This study shows that BOP1 and BOP2 regulate key organogenic events in the proximal region of cotyledon and leaf primordia. Direct activation of AS2 transcription by BOP1 and BOP2 is found to be required for KNOX1 gene repression at the leaf base and may represent a conserved mechanism for coordinating leaf morphogenesis with patterning along the adaxial-abaxial and the proximal-distal axes.
Abstract
Continuous organ formation is a hallmark of plant development that requires organ-specific gene activity to establish determinacy and axial patterning, yet the molecular mechanisms that coordinate these events remain poorly understood. Here, we show that the organ-specific BTB-POZ domain proteins BLADE-ON-PETIOLE1 (BOP1) and BOP2 function as transcriptional activators during Arabidopsis thaliana leaf formation. We identify as a direct target of BOP1 induction the ASYMMETRIC LEAVES2 (AS2) gene, which promotes leaf cell fate specification and adaxial polarity. We find that BOP1 associates with the AS2 promoter and that BOP1 and BOP2 are required for AS2 activation specifically in the proximal, adaxial region of the leaf, demonstrating a role for the BOP proteins as proximal-distal as well as adaxial-abaxial patterning determinants. Furthermore, repression of BOP1 and BOP2 expression by the indeterminacy-promoting KNOX gene SHOOTMERISTEMLESS is critical to establish a functional embryonic shoot apical meristem. Our data indicate that direct activation of AS2 transcription by BOP1 and BOP2 is vital for generating the conditions for KNOX repression at the leaf base and may represent a conserved mechanism for coordinating leaf morphogenesis with patterning along the adaxial-abaxial and the proximal-distal axes.
INTRODUCTION
Higher plant development is a continuous process of organogenesis sustained by pluripotent stem cell reservoirs at the growing tips, the apical meristems. Establishment and maintenance of the shoot apical meristem (SAM) depends on the activity of class I KNOTTED1-like homeobox (KNOX) genes that are highly expressed in meristem cells (Lincoln et al., 1994; Dockx et al., 1995; Long et al., 1996; Belles-Boix et al., 2006). Lateral organs such as leaves initiate as small groups of homogeneous founder cells on the flanks of the SAM, and during early morphogenesis, these primordia become patterned along three axes of polarity: the adaxial-abaxial, proximal-distal, and medial-lateral axes. Subsequent growth and differentiation result in the elaboration of a mature three-dimensional leaf structure with a proximal petiole and a distal blade in simple-leaved species such as Arabidopsis thaliana. In contrast with adaxial-abaxial polarity, about which much is known, the molecular mechanisms that specify proximal-distal polarity in plants are very poorly understood.
Lateral organ morphogenesis and patterning processes require extensive genome reprogramming. Initiation of a determinant leaf structure is closely associated with repression of KNOX gene expression in the organ founder cells (Jackson et al., 1994; Long et al., 1996). In Arabidopsis, the Myb gene ASYMMETRIC LEAVES1 (AS1) is expressed in lateral organ primordia in a reciprocal pattern to that of the class I KNOX gene SHOOTMERISTEMLESS (STM), and STM represses AS1 expression in the SAM (Byrne et al., 2000). AS1 and the LOB domain protein AS2 in turn stably silence the class I KNOX genes BP and KNAT2 in developing leaf primordia (Byrne et al., 2000, 2002; Ori et al., 2000; Semiarti et al., 2001; Guo et al., 2008) by forming a complex with the chromatin-remodeling factor HIRA (Phelps-Durr et al., 2005). In addition, AS1, which is expressed throughout young leaf primordia, and AS2, which is specifically expressed in the adaxial domain of lateral organs, promote adaxial leaf identity and gene expression (Lin et al., 2003; Xu et al., 2003; Li et al., 2005; Ueno et al., 2007). Yet despite the critical roles AS1 and AS2 play in lateral organ morphogenesis, little is known about the upstream factors that induce their specific expression in organ primordia.
The BLADE-ON-PETIOLE1 (BOP1) and BOP2 genes play important roles in regulating leaf morphogenesis and patterning (Ha et al., 2003, 2007). bop1-1 dominant-negative and bop1 bop2 null mutants form ectopic outgrowths of blade tissue along the petioles of cotyledons and leaves, suggesting a role in proximal-distal pattern formation, and the BOP genes negatively regulate expression of the class I KNOX genes BP, KNAT2, and KNAT6 in developing leaf primordia (Ha et al., 2003, 2007). BOP1 and BOP2 upregulate the expression of three members of the AS2/LOB family (Iwakawa et al., 2002; Shuai et al., 2002; Matsumura et al., 2009), AS2, ASL4/LOB, and ASL1/LBD36, in leaf primordia (Ha et al., 2007). They also promote adaxial organ identity, positively regulating the expression of the adaxial polarity genes PHB and PHV as well as negatively regulating the expression of the abaxial polarity genes FIL and KAN1 (Ha et al., 2007). BOP1 and BOP2 belong to a family of BTB/POZ domain and ankyrin repeat-containing proteins that includes the plant defense response regulator NPR1 and are expressed in a small group of proximal, adaxial lateral organ cells adjacent to the meristem-organ boundary (Ha et al., 2004; Hepworth et al., 2005). Although NPR1 is known to be a transcriptional regulator that mediates salicylic acid (SA)–induced gene expression in association with TGA family transcription factors (Cao et al., 1997; Zhang et al., 1999; Despres et al., 2000, 2003), the regulatory mechanism through which BOP1 and BOP2 influence organ morphogenesis is unknown.
