et al., 1999; Manning et al., 2006; Shindo et al., 2006; Makarevitch et al., 2007). More importantly, a recent study showed that altering the transcription of a few regulatory genes through epigenetic variations is associated with growth vigor in hybrids (Ni et al., 2009). Exploring the inherited natural variation of epigenetic patterns between genetically diverse strains at a genome-wide level will allow us to assess their importance in phenotypic plasticity and might have major consequences for biological research and agriculture (Richards, 2008).
As one of the most important crops worldwide, rice (Oryza sativa) has become a model plant for genomic research. The availability of the complete genomic sequence for the japonica (cv Nipponbare) and indica (cv 93-11) subspecies of rice (Goff et al., 2002; Yu et al., 2002) has already enabled the investigation of transcriptome activity in a range of tissues and developmental stages (Li et al., 2006; Nobuta et al., 2007), the high-resolution mapping of epigenetic modifications for chromosomes (Li et al., 2008b), and the genome-wide identification of genetic variation in gene expression between rice subspecies and their hybrids (Zhang et al., 2008a; Wei et al., 2009). The high growth vigor of hybrids between Nipponbare and 93-11 also provides a good opportunity for the investigation of the molecular basis of heterosis in rice (Zhang et al., 2008a). While these studies delivered valuable insights for understanding the dynamics of the rice genome, they are limited to only a portion of the transcriptome (two-thirds of all annotated rice genes investigated; Zhang et al., 2008a) and epigenome (two chromosomes surveyed; Li et al., 2008b) of the total rice genome. Furthermore, the currently available data for rice transcriptomes and epigenomes is of limited utility to reveal the complex interactions among them by direct comparative analyses because these data have been obtained under different conditions in separate studies. Recently developed high-throughput sequencing technologies have been successfully used to study epigenomes and transcriptomes (Zhu, 2008; Simon et al., 2009; Wang et al., 2009a, 2009b). Applying these technologies to conduct a global survey of both epigenetic and transcriptional variation between different rice subspecies and their hybrids in one study would provide new insights into genome activity during evolution and the complex regulatory machinery for differential gene expression in hybrids.
Here, we describe the integrated comparative analysis of the epigenome and overall transcriptional output (including mRNA and small RNAs) for two rice subspecies and their reciprocal hybrids. We found a hierarchical relationship of gene expression and epigenetic modifications in the rice genome. Importantly, we identified abundant epigenetic and transcriptional variations among hybrids and parental inbred lines and found that these two types of polymorphisms correlate. Furthermore, we found that the small RNA transcriptome differed in composition and expression between hybrid offspring and their inbred parents. Together, our highly integrated data sets provide fresh insights into the dynamic variations and complex relationships between the transcriptome and epigenome in rice and enable a better understanding of the interactions of two different genomes.
RESULTS
Integrated Maps of the Transcriptomes and Epigenomes of Two Rice Subspecies and Their Reciprocal Hybrids
To gain insight into natural epigenetic variation and its relationships to changes in transcript abundance in the genomes of two rice subspecies, we generated genome-wide integrated maps of mRNA transcripts, DNA methylation, and three select histone modifications (H3K4me3, H3K9ac, and H3K27me3) by employing high-throughput Illumina/Solexa 1G sequencing. We used shoots of four-leaf stage seedlings of two rice subspecies (O. sativa ssp japonica cv Nipponbare and O. sativa ssp indica cv 93-11) and their reciprocal hybrids for our studies. Both reciprocal hybrids used in this study show significant growth vigor compared with their parents (see Supplemental Figure 1 online). Random hexamer-primed cDNA synthesis from purified poly(A) mRNA, digestion of genomic DNA with the methylation-sensitive restriction enzyme McrBC, and chromatin immunoprecipitation (ChIP) with specific antibodies were used to generate the respective sequencing libraries following previously published procedures (Gendrel et al., 2005; Barski et al., 2007; Mikkelsen et al., 2007; Nagalakshmi et al., 2008; Wilhelm et al., 2008; Wang et al., 2009a). Sequence reads were aligned with the well-annotated rice genome (The Institute for Genomic Research, TIGR version 5.0) (Yuan et al., 2005) using the MAQ algorithm (Li et al., 2008a) to determine the frequency of reads matching each genomic base pair position. To exploit sequencing data for repetitive regions, all reads that could be mapped to multiple locations in the genome were assigned to one position at random and were retained for further analyses. In total, >277 million genome-matched reads were obtained from 20 sequencing libraries from four rice genotypes for genome-wide analyses of gene expression and epigenetic modifications (see Supplemental Table 1 online).
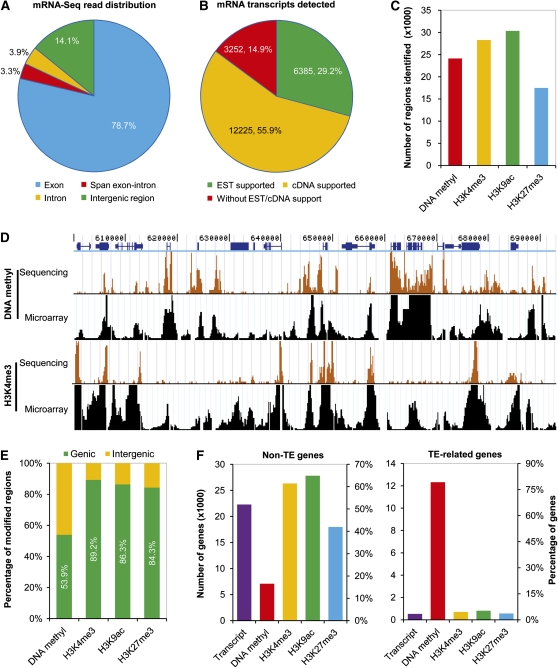
We first characterized the transcriptome and epigenome in rice using sequencing data from Nipponbare. As expected, most (78.7%) of the mRNA-Seq (mRNA-sequencing) reads were found to map to annotated exons in the rice genome (Figure 1A). We quantified the transcript levels by adding the number of reads that mapped to exonic regions of annotated gene models and normalized them with the predicted mRNA length (in kilobase pairs). For genes with multiple transcripts of different lengths or of the same length, we used the longest transcript or a randomly selected transcript for further analyses, respectively. Reads mapped across splice boundaries were also excluded for further analyses. Based on the read coverage of annotated introns in the rice genome, we set an empirical cutoff value to provide a conservative assessment of transcript diversity and active genes in our mRNA-Seq libraries (see Supplemental Figure 2A online). Using this criterion, the transcripts of more than half of the currently annotated nontransposable element protein coding genes (non-TE genes) were detected (21,351, 52.0%), whereas only a few transposable element–related genes (TE-related genes) were found to be expressed above background transcriptional activity (511, 3.4%) (Figure 1F). We found that the detectable expression for a total of 21,862 genes in our mRNA-Seq libraries of Nipponbare was comparable to the number of active genes detected in MPSS libraries derived from rice using similar tissues (Nobuta et al., 2007). We also compared the gene expression levels determined by our mRNA-Seq with previously published microarray data and found a general consistency between both methods (see Supplemental Figure 3 online). Over 85% of the genes that were found to be transcribed in this study were supported by ESTs or full-length cDNAs (Figure 1B). On the other hand, of all annotated rice genes, most of the genes supported by full-length cDNAs (12,225, 80.7%) or ESTs (6385, 69.6%), but only a small fraction of genes lacking cDNA or EST support (3252, 10.2%), had detectable transcripts in our study (see Supplemental Figure 2B online). These data indicate an overall agreement between our mRNA-Seq approach and other experimental methods.
Figure 1.
Integrated Profiling of the Rice Transcriptome and Epigenome by High-Throughput Sequencing.
(A) Distribution of mapped reads for mRNA-Seq library in the rice genome.
(B) Proportion of mRNA transcripts identified by mRNA-Seq according to gene annotations supported by EST or full-length cDNA data.