In this study, we show that BOP1 and BOP2 regulate key organogenic events in the proximal region of Arabidopsis cotyledon and leaf primordia. We find that BOP1 and BOP2 proteins can dimerize in vivo and function as transcriptional coactivators when recruited to target gene promoters. We demonstrate that BOP1/2 activity is required for AS2 activation specifically in the proximal region of the leaf and that BOP1 is a direct upstream regulator of AS2 during leaf development that binds to sites in the AS2 promoter containing a bZIP binding motif. Furthermore, we show that STM represses BOP1 and BOP2 expression in the embryonic SAM and that the absence of BOP activity permits ectopic shoot meristem formation in stm seedlings. Our data suggest that BOP1 and BOP2 regulate leaf morphogenesis along the proximal-distal axis by directly inducing AS2 transcription at the leaf base, establishing the conditions for repression of class I KNOX gene expression to correctly pattern the petiole tissues.
RESULTS
BOP1 and BOP2 Function as Transcriptional Activators
To better understand the molecular mechanism of BOP1 and BOP2 activity, we took NPR1 as a paradigm for the family. NPR1 has a dual nuclear and cytoplasmic subcellular localization pattern (Despres et al., 2000), and nuclear-localized NPR1 provides transcriptional activation activity for defense gene induction (Rochon et al., 2006). A dual subcellular localization pattern has also been observed for BOP2 (Hepworth et al., 2005), and we likewise detected BOP1-green fluorescent protein fusions in both the nucleus and the cytoplasm (see Supplemental Figure 1 online). Based on these data, we hypothesized that BOP1 and BOP2, like NPR1, might function as transcriptional activators.
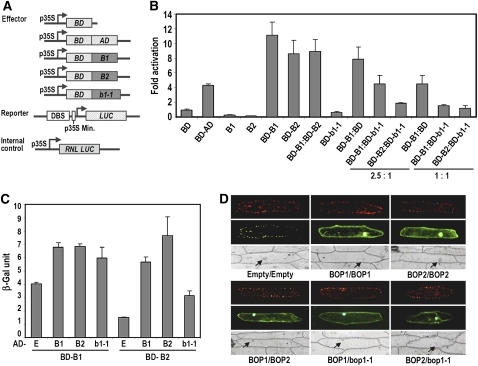
We tested whether the BOP proteins could stimulate transcription when tethered to DNA by assaying constructs consisting of the Gal4 DNA binding domain (BD) fused to either BOP1 (BD-BOP1) or BD-BOP2 (Figure 1A) in in vivo plant transcription assays (Huq et al., 2004). Transfection of unfused BOP1 or BOP2 did not generate luciferase activation beyond the baseline level of transcription (Figure 1B). By contrast, transfection with BD-BOP1 or BD-BOP2 activated transcription 11.2- and 8.6-fold above the baseline level, respectively (Figure 1B). Cotransfection of BD-BOP1 with BD-BOP2 produced similar fold induction values to those of BD-BOP1 or BD-BOP2 alone (Figure 1B). These results demonstrate that BOP1 and BOP2 have the capacity to activate transcription, dependent on their recruitment to the target promoter.
Figure 1.
Functional Analysis of the BOP Proteins.
(A) Schematics of the constructs used for the transactivation assays: AD:GAL4 activation domain, BD:GAL4 DNA binding domain, DBS:GAL4 binding site, LUC:Firefly Luciferase, RNL LUC:Renilla Luciferase, B1:BOP1, and B2:BOP2.
(B) Transcription activation assays of BOP1, BOP2, and bop1-1 recruited to DNA through the Gal4 BD (BD-B1:BD-BOP1, BD-B2:BD-BOP2, and BD-b1-1:BD-bop1-1). Gal4 BD-AD and BD were used as positive and negative controls, respectively. Wild-type BD-BOP1 or BD-BOP2 construct was mixed with mutant BD-bop1-1 construct at a 2.5:1 ratio or a 1:1 ratio. RNL LUC was used as internal control to measure the photon count ratio of the reporter LUC and RNL LUC for each sample. B1 and B2 indicate BOP1 and BOP2 protein expressed alone. Fold activation represents the relative luciferase units obtained for the given construct(s) divided by those obtained with the unfused Gal4 BD construct alone (n = 9). Error bars represent sd.
(C) β -Galactosidase assays quantifying reporter gene expression in the yeast two-hybrid interactions. Error bars represent sd (n = 9). E, empty vector.
(D) In planta bimolecular fluorescence complementation assays. Combinations of constructs are shown below the images. A yellow fluorescent protein (YFP) signal (green, middle panel) indicates a physical interaction between proteins. A 35Spro:RFP construct (red, top panel) was used to determine transformation efficiency. Arrows point to the locations of the nuclei under bright field (bottom panel).
Because bop1-1 plants exhibited dominant-negative phenotypes similar to those of bop1-4 bop2-11 plants (Ha et al., 2004), we assessed whether mutant bop1-1 protein could inhibit wild-type BOP transactivation activity. bop1-1 protein fused to the Gal4 BD (BD-b1-1) showed no transactivation activity (Figure 1B), indicating that the addition of four amino acids to the C terminus of the BOP1 protein completely abolished its transcriptional activation capacity. In the presence of bop1-1 protein, the transactivation capacity of wild-type BOP1 and BOP2 protein was strongly reduced (Figure 1B). Thus, bop1-1 protein either interferes with the transactivation function of the wild-type BOP proteins or itself functions as a transcriptional inhibitor.
Next, we tested for potential interactions between the BOP1, BOP2, and bop1-1 proteins. In the yeast two-hybrid system, BOP1 and BOP2 protein fused to the Gal4 DNA BD showed autoactivation activity when tested with the control empty vector (E) containing the activation domain (AD). Nonetheless, we could clearly detect pairwise interactions between the BOP1, BOP2, and bop1-1 proteins (Figure 1C). The BOP1 and BOP2 proteins showed equally strong interactions as homodimers and heterodimers (Figure 1C). Using bimolecular fluorescence complementation assays, we verified that both BOP1 and BOP2 form homodimers in planta and that BOP1 and BOP2 can also heterodimerize (Figure 1D). These interactions were observed in both the cytoplasm and the nucleus. bop1-1 protein interacted strongly in the nucleus and more weakly in the cytosol with both BOP1 and BOP2 proteins (Figure 1D). We conclude that BOP1 and BOP2 can physically interact to form homodimers and heterodimers and that bop1-1 mutant protein can interact with both wild-type proteins, potentially to attenuate their abilities to activate target gene transcription.