(C) Identification of genomic regions associated with DNA methylation (DNA methyl), H3K4me3, H3K9ac, and H3K27me3.
(D) Comparisons of DNA methylation and H3K4me3 modification patterns in a representative region on rice chromosome 4 using Solexa sequencing (this study) and microarray technology (Li et al., 2008b). The predicted coding sequences are shown in blue above.
(E) Frequencies of epigenetically modified regions in genic and intergenic regions.
(F) Number and percentage of non-TE genes and TE-related genes identified with expression or epigenetic modifications.
Genomic regions associated with DNA methylation and histone modifications were identified using model-based analysis of ChIP-Seq (MACS) software, which tests sliding windows across the genome and uses the Poisson distribution to identify the density of reads in enriched and nonenriched regions in order to call peaks (Zhang et al., 2008c) (Figure 1C). We found a strong enrichment of DNA methylation in heterochromatic regions, but by contrast, all three histone modifications studied here showed the same overall patterns of high enrichment in euchromatic regions (see Supplemental Figure 4 online). We compared the distribution patterns of DNA methylation and H3K4me3 modification obtained from our sequencing data with that from a previous microarray study (Li et al., 2008b) and found a similar overall distribution pattern of both epigenetic marks revealed by the two studies (Figure 1D; see Supplemental Figure 5 online). Mapping the positions of modified regions relative to genes revealed that the vast majority of regions that bear histone modifications (84.3 to 89.2%) and more than half of the DNA methylated regions (53.9%) were associated with annotated transcribed regions (or genic regions) (Figure 1E). Further inspection of the average read coverage in different genic regions showed that DNA methylation was distributed throughout transcribed regions but that histone modifications formed strong peaks within regions ∼ 1 kb downstream of the transcription start site (TSS) (see Supplemental Figure 6 online). Therefore, we focused our further analyses of epigenetic modifications only on those associated with genic regions. We evaluated the levels of histone modifications of each gene by directly counting the number of reads mapping within 1 kb downstream of the TSS. Additionally, we estimated the extent of DNA methylation for each gene by adding the number of reads located in the full-length transcribed region and normalized it with the predicted gene length (reads per kilobase). Out of 41,043 non-TE genes in the TIGR database, we detected 6783 (16.5%) genes that contained DNA methylation, whereas 25,207 (61.4%), 26,623 (64.9%), and 17,211 (41.9%) were found to be modified by H3K4me3, H3K9ac, and H3K27me3, respectively, when we used a threshold of read coverage defined by randomization (P value < 0.001). By contrast, of 15,232 TE-related genes in the TIGR database, 12,063 (79.2%) contained DNA methylation, whereas only several hundred (552 to 788, 3.6 to 5.2%) contained histone modifications. The proportions of genes with histone modifications resembled that of gene expression (Figure 1F).
We observed the same pattern of read distribution and identified a similar number of non-TE and TE-related genes with expression or modifications in the other three rice genotypes (i.e., 93-11, Nipponbare/93-11 F1 hybrid, and 93-11/Nipponbare F1 hybrid) (see Supplemental Figure 7 online). Altogether, we generated a comprehensive integrated genome-wide map of transcript abundance, DNA methylation, and three histone modifications of all annotated genes in two rice inbred lines and their reciprocal hybrids.
Gene Activity Correlates with a Hierarchy of DNA Methylation and Both Active and Repressive Histone Modifications in Transcribed Regions
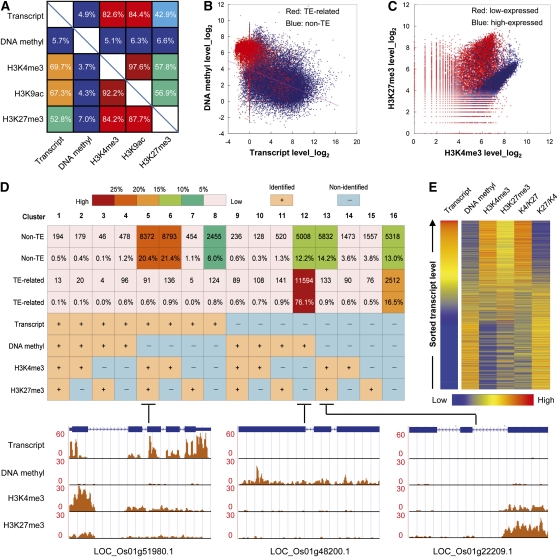
To analyze the relationships among epigenetic modifications and their correlations with gene activity, we first used our sequencing data for Nipponbare to determine relationships between concurrent gene expression and epigenetic modifications (Figure 2A). The genomic sequence of Nipponbare is publicly available and well annotated, which strengthens our findings. We found a very high frequency (92.2 and 97.6%) of concurrent modifications by H3K4me3 and H3K9ac, which are both transcriptionally activating epigenetic marks. Notably, we also detected a high frequency of concurrence between these two activating modifications and H3K27me3, a transcriptionally repressive mark. Further inspection showed that a large majority of genes with H3K27me3 also contained H3K4me3 (84.2%) or H3K9ac (87.7%), whereas a much smaller number of genes containing H3K4me3 (57.8%) or H3K9ac (56.9%) were comodified by H3K27me3. These strong differences in co-occurring histone modifications implicate the regulation of gene activity by disproportional modification with active and repressive epigenetic marks. As expected, we found a high frequency of concurrence between gene expression and two activating histone modifications, H3K4me3 and H3K9ac. Interestingly, we also detected a significant frequency of concurrence between gene expression and the repressive mark H3K27me3 (42.9% of all expressed genes were modified by H3K27me3, and 52.8% of all genes marked with H3K27me3 were expressed). We found that most genes with DNA methylation (84.3%) were transcriptionally silent, indicating a strong repressive effect of this epigenetic modification on gene activity.
Figure 2.
Relationships among Gene Expression and Epigenetic Modifications.
(A) Frequencies of concurrent epigenetic modifications and gene expression. Numbers indicate the percentage of genes that were detected with X and were also detected with Y.
(B) Correlation between DNA methylation and gene expression. Red, TE-related genes; blue, non-TE genes.
(C) Correlation between H3K4me3 and H3K27me3 modifications. Only data of genes with a transcript level <10 reads (red, low-expressed) or >100 reads (blue, high-expressed) per kilobase of predicted mRNA were plotted here.
(D) A permutation table for all combinations of DNA methylation, H3K4me3, and H3K27me3 modifications and gene expression. The number and percentage of TE-related or non-TE genes in each possible combination were calculated. Color scale indicates the proportion (from high to low) of genes. Typical examples from the predominant clusters visualized in the UCSC genome browser. Left, genes without DNA methylation and for which H3K4me3 modification was dominant were transcriptionally active. Middle, genes for which DNA methylation was dominant were depleted in H3K4me3 and H3K27me3 and were transcriptionally repressive. Right, genes without DNA methylation and for which H3K27me3 modification was dominant were transcriptionally repressive.
(E) Heat map of epigenetic modification levels and the ratio (K4/K27) between H3K4me3 and H3K27me3 levels (normalized by total mapped reads) on all annotated rice genes sorted by their expression level measured by mRNA-Seq. K4, H3K4me3; K27, H3K27me3.
We further examined the quantitative relationships among epigenetic modifications and gene activity using pairwise scatterplots. We observed a weak negative correlation between DNA methylation and transcript level (Figure 2B; see Supplemental Figure 8 online). As expected, we detected obvious positive correlations between the amounts of the two activating epigenetic marks studied (H3K4me3 and H3K9ac) and transcript abundance (see Supplemental Figure 8 online). Taking into account the very high levels of concurrence and correlation between H3K4me3 and H3K9ac (Figure 2A), we did not include H3K9ac in our further analyses. Interestingly, we also detected a weak positive correlation between the repressive mark H3K27me3 and gene expression (Pearson correlation = 0.302; see Supplemental Figure 8 online). In accordance with the concurrence of H3K27me3 and H3K4me3 (Figure 2A), we found a high positive correlation of the two epigenetic modifications in our scatterplots (Pearson correlation = 0.721), in which a bipeak distribution was observed (see Supplemental Figure 8 online). Further investigation revealed that the genes involved in the two distribution peaks were associated with high and low levels of expression, respectively (Figure 2C). Therefore, even in the presence of H3K27me3, a high level of H3K4me3 could still drive high expression of a gene. This indicates that both the absolute and relative levels of H3K27me3 and H3K4me3 modifications in transcribed regions correlate with gene expression in the rice genome.