Nuclear Localization of BOP1 Protein Is Necessary and Sufficient for Biological Function
To determine the biological relevance of BOP1 protein localization in the nucleus, we generated a translational fusion of BOP1 to the hormone binding domain of the rat glucocorticoid receptor (BOP1-GR) and transformed this construct into Landsberg erecta (Ler), bop1-1, and bop1-4 bop2-11 plants. Application of dexamethasone (Dex) hormone leads to translocation of the GR fusion protein from the cytoplasm to the nucleus (see Supplemental Figure 2 online). After a 4-h Dex treatment, BOP1-GR fusion protein was enriched in the nuclear fraction compared with non-Dex-treated fractions (see Supplemental Figure 2 online). These results show that BOP1:GR fusion protein is translocated into the nucleus in a steroid-dependent manner.
The development of 35Spro:BOP1-GR Ler plants was indistinguishable from that of wild-type Ler plants in the absence of hormone (Figures 2A and 2B). Following Dex treatment, 35Spro:BOP1-GR Ler plants displayed upward curling leaf phenotypes (Figures 2C and 2D) characteristic of BOP1 gain-of-function plants (Figure 2E). A subtle distinction between the two is that in 35Spro:BOP1-GR Ler plants, this upward curling can occur toward the tip of the blade, causing waving of the leaves (Figure 2D), whereas 35Spro:BOP1 Ler leaves show upward curling more prevalently along the margins (Figure 2E). By contrast, untransformed wild-type Ler plants grown in the presence of Dex showed no phenotypic changes (see Supplemental Figure 3 online). Thus, the upward curling leaf phenotypes observed in the 35Spro:BOP1-GR Ler plants is due to Dex-dependent ectopic activity of the BOP1-GR fusion protein.
Figure 2.
Dose Responsiveness of the 35Spro:BOP1-GR Phenotype.
(A) Ler.
(B) to (D) 35Spro:BOP1-GR Ler.
(E) 35Spro:BOP1 Ler.
(F) bop1-1.
(G) to (I) 35Spro:BOP1-GR bop1-1.
(J) 35Spro:BOP1 bop1-1.
Representative 16-d-old whole plants (left panel) and heteroblastic leaf series (right panel) from cotyledons up to leaf number two, four, or five are shown for each line after incubation in the indicated concentration of Dex. Bars = 10 mm in (A), (B), (F), and (G) and 5 mm in (C) to (E) and (H) to (J).
Similarly, the development of 35Spro:BOP1-GR bop1-1 transformants was indistinguishable from that of untransformed bop1-1 plants in the absence of hormone (Figures 2F and 2G). bop1-1 plants showed no phenotypic alterations in the presence of Dex (see Supplemental Figure 3 online), whereas 35Spro:BOP1-GR bop1-1 plants displayed partial to near-complete rescue of the rosette leaf phenotypes (Figures 2H and 2I). In the presence of 10 μ m Dex, the leaves of 35Spro:BOP1-GR bop1-1 plants were mostly wild-type in appearance or displayed a single ectopic outgrowth, and some also showed strongly upward curling leaf phenotypes (Figures 2H and 2I) characteristic of 35Spro:BOP1 bop1-1 (Figure 2J) plants. Similar results were obtained with 35Spro:BOP1-GR bop1-4 bop2-11 plants (see Supplemental Figure 4 online). These data show that BOP1-GR fusion protein is biologically active in a hormone dose-dependent manner and that nuclear localization of BOP1 protein is necessary and sufficient for its biological function.
Activation of AS2 and ASL4/LOB by BOP1 Induction
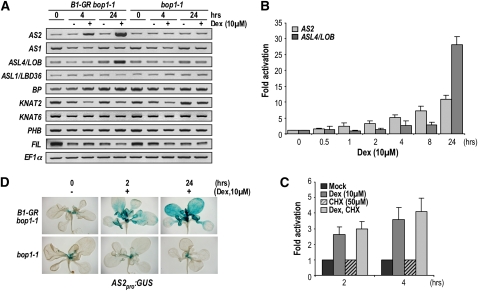
We hypothesized that if BOP1 functions as a transcriptional activator in vivo, then it might directly regulate some of its known downstream target genes (Ha et al., 2007). To identify direct targets of BOP1 regulation, we examined the effect of BOP1-GR activation on the expression of candidate ASL/LBD, class I KNOX, and adaxial-abaxial polarity genes (Figure 3A). We found that AS2 and ASL4/LOB mRNA levels were elevated within 4 and 24 h, respectively, of Dex treatment (Figure 3A). In a time course of Dex induction in 35Spro:BOP1-GR bop1-1 and bop1-1 plants, AS2 expression was induced as early as 1 h after Dex application and continuously increased over the 24-h time course (Figure 3B). By contrast, increased ASL4/LOB expression only became detectable 4 to 8 h after induction. These data are consistent with AS2 being an immediate target and ASL4/LOB being an indirect target of BOP1 activation.
Figure 3.
AS2 Is an Immediate Target of BOP1.
(A) Expression profiles of BOP1 downstream target genes. RT-PCR analysis of AS1, AS2, ASL4/LOB, ASL1/LBD36, BP, KNAT2, KNAT6, PHB, and FIL expression in 11-d-old bop1-1 or 35S:BOP1-GR bop1-1 plants mock treated or treated with Dex for 0, 4, or 24 h. The PCR products were visualized using ethidium bromide staining, and EF1 α was used as a control.