We generated a permutation table for all possible combinations of DNA methylation, H3K4me3, H3K27me3, and gene expression (designated as cluster 1 to cluster 16; Figure 2D; see Supplemental Figure 9 online). Comparisons of the proportion of genes in each cluster revealed a hierarchical regulation of gene activity. The majority of TE-related genes (92.6%) fell into only two clusters of transcriptional quiescence, showing that they were either DNA methylated (76.1%, cluster 12) or did not contain any of the three modifications analyzed (DNA methylation, H3K4me3, and H3K27me3) (16.5%, cluster 16). Non-TE genes exhibited more complex hierarchical relationships between gene expression and epigenetic modifications. For example, among all non-TE genes that showed DNA methylation (clusters 1 to 4 and clusters 9 to12), most of them were depleted in H3K4me3 and H3K27me3 (cluster 4 and cluster12) and were often transcriptionally silent (12.2% in cluster 12 versus 1.2% in cluster 4). Secondly, we found that transcriptional activity of genes without DNA methylation could be regulated by the combinatorial result of H3K4me3 and H3K27me3 (clusters 5 to 7 and clusters 13 to 15). Genes for which H3K4me3 modification was dominant were transcriptionally active (20.4% in cluster 5), whereas genes for which H3K27me3 modification was dominant were usually transcriptionally repressed (14.2% in cluster 13). Furthermore, non-TE genes that did not contain any detectable H3K4me3, H3K27me3, or DNA methylation were either transcriptionally active (6.0%, cluster 8) or repressed (13.0%, cluster 16) and were most likely regulated by other genetic or epigenetic factors.
To obtain a comprehensive understanding of global interaction patterns between transcription and epigenetic modifications, we further generated an integrated epigenomic map by sorting all genes based on their expression level (Figure 2E). As found before (Li et al., 2008b; Wang et al., 2009a), we detected antagonistic patterns between DNA methylation and H3K4me3, in which genes with low levels of DNA methylation had high levels of H3K4me3 and showed high levels of expression, whereas genes with high levels of DNA methylation had low levels of H3K4me3 and exhibited low levels of expression. The repressive mark H3K27me3 showed an intriguing pattern when compared with gene expression. Further inspection revealed that gene expression levels were positively correlated with the ratio of H3K4me3/H3K27me3 and were negatively correlated with the ratio of H3K27me3/H3K4me3, thereby facilitating our understanding of the repressive role of H3K27me3 on gene activity (Figure 2E). These analyses suggest that in rice, DNA methylation plays a role in dictating the transcriptional activity of genes and that the combinatorial interactions of histone modifications provide a fine-tuning of their expression levels.
Natural Epigenetic Variations Correlate with Changes in Transcript Abundance among Hybrids and Parental Lines
Comparative genomics has revealed a rapid and dramatic divergence of genomic sequences between indica and japonica rice subspecies (Ma and Bennetzen, 2004). Since all of our sequencing reads were aligned to the genome of the japonica subspecies, cv Nipponbare, polymorphisms in DNA sequences between subspecies could result in inaccurate estimates of the levels of expression or epigenetic modifications of some genes in the indica subspecies, cv 93-11. In an effort to minimize the impact of sequence polymorphisms on our conclusions, we identified all orthologous genomic regions between Nipponbare and 93-11 by comparing the published genomes of both subspecies and by using only those reads that mapped to colinear regions to identify natural epigenetic and transcriptional variations. Therefore, a total of only 20,638 genes (18,922 non-TE genes and 1716 TE-related genes) that were located in conserved genomic regions were included in our further analyses.
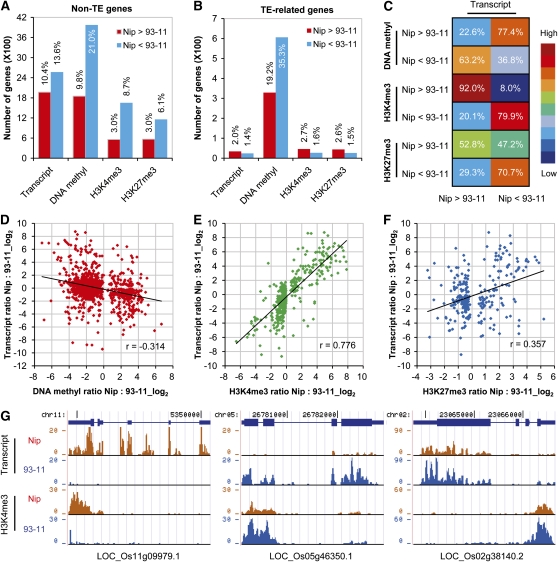
Using a rigorous significance test (Audic and Claverie, 1997), we examined the differences in gene expression, DNA methylation, and H3K4me3 and H3K27me3 modifications between Nipponbare and 93-11 for transcribed regions using a P value of < 0.001. A total of 4538 non-TE genes (24.0%) were found to show differential expression when both accessions were compared (false discovery rate [FDR] = 0.0045). We found that significantly more non-TE genes were differentially modified by DNA methylation than by H3K4me3 and H3K27me3 (Figure 3A). Moreover, we observed that more than half of the TE-related genes (54.5%) were differentially modified by DNA methylation. By contrast, only very few of the TE-related genes showed differences in H3K4me3 and H3K27me3 modifications (Figure 3B). In summary, these data suggest that variations in DNA methylation occur more often than for histone modifications between these two rice subspecies.
Figure 3.
Correlations between Epigenetic Natural Variations and Changes in Transcript Abundance between Two Parental Lines.
(A) and (B) Number and percentage of non-TE and TE-related genes exhibiting differences in gene expression or epigenetic modifications between Nipponbare (Nip) and 93-11. Red, number or percentage of genes for which the levels of expression or epigenetic modifications is higher in Nipponbare than in 93-11. Blue, number or percentage of genes for which the levels of expression or epigenetic modifications is lower in Nipponbare than in 93-11.
(C) Frequencies of concurrence between differences in gene expression and differences in epigenetic modifications. Numbers indicate the percentage of genes that were differentially modified and that were also differentially expressed. Color scale indicates the proportion (from high to low) of genes.
(D) to (F) Correlations between differential epigenetic modifications (P value < 0.001) and differential gene expression (P value < 0.001).
(G) Typical examples of genes showing correlations between differential H3K4me3 modification and differential gene expression.
To investigate the relationship between natural epigenetic variation and changes in transcript abundance, we counted the frequencies of concurrence between differential gene expression and differential modifications (Figure 3C). We found a very high level of concordance between the differences in H3K4me3 modification and the differences in gene expression. For instance, among genes containing a higher level of H3K4me3 in Nipponbare than in 93-11, 92.0% also showed higher expression in Nipponbare. Moreover, obvious opposite relationships between differential DNA methylation and differential gene expression were observed. For example, among genes with higher DNA methylation in Nipponbare than in 93-11, 77.4% exhibited lower expression in Nipponbare. Interestingly, we found that there was a good concurrence between difference in H3K27me3 modification and gene expression in 93-11, but only weakly so in Nipponbare (Figure 3C). This might indicate that there are differences in combinatorial H3K4me3/H3K27me3 modifications and their effects on gene expression between the two parental inbred lines.