(B) Induction of AS2 and ASL4/LOB expression in 11-d-old 35Spro:BOP1-GR bop1-1 plants mock treated or treated with Dex for the indicated times.
(C) Relative temporal expression of AS2 in 11-d-old 35Spro:BOP1-GR bop1-1 plants after 2 or 4 h of mock, Dex, CHX, or Dex+CHX treatment. Expression values are normalized to the respective mock treatment control. Mean transcript levels in (B) and (C) were determined by real-time quantitative RT-PCR analyses of three biological replicates, normalized to TUB4. Error bars represent sd.
(D) AS2pro:GUS activity in 11-d-old 35Spro:BOP1-GR bop1-1 plants treated with 10 μ M Dex for 0, 2, or 24 h.
Rapid activation of AS2 transcription suggested that it might occur independently of protein synthesis. We tested this hypothesis by analyzing AS2 expression in response to Dex induction in the presence of the protein synthesis inhibitor cycloheximide (CHX) (Figure 3C). The effect of CHX was monitored by examining the expression of IAA1 (see Supplemental Figure 5 online), which is strongly induced by CHX alone (Abel et al., 1995). Combined Dex plus CHX treatment resulted in the induction of AS2 mRNA similarly to Dex treatment alone (Figure 3C; see Supplemental Figure 5 online). Thus, the rapid induction of AS2 by BOP1 does not require protein synthesis, indicating that AS2 is likely a direct transcriptional target of BOP1.
We determined in which tissues BOP1 activates AS2 expression by examining the activity of a promoter AS2:GUS reporter gene fusion in response to BOP1-GR induction. This AS2pro:GUS construct, in which the GUS coding sequence was fused to 4776 bp of AS2 upstream sequence, was sufficient to complement the as2 phenotype. Consistent with previous reports (Iwakawa et al., 2007; Wu et al., 2008), the AS2pro:GUS transgenic lines all showed the same pattern of GUS activity on the adaxial side of cotyledons and leaves, as well as in root tips (see Supplemental Figure 6 online). After 2 h of BOP1 induction, AS2pro:GUS activity in 35Spro:BOP1-GR bop1-1 plants was elevated in developing leaves and expanded more distally into the blade of mature leaves (Figure 3D). After 24 h of induction, ectopic GUS activity was observed throughout the blades of mature leaves and the base of the cotyledons. No change in AS2pro:GUS activity was observed in bop1-1 plants following 24 h of Dex treatment (Figure 3D). These data indicate that BOP1 is sufficient to activate AS2 transcription in vegetative tissues and that the effect of BOP1 on the AS2 promoter is likely to be direct.
BOP1 Directly Associates with the AS2 Promoter
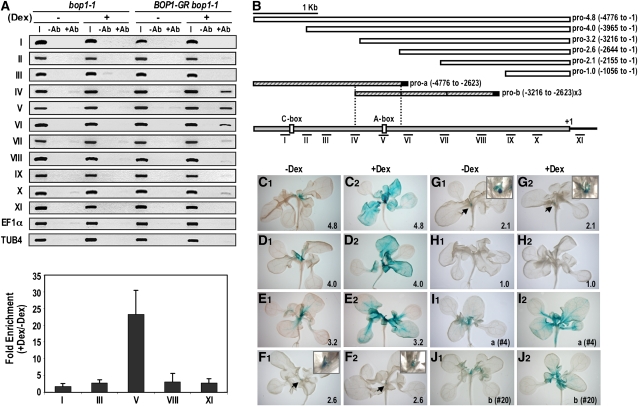
To determine whether BOP1 directly associates with AS2 regulatory sequences, we performed chromatin immunoprecipitation (ChIP) using BOP1-GR bop1-1 transgenic plants. After determining that full-length BOP1-GR fusion protein in the nuclear fraction was specifically recognized by an anti-GR antibody (see Supplemental Figure 2 online), we performed ChIP assays using 11 sets of primers spanning 5 kb of the AS2 upstream genomic sequence and coding region. Compared with the input and mock control, Dex-treated BOP1-GR bop1-1 samples showed strong enrichment at sites IV, V, and VI in the promoter region (Figures 4A and 4B). Using quantitative PCR, we detected 25-fold enrichment of site V after Dex treatment (Figure 4A). ChIP assays performed on untransformed bop1-1 plants showed no significant enrichment of the promoter regions tested, nor did the control EF1 α and TUB4 genomic regions. These data demonstrate that BOP1 associates in vivo with specific regulatory sites located in the AS2 promoter and together with the transactivation results suggest that BOP1 functions in a transcriptional activator complex that acts directly at the AS2 promoter.
Figure 4.
Identification of the Genomic Region Responsible for AS2 Regulation by BOP1.
(A) Detection of AS2 genomic fragments in bop1-1 and 35Spro:BOP1-GR bop1-1 seedlings after ChIP using anti-GR antiserum. Roman numerals denote 200- to 500-bp genomic fragments amplified from the AS2 promoter and coding sequences (top). Enrichment of genomic fragments in bop1-1 and 35Spro:BOP1-GR bop1-1 samples after Dex treatment was quantified by real-time quantitative RT-PCR (bottom) after normalization to the unrelated TUB4 control sequence. Error bars represent sd. I, input; −Ab, without antibody, +Ab, with antibody.
(B) Diagram of the AS2 promoter (gray). White boxes indicate the 5 ′ regulatory fragments used in the GUS reporter constructs. Hatched boxes in pro-a and pro-b contain the region from −4776 to −2623 and a three tandem repeat of the region from −3216 to −2623, respectively, fused to a 35S minimal promoter (black boxes). The predicted A-box (−2818) and C-box (−4254) bZIP protein binding sites are indicated. The positions of the promoter regions analyzed by ChIP are shown at the bottom.