Furthermore, we quantitatively examined the correlations between differences in gene expression and differences in epigenetic modifications. Only genes with significant differences in both transcription and epigenetic modification were selected for further analysis using pairwise scatterplots. We found that H3K4me3 was strongly positively correlated with differential gene expression (Pearson correlation = 0.776) (Figures 3E and 3G), whereas DNA methylation was negatively correlated with differential gene expression, albeit only weakly (Pearson correlation = −0.314) (Figure 3D). We also observed a weak correlation between differential H3K27me3 modification and differential gene expression (Figure 3F). Furthermore, we detected the same overall correlation patterns between differential gene expression and differential epigenetic modifications when parental inbred lines and their hybrid offspring were compared (see Supplemental Figure 10 online).
Variations in Gene Expression and Epigenetic Modifications Show Distinct Patterns in Reciprocal Hybrids
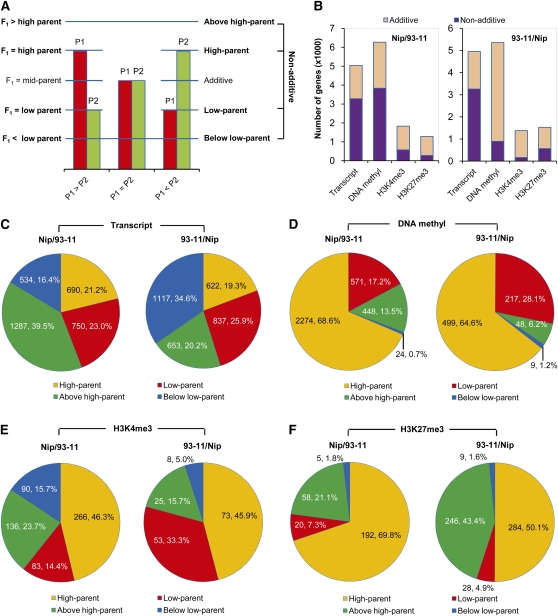
Variation of gene expression in hybrids can be classified as additive or nonadditive relative to their inbred parents, which means that hybrids can show a transcript level equal to, or deviating from, the mid-parent value, which is the average of the two parents. Four subdivided patterns of gene action have frequently been used to describe nonadditive gene expression. High-parent or low-parent patterns occur whenever the gene expression level in hybrids is similar to the higher parent or to the lower parent, respectively. Above high-parent or below low-parent patterns occur whenever the expression level in hybrids is above the higher parent or below the lower parent, respectively (Birchler et al., 2003; Springer and Stupar, 2007; Figure 4A). Based on these modes, we characterized the epigenetic variation and transcriptional polymorphisms of genes in reciprocal hybrids.
Figure 4.
Variation Patterns of Gene Expression and Epigenetic Modifications in Reciprocal Hybrids.
(A) Modes of gene action in hybrid. Red bar, gene expression levels in parent 1 (P1). Green bar, gene expression levels in parent 2 (P2). Blue horizontal line, gene expression levels in hybrid cross between P1 and P2. x axis, comparisons of gene expression levels between P1 and P2. y axis, comparisons of gene expression levels between parents and hybrid and the patterns for each case.
(B) Additive and nonadditive variation in gene expression and epigenetic modifications in hybrids. Additive, hybrids show a transcript or modification level equal to the mid-parent value (average of the two parents). Nonadditive, hybrids show a transcript or modification level deviating from the mid-parent value.
(C) Subdivided patterns of nonadditive variation in gene expression (high-parent, above high-parent, low-parent, and below low-parent) in reciprocal hybrids. High-parent or low-parent patterns, the gene expression level in hybrids is similar to the higher parent or to the lower parent, respectively. Above high-parent or below low-parent patterns, the gene expression level in hybrids is above the higher parent or below the lower parent, respectively.
(D) to (F) Subdivided patterns of non-additive variation in DNA methylation and histone modifications in hybrids.
From 20,638 genes analyzed, 5044 (24.4%, FDR = 0.0041) and 4951 (24.0%, FDR = 0.0042) were identified with differential expression in reciprocal hybrids, most of which showed a nonadditive expression pattern (Figure 4B). Moreover, 6274 (30.4%, FDR = 0.0033) and 5354 (25.9%, FDR = 0.0039) genes were identified that showed differential DNA methylation in reciprocal hybrids. Most genes exhibited a nonadditive pattern in Nipponbare/93-11, whereas significantly more genes showed an additive pattern in 93-11/Nipponbare. By contrast, fewer genes were found to have differential histone modifications in both hybrids, most of which exhibited an additive pattern (Figure 4B). We also found that TE-related genes showed a high frequency of variation in DNA methylation but less variation in transcription and H3K4me3 or H3K27me3 modifications in hybrids (see Supplemental Figure 11 online). These data indicate that variations in DNA methylation occur more often than histone modifications in hybrids.
We further examined nonadditive patterns of gene expression and epigenetic modifications in hybrids. Although almost the same number of genes exhibited a nonadditive expression pattern when both hybrids were compared (3261 in Nipponbare/93-11 and 3229 in 93-11/Nipponbare; Figure 4B), we found that only 1950 of these genes were shared among both hybrids and that different proportions of genes were associated with four subdivided variation patterns (Figure 4C). Among all genes with nonadditive expression, the largest group of genes (39.5%) showed above high-parent pattern in Nipponbare/93-11, whereas the largest single group of genes (34.6%) represented a below low-parent pattern in 93-11/Nipponbare (Figure 4C). This might indicate cytoplasmic effects on gene expression in hybrids. We identified similar proportions of genes showing high-parent or low-parent expression between reciprocal hybrids (Figure 4C). By contrast, although the total number of genes identified with nonadditive variation of DNA methylation is significantly different between reciprocal hybrids (Figure 4B), similarly large proportions of genes (68.6% in Nipponbare/93-11 and 64.6% in 93-11/Nipponbare) exhibited high-parent effects in both genotypes, whereas only very few genes showed an below low-parent effect (Figure 4D). Moreover, among a small number of genes identified with nonadditive variation of histone modifications (Figure 4B), we found that most also showed high-parent variation (Figures 4E and 4F). These data suggested that the epigenetic modifications studied here are in general upregulated in hybrids. We also performed additional comparative analyses of our data for all annotated rice genes (56,278, including those located in nonorthologous regions) among different genotypes and found that the general trends of variation in gene expression and epigenetic modifications are identical compared with orthologous regions (see Supplemental Figure 12 online). Moreover, from this overall comparison, we identified 2800 genes showing the same direction of nonadditive expression in both reciprocal hybrids and investigated their distribution among the Gene Ontology functional categories. We found that that these genes are enriched significantly in pathways for energy metabolic processes (see Supplemental Figure 13 online), of which six key components involved the in Calvin cycle (carbon metabolism) were all upregulated significantly relative to mid-parent in both reciprocal hybrids (see Supplemental Figure 14 online).
Allelic Bias of Gene Expression and Epigenetic Modifications in Hybrids Correlates with Parental Differences
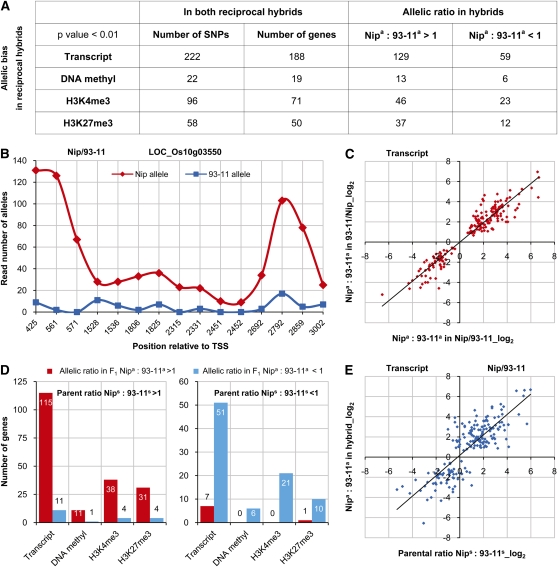
To assess differential allelic expression or epigenetic modifications in hybrids, we identified single nucleotide polymorphisms (SNPs) by comparing the sequencing reads at each base position in exons (for gene expression) or genic regions (for epigenetic modifications) of 20,638 homologous genes between Nipponbare and 93-11 and used these SNPs as markers to discriminate allele-specific sequencing reads. Allele-specific biases were evaluated using a binomial test with the null hypothesis that two parental alleles are uniformly expressed or modified in the hybrid. To ensure accuracy and reliability, only SNPs identified with a significant allele-specific bias at a P value cutoff of 0.01 in both reciprocal hybrids were included in our further analyses. Since many genes with a low transcript or epigenetic modification level were excluded by this threshold, our approach provides a very conservative estimate of allelic bias in the rice genome. Using these criteria, among 2205 SNPs (1754 genes) detected in both reciprocal hybrids, 398 (328 genes) were identified that showed an allelic bias for accumulated transcripts or epigenetic modifications in both reciprocal hybrids with an average FDR of 0.06 (Figures 5A and 5B; see Supplemental Figure 15 online). Most of the observed biases were associated with one detectable SNP. Among those genes with more than one SNP identified, two showed a contradictory allelic expression bias for different SNPs and were also excluded from further analyses.
Figure 5.
Allelic Bias of Gene Expression and Epigenetic Modifications in Reciprocal Hybrids.
(A) Detection of allelic bias in gene expression and epigenetic modifications in hybrids using SNPs between parental lines. Only SNPs identified with a significant allelic bias at a P value cutoff of 0.01 in both reciprocal hybrids were included. Nipa, Nipponbare allele; 93-11a, 93-11 allele.
(B) An example of a gene exhibiting allelic expression bias in Nip/93-11 hybrid. Read number of alleles detected by SNPs at each position was plotted.
(C) Correlation of allelic expression bias between reciprocal hybrids.
(D) Allelic expression bias in hybrids according to their differences between parents. Nips, gene expression level in Nipponbare represented by the number of reads covering a SNP; 93-11s, gene expression level in 93-11 represented by the number of reads covering a SNP.
(E) Correlation between differential parental expression (represented by the number of reads covering a SNP) and allelic expression bias in Nip/93-11 hybrid.
To test whether a parent-of-origin effect was responsible for the allelic bias in hybrids, we compared the ratio of two parental alleles between reciprocal hybrids. We found that almost all genes identified in our study exhibited the same direction of allele-specific bias in expression or epigenetic modifications in both hybrids, suggesting that there is no significant parent-of-origin effect for the action of parental alleles in hybrids (Figure 5C; see Supplemental Figure 16 online). Furthermore, we investigated the effect of differences in gene expression or epigenetic modifications (here represented by the number of reads covering a SNP position) between both parental lines on the allele-specific patterns in their reciprocal hybrids. We found that a large majority of genes with a higher level of gene expression or epigenetic modifications in Nipponbare also exhibited Nipponbare allele bias in hybrids and vice versa (Figure 5D). We further quantitatively examined the relationship between parental differences and allelic bias and found that differential allelic expression or epigenetic modifications in reciprocal hybrids was highly positively correlated with differences in gene expression or epigenetic modifications (represented by the number of reads covering a SNP position) between parents (Figure 5E; see Supplemental Figures 17A and 18 online). Moreover, similar correlation patterns were also observed when using the genes' transcript or epigenetic modification data (see Supplemental Figure 17B online). Together, these data suggest a differential contribution of parental alleles toward gene activity and epigenetic modifications in hybrids. Analyzing functional categories indicated that those genes exhibiting allelic expression bias in both reciprocal hybrids (see Supplemental Data Set 1 online) were relatively enriched in pathways for energy metabolic processes (see Supplemental Figure 19 online).
Small RNAs Show Differences in Composition between Two Parental Inbred Lines and Are Predominantly Downregulated in Hybrid Offspring
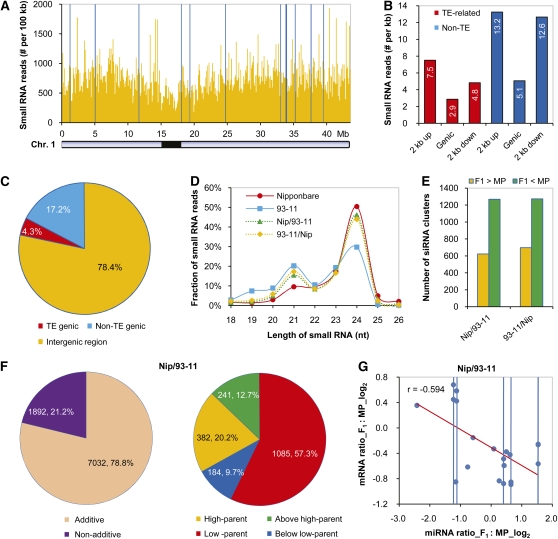
We surveyed the small RNA transcriptomes of two parental rice inbred lines and their reciprocal hybrid offspring using the same biological material as for our epigenetic and mRNA analyses. Small RNAs were gel purified from total RNA and ligated with specific oligonucleotide adapters to generate libraries for high-throughput Illumina 1G sequencing. The raw sequencing reads were processed computationally to remove the adapter sequences and were then aligned to the well-annotated japonica rice genome sequence. After removing rRNA, tRNA, and sn/snoRNA, >20 million genome-matched reads were obtained from four libraries of parents and hybrids (see Supplemental Table 1 online). We first investigated the distribution of genomic sequences matching small RNA reads across the rice genome in Nipponbare. We observed a wide distribution of small RNAs on rice chromosomes and a relatively low abundance of small RNAs in the pericentromeric region (Figure 6A). Further investigation revealed that genomic sequences matched by small RNA reads tended to cluster 2 kb upstream or 2 kb downstream of the transcribed regions of annotated rice genes and that non-TE genes showed a higher enrichment of small RNAs than the TE-related genes (Figure 6B). As expected, we found that a large majority of rice small RNA reads (78.4%) were associated with intergenic regions (Figure 6C; Nobuta et al., 2007). We observed differences in the distributions of small RNA size between both parents (Figure 6D). Of all 18- to 26-nucleotide reads, 24-nucleotide small RNAs were more predominant in Nipponbare (50.5%) than in 93-11 (29.8%), whereas 21-nucleotide small RNAs exhibited an opposite distribution (9.6% in Nipponbare versus 20.2% in 93-11). This means that rice might show a subspecies-specific distribution of small RNAs. We observed almost the same distribution patterns of small RNA size classes between the reciprocal hybrids, in which the proportions of both 21- and 24- nucleotide small RNAs were close to the high parent (Figure 6D).
Figure 6.
Diversity of Small RNAs in Composition and Expression between Parents and Hybrids.
(A) Distribution of small RNAs on rice chromosome 1. The y axis represents the added small RNA reads per 100 genomic region. Blue vertical lines, >2000 reads per 100-kb genomic regions. Black bar, pericentromeric region.
(B) Classes of genomic features (the genic or transcribed regions of annotated rice genes, 2 kb upstream or 2 kb downstream of the genic regions) matched by rice small RNAs. The average number of small RNA reads per kilobase region of each genomic feature is counted.
(C) Distribution of small RNAs (mapped reads after the removal of rRNAs, tRNAs, snRNA, and snoRNA) among genic (TE- and non-TE) and intergenic regions in the rice genome.
(D) Small RNA length distribution in two parental inbred lines and their reciprocal hybrids.
(E) Differential expression of siRNA clusters between F1 hybrids and their parents. MP, mid-parent value.
(F) Expression patterns of siRNA clusters in Nip/93-11 hybrid.
(G) Negative correlation between the expression of 14 miRNAs and their 20 targets that are both exhibiting nonadditive variation in both reciprocal hybrids. Multiple targets of a single miRNA are indicated by a vertical line.