(C) to (J) Functional analysis of the AS2 regulatory region. Representative expression patterns of 35Spro:BOP1-GR bop1-1 plants carrying the AS2pro:GUS reporter constructs diagramed in (B) are shown. Promoter activity was monitored after incubation in the presence (+) or absence (−) of 10 μ M Dex for 24 h. Arrows indicate the SAM region, and magnified views of the shoot apices are shown in the boxes. The label in the lower right-hand corner indicates the AS2 promoter deletion construct used.
AS2 Promoter Deletion Analysis
To confirm that the genomic region enriched in the ChIP assay was responsible for the specific regulation of AS2 by BOP1 protein, we generated a series of β -glucuronidase (GUS) constructs containing deletions of the AS2 5 ′ regulatory region (Figure 4B). In the absence of Dex, BOP1-GR bop1-1 plants carrying the pro-4.0 and pro-3.2 deletion constructs showed the same pattern of GUS activity (Figures 4D1 and 4E1) as plants carrying the pro-4.8 full-length promoter construct (Figure 4C1; see Supplemental Figure 6 online), indicating that the 3.2-kb upstream region contains all the cis-acting elements necessary for proper AS2 expression. By contrast, plants carrying either the pro-2.6 or the pro-2.1 construct displayed very weak GUS activity, restricted to the adaxial domain of young leaf primordia and the early stage of ectopic outgrowth along the bop1-1 petioles (Figures 4F1 and 4G1). Plants carrying the pro-1.0 construct displayed no GUS activity (Figure 4H1). The upstream region between 2.6 and 3.2 kb corresponds to sites IV, V, and VI with which BOP1 associates in the ChIP assays (Figures 4A and 4B), confirming the in vivo requirement for these BOP1 binding elements in the AS2 promoter.
We next determined which regions of the AS2 promoter responded to BOP1 induction by Dex treating BOP1-GR bop1-1 plants carrying the various AS2 promoter deletion constructs. After 24 h of Dex treatment, transgenic plants carrying the pro-4.8, pro-4.0, or pro-3.2 construct displayed ectopic GUS activity throughout the leaf blades, indicating a strong response to BOP1 activation (Figures 4C2 to 4E2). Lines carrying shorter constructs showed no Dex-dependent induction (Figures 4F2 to 4H2). These data show that the region between 2.6 and 3.2 kb upstream of the AS2 start site is required for the response of AS2 to BOP1 activation.
We generated additional promoter constructs to confirm the importance of this BOP1-responsive region. The first contained the most distal region of the AS2 promoter, from 2.6 to 4.8 kb (pro-a). The second contained three tandem repeats of the 2.6- to 3.2-kb region (pro-b). We analyzed BOP1-GR bop1-1 lines with pro-a or pro-b driven GUS activity to determine their responsiveness to BOP1 activation. Thirteen lines carrying the pro-a construct showed weak GUS activity in young leaf primordia (Figure 4I1). After Dex treatment, nine of these lines displayed expanded GUS staining into the proximal region of the maturing leaf blades (Figure 4I2). Three lines carrying the pro-b construct showed weak GUS activity in young leaf primordia (Figure 4J1), and after Dex treatment, all three showed expanded GUS activity into the proximal leaf blade (Figure 4J2). These data demonstrate that the 2.6- to 3.2-kb region upstream of the AS2 coding sequence is necessary and sufficient for AS2 induction by BOP1 protein.
AS2 Activation by BOP1 and BOP2 in the Proximal Leaf Domain
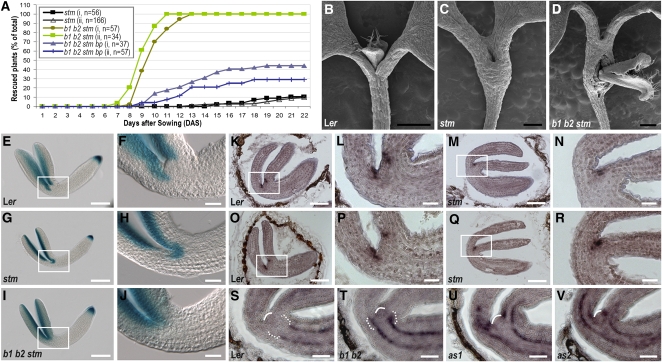
To determine in which tissues BOP1 protein activates AS2 expression in vivo, we examined the activity of a AS2pro:GUS reporter gene construct in bop1-1 and bop1-4 bop2-11 plants. Previous experiments detected AS2 transcripts in embryonic protoderm cells on the adaxial side of cotyledon primordia beginning at the heart stage (Iwakawa et al., 2002, 2007; Wu et al., 2008). This expression pattern was observed until the torpedo stage in AS2pro:GUS embryos. Bent-cotyledon stage embryos displayed strong GUS staining in the adaxial domain of the cotyledons (Figures 5A and 5D). As the embryos fully matured, AS2pro:GUS activity appeared weaker in this domain, and after germination, GUS staining was no longer detected in the cotyledons.
Figure 5.
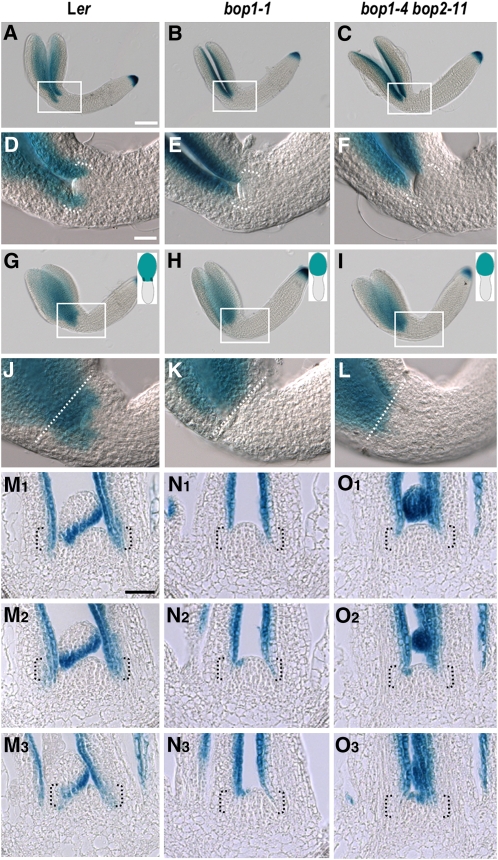
Polar Regulation of AS2 Expression by BOP1 and BOP2.