Since the processing of miRNA from a precursor is not always exact, and some small RNAs are derived from the precursor but shifted from the annotated miRNA, we estimated the expression level of each rice miRNA by summing all the small RNA reads that correspond to the precursors of each known rice miRNA (Nobuta et al., 2009) (see Supplemental Data Set 2 online). The remaining small RNA reads (mostly siRNA) from all four genotypes were pooled and used to identify siRNA clusters (i.e., genomic regions matched by small RNAs) by clustering adjacent small RNA reads. The expression level of a siRNA cluster was estimated by adding up the number of small RNA reads in it. Pairwise comparisons of each miRNA and siRNA cluster from four rice genotypes were performed to identify variations in small RNA abundance among parents and reciprocal hybrids. As described above, we determined the significance of differences of small RNAs by a rigorous significance test using a P value of < 0.001 (Audic and Claverie, 1997). Of 235,257 siRNA clusters identified, 13,284 (5.6%) were found to show differential expression above the threshold between both parents (FDR = 0.018). We observed that more siRNA clusters were downregulated than upregulated in both reciprocal hybrids compared with mid-parent value (Figure 6E), which points to a suppression of siRNAs in hybrids (see Supplemental Figure 20 online). When compared with parents, 8924 siRNA clusters showed differential expression in Nipponbare/93-11 (FDR = 0.026), and 8840 siRNA clusters showed differential expression in 93-11/Nipponbare (FDR = 0.027), most of which showed additive patterns in both reciprocal hybrids (Figure 6F; see Supplemental Figure 21A online). Further investigation of siRNA clusters with nonadditive variation patterns revealed that most of them (57.3 and 58.3%) exhibited a low-parent pattern in reciprocal hybrids (Figure 6F; see Supplemental Figure 21A online), suggesting a dominant-negative regulation of siRNA transcription in hybrids. We also used total small RNA reads associated with 2-kb regions upstream of the TSS of each gene in orthologous genomic regions for comparisons between hybrids and parental lines and observed the same pattern of variation in hybrids (see Supplemental Figure 21B online).
Among 414 known rice miRNAs, 58 showed the same pattern of variation relative to mid-parent in both reciprocal hybrids (FDR = 0.007), 22 of which were upregulated and 35 were downregulated. We found that the target genes of 14 miRNAs also showed the same pattern of variation relative to mid-parent in both reciprocal hybrids and that there is a clear negative correlation between the expression level of miRNAs and their target genes (Figure 6G; see Supplemental Figure 22 online).
DISCUSSION
In this study, we generated comprehensive and integrated high-resolution maps of genomic distributions of DNA methylation and histone modifications, as well as of mRNA and small RNA transcriptomes in two subspecies of rice, and in their reciprocal hybrids. Our data should therefore be a useful resource for investigations of the molecular mechanisms of heterosis in rice and other plant species.
We observed remarkably concurrent modifications of the activating H3K4me3 mark and the repressive H3K27me3 mark in the rice genome and found that the transcription level correlated with both absolute and relative levels of these two opposing modifications, as reported for animals (Bernstein et al., 2006; Roh et al., 2006; Barski et al., 2007; Mikkelsen et al., 2007). We categorized complex interactions between a dynamic epigenome and transcriptome in rice by associating gene activity with a hierarchy of DNA methylation and concurrent histone modifications (H3K4me3/H3K27me3) in transcribed regions (Figure 2D). Genes that were heavily DNA methylated were usually transcriptionally repressed, and they were often devoid of histone modifications, as observed for the large majority of TE-related genes but also some non-TE genes. By contrast, genes without or with a relatively low level of DNA methylation were transcriptionally permissive. This explains the negative correlation between the transcription level and DNA methylation we observed (Figure 2B; see Supplemental Figure 8 online), as well as the concurrence of this repressive mark with the expression of some genes (Zhang et al., 2006; Zilberman et al., 2007). Moreover, the concurrent modifications H3K4me3 and H3K27me3 were associated with a dynamic gene expression pattern, in which genes that were dominated by H3K4me3 correlated with higher transcriptional activity, whereas genes that were dominated by H3K27me3 correlated with lower transcriptional activity (Figure 2E).
Recently, inconsistent results have been reported on the relationship between variation in gene expression and differential DNA methylation in transcribed regions (Vaughn et al., 2007; Zhang et al., 2008b). Our study revealed a weak negative correlation between differences in DNA methylation and differences in the expression of some genes (Figures 3C and 3D; see Supplemental Figure 10 online). This might indicate a potential contribution of this repressive epigenetic mark to transcriptional variation among different genotypes, which deserves further investigations in different tissues and at different developmental stages. By contrast, a strong positive correlation between the differences in gene expression and H3K4me3 modification was observed (Figures 3C and 3E; see Supplemental Figure 10 online), implicating a strong quantitative correlation genome-wide.
In hybrids, diversity in gene expression can be the result of variation in cis-acting elements (e.g., promoter regions) or trans-acting regulatory factors (e.g., transcription factors) (Springer and Stupar, 2007). It has been suggested that nonadditive variation is generally controlled by trans-acting factors, whereas additive variation could result from both cis- and trans-acting effects (Swanson-Wagner et al., 2006; Springer and Stupar, 2007; Guo et al., 2008). In our study, significantly more genes exhibited nonadditive than additive expression patterns in both reciprocal hybrids (Figure 4B; see Supplemental Figure 12 online), suggesting predominantly trans-acting effects in the transcriptional diversity of these genes in hybrids.
We observed inconsistent above high-parent and below low-parent expression patterns between reciprocal hybrids (Figure 4C). Since reciprocal hybrids share the same nuclear genetic background, this discrepancy in variation implies a cytoplasmic effect on the direction of divergence of gene expression in hybrids. Moreover, DNA methylation was found to exhibit inconsistent additive or nonadditive patterns between reciprocal hybrids (Figure 4B), implying cytoplasmic effects on the extent of variation in DNA methylation in hybrids. Since there are no obvious phenotypic differences between reciprocal hybrids used in our study (see Supplemental Figure 1 online), these cytoplasmic effects in hybrids could cause confusion in the identification of genetic or epigenetic variation that underlies the phenotypic difference between hybrids and parents and can be excluded by investigating genes with the same patterns of variation in reciprocal hybrids. In our study, a total of 2800 genes exhibiting the same direction of differential expression relative to mid-parent values in both reciprocal hybrids were identified from all annotated rice genes. We found that these genes were significantly enriched for metabolic functions (e.g., Calvin cycle), which supports the idea that these genes are involved in the growth vigor of hybrids, which deserves further investigation.
Following previous reports (Mikkelsen et al., 2007; Guo et al., 2008), we also investigated whether an allelic bias in gene expression or epigenetic modifications was present in hybrids. The patterns uncovered here were clear and showed an allelic bias that exhibited the same direction between reciprocal hybrids and a positive correlation with differences between parents. This indicates that parental alleles tend to preserve their characteristic states of activity and chromatin modifications in hybrids. However, the action of both parental alleles in hybrids could be affected by cytoplasm from maternal lines, which makes the cytoplasm different between the reciprocal hybrids. This means that although parental alleles keep the same proportion of expression or modifications in both reciprocal hybrids as that between parents (no significant parent-of-origin effect), the respective gene as a whole could be expressed differently between reciprocal hybrids (cytoplasmic effect).
A previous study showed that unlike a concentrated distribution across pericentromeric regions in Arabidopsis, small RNAs in rice exhibit a wide distribution on chromosomes (Nobuta et al., 2007). In addition to a chromosome-wide distribution pattern of rice small RNAs, we observed that they were relatively less abundant in centromeric regions, which are generally devoid of protein-coding genes. However, when we associated small RNAs with genes, our observations for rice are in accord with findings for Arabidopsis (Kasschau et al., 2007), in which small RNA populations cluster in and around genes, and in which no general correlation between the transcription level of genes and the abundance of small RNAs around them has been observed.