(A) to (F) show a frontal view, and (G) to (L) show a sagittal view. (D) to (F) and (J) to (L) are magnified views of the regions boxed in (A) to (C) and (G) to (I), respectively. A schematic of the sagittal view of the embryo on its side with one cotyledon atop the other is shown for (G) to (L). In (J) to (L), the dotted line marks the boundary between the cotyledon and cotyledonary petiole. In (D) to (F) and (M) to (O), brackets denote the proximal organ region. Bars = 100 μ m in (A) to (C), (G) to (I), and (M) to (O) and 25 μ m in (D) to (F) and (G) to (L).
(A), (D), (G), and (J) GUS activity in AS2pro:GUS Ler bent-cotyledon stage embryos.
(B), (E), (H), and (K) GUS activity in AS2pro:GUS bop1-1 bent-cotyledon stage embryos.
(C), (F), (I), and (L) GUS activity in AS2pro:GUS bop1-4 bop2-11 bent-cotyledon stage embryos.
(M) Serial sections of a 10-d-old AS2pro:GUS Ler shoot apex.
(N) Serial sections of a 10-d-old AS2pro:GUS bop1-1 shoot apex.
(O) Serial sections of a 10-d-old AS2pro:GUS bop1-4 bop2-11 shoot apex.
Up until the torpedo stage, no difference in the AS2 expression pattern between wild-type and bop embryos was detected. At the early bent-cotyledon stage, wild-type embryos displayed AS2 promoter-driven GUS activity from the base to the tip of the cotyledons (Figures 5A and 5D). By contrast, in bop1-1 and bop1-4 bop2-11 embryos, AS2pro:GUS activity was not detected in the proximal domain of the cotyledons near the SAM (Figures 5B, 5C, 5E, and 5F). Sagittal views confirm that GUS staining was absent from the proximal domain of bop embryonic cotyledons (Figures 5G to 5L), the region in which the bop1-1 phenotype is detected after germination.
Next, we analyzed AS2pro:GUS activity at the vegetative stage. Compared with wild-type seedlings (Figure 5M; see Supplemental Figures 6A and 6B online), GUS activity was reduced or absent in the leaf base of both bop1-1 and bop1-4 bop2-11 seedlings (Figures 5N and 5O; see Supplemental Figures 6C and 6D online). This region corresponds to the BOP1 and BOP2 expression domains (see Supplemental Figure 7 online), demonstrating that BOP1 and BOP2 are required during both embryonic and vegetative development to induce AS2 expression in the proximal domain of cotyledons and rosette leaves.
Requirement of AS2 for bop and as Leaf Phenotypes
Our data indicate that BOP1 and BOP2 are required to activate AS2 expression in the proximal domain of cotyledons and leaves, the region in which the bop ectopic outgrowth phenotype is manifested. We tested whether the absence of proximal AS2 activity is responsible for the bop phenotype by generating transgenic plants expressing AS2 under the control of 6.0 kb of BOP1 upstream sequence. When fused to a GUS reporter, this BOP1 promoter sequence drives strong GUS activity at the base of the cotyledons and leaves, recapitulating the native BOP1 expression domain (Figure 6J). We then determined whether driving AS2 in the BOP1 expression domain could rescue the bop proximal ectopic leaf outgrowth phenotypes (Figures 6A and 6C). Transformation of BOP1pro:AS2 into bop1-4 bop2-11 and bop1-1 plants led to the elevation of AS2 expression levels (Figure 6I) and a dramatic reduction in the amount of ectopic outgrowth along the petioles (Figures 6B and 6D). Eleven percent of bop1-1 lines (n = 37) and 5% of bop1-4 bop2-11 lines (n = 230) showed phenotypic complementation (see Supplemental Table 1 online). Thus, the ectopic outgrowth in bop leaves is directly attributable to the loss of AS2 expression in the proximal region of the primordia, indicating that BOP1 and BOP2 act through AS2 to suppress meristematic activity and/or class I KNOX gene expression in this region.
Figure 6.
Rescue of the bop Phenotype by a BOP1pro:AS2 Transgene.
(A) bop1-4 bop2-11 rosette.
(B) BOP1pro:AS2 bop1-4 bop2-11 rosette.
(C) bop1-1 rosette.
(D) BOP1pro:AS2 bop1-1 rosette.
(E) as2-1 rosette.
(F) BOP1pro:AS2 as2-1 rosette.
(G) as1-1 rosette.
(H) BOP1pro:AS2 as1-1 rosette.
(I) AS2 expression in wild-type Ler, bop1 bop2 (b1 b2), bop1-1 (b1-1), and BOP1pro:AS2 plants. Mean transcript levels were determined by real-time quantitative PCR analyses and normalized to EF1 α. Error bars represent sd.
(J) BOP1pro:GUS activity in a Col seedling.
Arrowheads indicate ectopic organ outgrowths in (A) and (C), rescued leaf morphology in (B), (D), and (F), and lobed leaf morphology in (E). The arrow in (J) indicates the leaf base around the SAM. Bars = 10 mm.