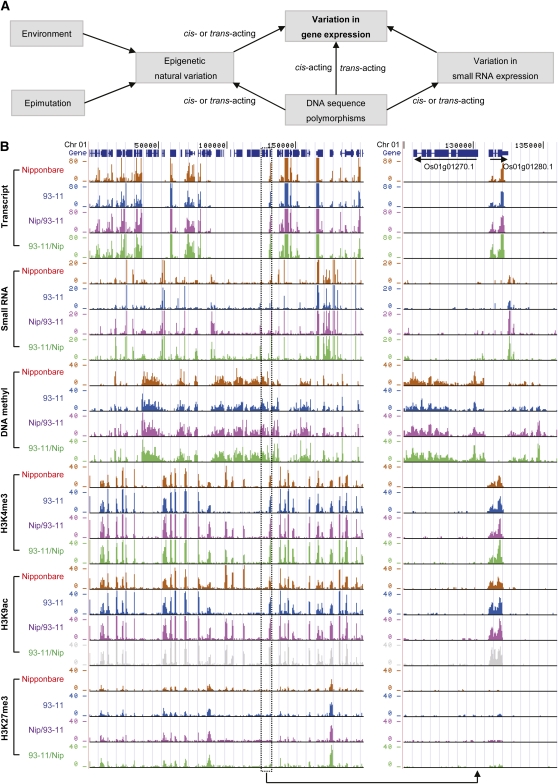
In plants, a series of pathways have evolved to generate various endogenous small RNAs that are often characterized by specific sizes (Vazquez, 2006). A discrepancy in the size distribution of small RNAs between two rice subspecies observed in our study possibly suggests a divergence of small RNA biogenesis during the evolution of the rice genome. Unlike our observations for mRNA transcription, small RNAs showed consistent patterns of variation between reciprocal hybrids, in which more small RNAs were downregulated than upregulated regardless of the difference between parents. Small RNAs are thought to mostly function as transcriptional or posttranscriptional gene repressors (Mallory and Vaucheret, 2006), which means the downregulation and upregulation of small RNAs in hybrids could result in upregulation and downregulation of their target genes, respectively (Ha et al., 2009). This intriguing observation adds another layer of regulatory machinery to the already complex web of gene regulation in hybrids and possibly indicates an important role during heterosis (Figure 7A).
Figure 7.
Interactions among Epigenetic Modifications, Small RNA Transcription, and Gene Expression and Their Variation in Hybrids.
(A) Description of the source of gene expression variation in hybrids.
(B) A representative genomic region on rice chromosome 1 showing integrated maps of predicted gene models with mRNA, small RNA, and epigenetic landscapes in two rice subspecies and their reciprocal hybrids.
In summary, we have described global integrated epigenetic and transcriptional maps and their interactional relationships for the rice genome and have conducted a pilot study to dissect intraspecies divergence of epigenomes and transcriptomes and their variation in hybrids. The entire data set has been uploaded to the National Institutes of Health Gene Expression Omnibus database (see accession numbers below) and is also available and can be accessed on our website based on the UCSC genome browser (Rice Hybrids Transcriptome and Epigenome Browser, http://www.pyc.pku.edu.cn/) (Figure 7B). We hope that the resources generated here will aid future studies to shed more light on the genetic and epigenetic basis of gene action in different genetic backgrounds that leads to significant phenotypic variability between parental inbred lines and their hybrids during specific developmental stages.
METHODS
Plant Materials and Growth Conditions
Rice cultivar Nipponbare (Oryza sativa ssp japonica) and 93-11 (O. sativa ssp indica) and their reciprocal F1 hybrids (Nipponbare/93-11 and 93-11/Nipponbare) were used for all experiments in this study. Seeds were grown in soil under 16-h-light/8-h-dark conditions at 28 ° C in a greenhouse. After 4 weeks, seedling shoots at the four-leaf stage were harvested, frozen in liquid nitrogen, and stored at –80 ° C for DNA and total RNA isolation or processed directly after harvesting for ChIP assay.
Sequencing Library Construction
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions and was treated with RNase-free DNase I (New England Biolabs) to remove any contaminating genomic DNA. mRNA was extracted from total RNA using Dynabeads oligo(dT) (Invitrogen Dynal) following the manufacturer's directions. First- and second-strand cDNA were generated using Superscript II reverse transcriptase (Invitrogen) and random hexamer primers. Double-stranded cDNA was fragmented by nebulization and used for mRNA library construction following the standard Illumina protocol. Small RNAs were gel isolated from total RNA and were used to create libraries for Illumina sequencing as described previously (Mi et al., 2008; Wang et al., 2009a). Genomic DNA extraction, methylated genomic DNA enrichment, and construction of Illumina sequencing libraries were performed as described previously (Li et al., 2008b; Wang et al., 2009a). Chromatin from seedling shoots was immunoprecipitated with antibodies against H3K4me3 (Upstate), H3K9ac (Upstate), or H3K27me3 (Upstate) as described previously (Gendrel et al., 2005). The eluted ChIP DNA was used to generate Illumina sequencing libraries following the manufacturer's protocol.
Accession Number
The original data set is deposited in the National Institutes of Health Gene Expression Omnibus database under accession number GSE19602.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Heterosis in Reciprocal Crosses between Rice Cultivars Nipponbare and 93-11.
Supplemental Figure 2. Detection of Gene Expression in Rice (cv Nipponbare) by mRNA-Seq.
Supplemental Figure 3. Scatterplot Comparing Gene Expression Levels Based on Microarray Hybridization Signals (y Axis; Zhang et al., 2008a) with Gene Expression Levels Based on mRNA Sequencing (x Axis; This Study).
Supplemental Figure 4. Distribution of DNA Methylation, H3K4me3, H3K9ac, H3K27me3, and Annotated Genes on Rice Chromosome 1.
Supplemental Figure 5. Comparisons of the Distribution Patterns of DNA Methylation and H3K4me3 Modification on Chromosomes 4 and 10 Obtained from Sequencing Data (This Study) with That from Previously Published Microarray Data (Li et al., 2008b).
Supplemental Figure 6. Distribution of DNA Methylation, H3K4me3, H3K9ac, and H3K27me3 Levels within Non-TE Genes and TE-Related Genes.
Supplemental Figure 7. Number and Percentage of Non-TE Genes and TE-Related Genes Identified with Expression or Epigenetic Modifications in 93-11, Nip/93-11, and 93-11/Nip.
Supplemental Figure 8. Pairwise Correlations among Epigenetic Modifications and Gene Expression.
Supplemental Figure 9. Permutation Tables for All Combinations of DNA Methylation, H3K4me3, and H3K27me3 Modifications and Gene Expression in 93-11, Nip/93-11, and 93-11/Nip.
Supplemental Figure 10. Correlations between Variation of Epigenetic Modifications and Variation of Gene Expression in Reciprocal Hybrids.
Supplemental Figure 11. Number of Non-TE Genes and TE-Related Genes Exhibiting Variation in Gene Expression or Epigenetic Modifications in Nip/93-11 and 93-11/Nip.
Supplemental Figure 12. Variation Patterns of Gene Expression and Epigenetic Modifications in Reciprocal Hybrids Investigated from All Annotated Rice Genes (56,278 Genes).
Supplemental Figure 13. Functional Categories of 2800 Genes Showing the Same Direction of Nonadditive Expression in Both Reciprocal Identified from All Annotated Rice Genes.
Supplemental Figure 14. Upregulation of Six Key Components Involved in Calvin Cycle (Carbon Metabolism) Relative to Mid-Parent in Both Reciprocal Hybrids.
Supplemental Figure 15. An Example of a Gene Exhibiting Allelic Expression Bias in 93-11/Nip Hybrid.
Supplemental Figure 16. Correlation of Allelic Bias in Epigenetic Modifications between Reciprocal Hybrids.
Supplemental Figure 17. Correlation between Differential Parental Expression and Allelic Expression Bias in Hybrids.
Supplemental Figure 18. Correlation between Differential Parental Epigenetic Modifications (Represented by the Number of Reads Covering a SNP) and Allelic Bias of Epigenetic Modifications in Nip/93-11 and 93-11/Nip.
Supplemental Figure 19. Functional Categories of 188 Genes Exhibiting Allelic Expression Bias in Both Reciprocal Hybrids.
Supplemental Figure 20. Suppression of Small RNAs in Hybrids.