Next, we assessed the ability of the BOP1pro:AS2 construct to rescue the as1 and as2 leaf phenotypes. BOP1pro:AS2 as1-1 plants (Figure 6H) were indistinguishable from as1-1 plants (Figure 6G), indicating that BOP1pro:AS2 activity had no detectable effect on the as1 phenotype. By contrast, BOP1pro:AS2 as2-1 plants displayed rescue of the as2-1 leaf phenotype (Figures 6E and 6F; see Supplemental Table 1 online). This result indicates that the expression of AS2 exclusively in the BOP1 domain at the proximal end of developing leaf primordia is sufficient to confer wild-type leaf morphology.
Relationship between the BOP and STM Pathways
Mutations in AS2 (and AS1) suppress the stm embryonic and vegetative SAM phenotypes, an effect that in the case of AS1 is BP dependent (Byrne et al., 2000, 2002). Finding that BOP1 and BOP2 induce AS2 expression near the boundary between the SAM and leaf primordia, we used genetic analysis to investigate whether BOP1 and BOP2 affect the STM pathway.
Plants homozygous for the strong stm-11 allele lacked an embryonic SAM (Long et al., 1996; Carles et al., 2004) and formed cotyledons that were fused at their base (Figure 7C), unlike wild-type plants (Figure 7B), yet 20 d after germination, 10% of stm-11 plants (n = 56 for experiment i and 166 for experiment ii) developed rosette leaves from either the shoot apex or the region between the fused cotyledonary petioles (Figure 7A). At germination, bop1-4 bop2-11 stm-11 seedlings appeared identical to stm-11 seedlings. However, 6 to 7 d later, some triple mutant seedlings began to develop leaves from the fused cotyledonary petiole region (Figure 7D), and by 13 d after germination, 100% of bop1 bop2 stm plants (n = 57 for i and 34 for ii) had formed a shoot meristem and produced true leaves from this region (Figure 7A). Compared with dome-shaped wild-type SAMs, which consisted of small, highly cytoplasmic cells organized into discrete cell layers, the flattened shoot apex region of stm-11 seedlings contained large, vacuolated cells (see Supplemental Figure 8 online). Sections through the fused cotyledonary petiole region of bop1 bop2 stm seedlings revealed within the differentiated tissue the presence of domes of small, highly cytoplasmic cells resembling those of wild-type shoot meristems, albeit somewhat less organized (see Supplemental Figure 8 online). Thus, the absence of the BOP proteins in stm-11 plants permits the formation of a functional shoot meristem at the base of the fused cotyledons.
Figure 7.
Genetic Interactions between BOP and the STM-AS2 Pathway.
(A) Rescue of stm-11 phenotypes by bop1-4 bop2-11 and bp-1. Plants with organogenesis from the fused cotyledon region were counted as rescued. (i) and (ii) represent separate experiments.
(B) to (D) Scanning electron micrographs of 7-d-old Ler (B), stm-11 (C), and bop1-4 bop2-11 stm-11 (D) plants.
(E) to (J) AS2pro:GUS activity in bent-cotyledon stage embryos.
(E) and (F) Ler.
(G) and (H) stm-11.
(I) and (J) bop1-4 bop2-11 stm-11.
(K) to (N) BOP1 expression in bent-cotyledon stage embryos.
(K) and (L) Ler.
(M) and (N) stm-11.
(O) to (R) BOP2 expression in bent-cotyledon stage embryos.
(O) and (P) Ler.
(Q) and (R) stm-11.
(S) to (V) BP expression in bent-cotyledon stage embryos.
(S) Ler.
(T) bop1-4 bop2-11.
(U) as1-1.
(V) as2-1.
(F), (H), (J), (L), (N), (P), and (R) are magnified views of the regions boxed in (E), (G), (I), (K), (M), (O), and (Q), respectively. In (S) to (V), white arches denote the SAM, and brackets indicate the cotyledonary petiole region. Bars = 0.5 mm in (B) to (D), 100 μ m in (E), (G), (I), (K), (M), (O), and (Q), and 25 μ m in (F), (H), (J), (L), (N), (P), and (R) to (V).
In as1 stm plants, BP can replace STM to promote embryonic and vegetative SAM formation (Byrne et al., 2002). To determine if BP is likewise necessary for the meristematic activity observed in bop1 bop2 stm plants, we constructed bop1-4 bop2-11 stm-11 bp-1 plants. We found that compared with 100% of bop1-4 bop2-11 stm-11 seedlings, only 30 to 40% of bop1-4 bop2-11 stm-11 bp-1 seedlings initiated a postembryonic shoot meristem (n = 37 for i and 57 for ii) (Figure 7A). These data indicate that, in the absence of BOP activity, BP can substitute for STM to establish a functional vegetative shoot meristem.
AS1 expression expands ectopically into the apical region of stm embryos, indicating that AS1 is negatively regulated by STM (Byrne et al., 2000). Genetic models propose that AS2 is also negatively regulated by STM (Byrne et al., 2002); however, this has not been demonstrated at the molecular level. To determine the relationship between STM and AS2, we examined AS2pro:GUS activity during stm-11 embryo development. Unlike AS1, AS2 was not ectopically expressed in stm-11 embryos until the torpedo stage. In bent-cotyledon stage wild-type embryos, AS2 expression was restricted to the adaxial region of the cotyledons (Figures 7E and 7F). In stm-11 embryos, AS2 was ectopically expressed at the junction between the fused cotyledons, where the SAM would normally have formed (Figures 7G and 7H; see Supplemental Table 2 online). Because ectopic activation of AS2 was observed only after the stm phenotype was manifested, this result indicates that AS2 expression is not directly regulated by STM, but rather by another factor functional in the SAM.