Supplemental Figure 21. Expression Patterns of Small RNAs in Hybrids.
Supplemental Figure 22. Negative Correlation between the Expression of 14 miRNAs and Their 20 Targets That Are Both Exhibiting Nonadditive Variation in Both Reciprocal Hybrids.
Supplemental Table 1. Summary Statistics for Illumina 1G Sequencing Libraries.
Supplemental Methods. Data Processing and Analysis.
Supplemental Data Set 1. List of Genes Showing Allelic Expression Bias in Both Reciprocal Hybrids.
Supplemental Data Set 2. Number of Small RNA Reads Associated with Known Rice miRNAs.
Supplementary Material
Acknowledgments
The studies conducted at Yale University were supported by grants from the National Institutes of Health (GM047850), the National Science Foundation Plant Genome Program (DBI0922604), and the National Science Foundation 2010 program (MCB-0929100). Studies conducted at Peking University were supported by grants from the Ministry of Science and Technology of China (2009DFB30030) and the Ministry of Agriculture of China (2009ZX08012-021B). Studies conducted at the National Institute of Biological Sciences were supported by special funds from the Ministry of Science and Technology of China and Beijing Commission of Science and Technology. We thank William Terzaghi, Jeffery Q. Shen, and Ligeng Ma for their suggestions and comments on the manuscript.
References
- Audic S., Claverie J.M. (1997). The significance of digital gene expression profiles. Genome Res. 7: 986–995 [DOI] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Benfey P.N., Mitchell-Olds T. (2008). From genotype to phenotype: Systems biology meets natural variation. Science 320: 495–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S.L. (2007). The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Birchler J.A., Auger D.L., Riddle N.C. (2003). In search of the molecular basis of heterosis. Plant Cell 15: 2236–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. (2002). DNA methylation patterns and epigenetic memory. Genes Dev. 16: 6–21 [DOI] [PubMed] [Google Scholar]
- Chan S.W., Henderson I.R., Jacobsen S.E. (2005). Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6: 351–360 [DOI] [PubMed] [Google Scholar]
- Cubas P., Vincent C., Coen E. (1999). An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401: 157–161 [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M., Zamore P.D. (2009). Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Goll M.G., Bestor T.H. (2005). Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74: 481–514 [DOI] [PubMed] [Google Scholar]
- Guo M., Yang S., Rupe M., Hu B., Bickel D.R., Arthur L., Smith O. (2008). Genome-wide allele-specific expression analysis using Massively Parallel Signature Sequencing (MPSS) reveals cis- and trans-effects on gene expression in maize hybrid meristem tissue. Plant Mol. Biol. 66: 551–563 [DOI] [PubMed] [Google Scholar]
- Ha M., Lu J., Tian L., Ramachandran V., Kasschau K.D., Chapman E.J., Carrington J.C., Chen X., Wang X.J., Chen Z.J. (2009). Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc. Natl. Acad. Sci. USA 106: 17835–17840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Jacobsen S.E. (2007). Epigenetic inheritance in plants. Nature 447: 418–424 [DOI] [PubMed] [Google Scholar]
- Kasschau K.D., Fahlgren N., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C. (2007). Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. (2007). The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li H., Ruan J., Durbin R. (2008a). Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wang X., Stolc V., Li X., Zhang D., Su N., Tongprasit W., Li S., Cheng Z., Wang J., Deng X.W. (2006). Genome-wide transcription analyses in rice using tiling microarrays. Nat. Genet. 38: 124–129 [DOI] [PubMed] [Google Scholar]
- Li X., et al. (2008b). High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell 20: 259–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Bennetzen J.L. (2004). Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101: 12404–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I., Stupar R.M., Iniguez A.L., Haun W.J., Barbazuk W.B., Kaeppler S.M., Springer N.M. (2007). Natural variation for alleles under epigenetic control by the maize chromomethylase zmet2. Genetics 177: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Vaucheret H. (2006). Functions of microRNAs and related small RNAs in plants. Nat. Genet. 38(Suppl.): S31–S36 [DOI] [PubMed] [Google Scholar]
- Manning K., Tor M., Poole M., Hong Y., Thompson A.J., King G.J., Giovannoni J.J., Seymour G.B. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Mi S., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5 ′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., Snyder M. (2008). The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z., Kim E.D., Ha M., Lackey E., Liu J., Zhang Y., Sun Q., Chen Z.J. (2009). Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K., McCormick K., Nakano M., Meyers B.C. (2009). Bioinformatics analysis of small RNAs in plants using next generation sequencing technologies. Methods Mol. Biol. 592: 89–106 [DOI] [PubMed] [Google Scholar]
- Nobuta K., Venu R.C., Lu C., Beló A., Vemaraju K., Kulkarni K., Wang W., Pillay M., Green P.J., Wang G.L., Meyers B.C. (2007). An expression atlas of rice mRNAs and small RNAs. Nat. Biotechnol. 25: 473–477 [DOI] [PubMed] [Google Scholar]
- Richards E.J. (2008). Population epigenetics. Curr. Opin. Genet. Dev. 18: 221–226 [DOI] [PubMed] [Google Scholar]
- Roh T.Y., Cuddapah S., Cui K., Zhao K. (2006). The genomic landscape of histone modifications in human T cells. Proc. Natl. Acad. Sci. USA 103: 15782–15787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C., Lister C., Crevillen P., Nordborg M., Dean C. (2006). Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev. 20: 3079–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S.A., Zhai J., Nandety R.S., McCormick K.P., Zeng J., Mejia D., Meyers B.C. (2009). Short-read sequencing technologies for transcriptional analyses. Annu. Rev. Plant Biol. 60: 305–333 [DOI] [PubMed] [Google Scholar]
- Springer N.M., Stupar R.M. (2007). Allelic variation and heterosis in maize: How do two halves make more than a whole?. Genome Res. 17: 264–275 [DOI] [PubMed] [Google Scholar]
- Swanson-Wagner R.A., Jia Y., DeCook R., Borsuk L.A., Nettleton D., Schnable P.S. (2006). All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc. Natl. Acad. Sci. USA 103: 6805–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn M.W., et al. (2007). Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 5: e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F. (2006). Arabidopsis endogenous small RNAs: Highways and byways. Trends Plant Sci. 11: 460–468 [DOI] [PubMed] [Google Scholar]
- Wang X., Elling A.A., Li X., Li N., Peng Z., He G., Sun H., Qi Y., Liu X.S., Deng X.W. (2009a). Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M. (2009b). RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G., et al. (2009). A transcriptomic analysis of super hybrid rice LYP9 and its parents. Proc. Natl. Acad. Sci. USA 106: 7695–7701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm B.T., Marguerat S., Watt S., Schubert F., Wood V., Goodhead I., Penkett C.J., Rogers J., Bahler J. (2008). Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453: 1239–1243 [DOI] [PubMed] [Google Scholar]
- Yu J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
- Yuan Q., Ouyang S., Wang A., Zhu W., Maiti R., Lin H., Hamilton J., Haas B., Sultana R., Cheung F., Wortman J., Buell C.R. (2005). The institute for genomic research Osa1 rice genome annotation database. Plant Physiol. 138: 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.Y., He H., Chen L.B., Li L., Liang M.Z., Wang X.F., Liu X.G., He G.M., Chen R.S., Ma L.G., Deng X.W. (2008a). A genome-wide transcription analysis reveals a close correlation of promoter INDEL polymorphism and heterotic gene expression in rice hybrids. Mol. Plant 1: 720–731 [DOI] [PubMed] [Google Scholar]
- Zhang X., Shiu S., Cal A., Borevitz J.O. (2008b). Global analysis of genetic, epigenetic and transcriptional polymorphisms in Arabidopsis thaliana using whole genome tiling arrays. PLoS Genet. 4: e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Ecker J.R. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nussbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008c). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.K. (2008). Epigenome sequencing comes of age. Cell 133: 395–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Gehring M., Tran R.K., Ballinger T., Henikoff S. (2007). Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39: 61–69 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.