Given that AS2 is an immediate target of BOP regulation, we compared the BOP and AS2 embryo expression patterns. In wild-type embryos, BOP1 (Figures 7K and 7L) and BOP2 (Figures 7O and 7P) were expressed in two foci in the proximal, adaxial region of the developing cotyledons. In stm-11 embryos, BOP expression was detected at the junction between the fused cotyledons (Figures 7M, 7N, 7Q, and 7R), coincident with the domain of ectopic AS2 expression. However, in bop1-4 bop2-11 stm-11 embryos, AS2pro:GUS was not ectopically activated in this central region (Figures 7I and 7J). These data indicate that STM directly or indirectly represses BOP1 and BOP2 expression in the wild-type embryonic SAM and that misexpression of BOP1/2 at the junction between the fused cotyledons in stm embryos is responsible for the ectopic activation of AS2 in this domain.
Rescued bop1 bop2 stm seedlings formed ectopic shoot meristems from the fused cotyledonary petiole region (Figure 7D), whereas as1 stm and as2 stm seedlings formed meristems at their shoot apices like as1 and as2 single mutants (Byrne et al., 2000, 2002). We reasoned that because BP can substitute for STM in as1 stm and as2 stm seedlings and is also partially responsible for the rescued phenotypes of bop1 bop2 stm seedlings (Figure 7A), the difference in phenotypic rescue between as1 stm, as2 stm, and bop1 bop2 stm seedlings might be due to the differential misregulation of BP expression. Thus, we compared the BP expression patterns in as1, as2, and bop1 bop2 embryos. In wild-type embryos, BP mRNA was detected in two stripes in the procambium layer of the hypocotyl (Figure 7S; see Supplemental Figure 9A online). In as1-1 and as2-1 embryos, ectopic BP expression was observed in the basal region of the cotyledons and flanking the SAM (Figures 7U and 7V). By contrast, ectopic BP expression was not observed flanking the shoot apex in bop1-4 bop2-11 embryos (Figure 7T). However, BP expression was expanded into the presumptive junction between the hypocotyl and cotyledon base (Figure 7T; see Supplemental Figures 9B and 9C online), the area corresponding to the region of de novo shoot meristem generation in stm-11 bop1-4 bop2-11 seedlings (Figure 7D). Region-specific misregulation of BP expression can therefore account for the differences between the as1 stm, as2 stm, and bop1 bop2 stm embryo phenotypes, although interestingly, BOP-dependent BP suppression appears to be regulated nonautonomously.
DISCUSSION
The BOP1 and BOP2 proteins contain a conserved BTB/POZ domain, which has been shown to mediate protein–protein interactions and to confer transcription activation capacity to the related NPR1 protein (Rochon et al., 2006). We find that BOP1 and BOP2, like NPR1, can function as transcriptional activators when recruited to target DNA. Because they lack known DNA binding sequences, it seems likely that BOP1 and BOP2 act as transcriptional coactivators in vivo when recruited to the promoter region of target genes through their interaction with DNA binding proteins. BOP1 and BOP2 can physically interact with one another in vitro and in vivo, and bop1-1 mutant protein can interact with both wild-type proteins. In addition, bop1-1 protein strongly reduces the transactivation capability of the BOP1 and BOP2 proteins. Based on these data, we propose that the dominant-negative phenotype of the bop1-1 plants is caused by the interference of bop1-1 mutant protein with wild-type BOP2 protein activity, causing bop1-1 plants to resemble bop1 bop2 double mutant plants.
NPR1 acts as a coactivator to regulate Arabidopsis defense gene expression (Cao et al., 1994). This coactivator function of NPR1 is mediated by SA-dependent protein modification, whereas recruitment of the protein to target promoters is autonomous and SA independent. In contrast with NPR1, BOP1 and BOP2 display transactivation activity independent of SA or other stimuli. Interestingly, the BOP proteins lack all or part of two domains of NPR1, one located from amino acids 22 to 44 and the other located from amino acids 463 to 513 (see Supplemental Figure 10 online), that repress transactivation activity in the absence of SA (Rochon et al., 2006). BOP1 and BOP2 also lack two Cys residues (Cys-521 and Cys-529) that modulate transactivation in SA-stimulated cells (Rochon et al., 2006). These specific structural differences between the BOP proteins and NPR1 may largely account for the lack of responsiveness of the BOP proteins to SA.
Recently it has been reported that S-nitrosylation on residue Cys-156 of NPR1 induced its oligomerization upon SA induction to maintain protein homeostasis (Tada et al., 2008). The BOP proteins lack the Cys residue that corresponds to Cys-156 of NPR1, whereas four other Cys residues in the BTB/POZ domain important for oligomerization (Mou et al., 2003) appear to be conserved (see Supplemental Figure 10 online). Because the BOP proteins localize to both the nucleus and the cytosol without any stimulus, the absence of the Cys-156 residue can be considered one structural feature that releases their subcellular localization from SA regulation. Nuclear localization of BOP1 protein is necessary and sufficient for its biological function; thus, cytoplasmic retention of BOP1 protein might maintain protein homeostasis. However, it remains to be seen whether regulated nuclear transport exists for the BOP proteins and whether they respond to a developmental signal(s) as has been previously proposed (Hepworth et al., 2005).
BOP1 and BOP2 are positive regulators of AS2 transcription. Among the known BOP1 and BOP2 target genes, only the expression of AS2 was rapidly induced by 35Spro:BOP1-GR, indicating that it is an early target of the BOP pathway. Induction of AS2 expression by BOP1 did not require protein synthesis, consistent with AS2 being a direct target of BOP1. By contrast, ASL4/LOB showed delayed induction compared with that of AS2. Because AS2 positively regulates ASL4/LOB expression (Byrne et al., 2002), ASL4/LOB regulation by BOP protein could be mediated through AS2. The regulation of adaxial-abaxial polarity genes by BOP1 and BOP2 (Ha et al., 2007) may also be mediated partially or fully through AS2. AS2 being a major target of BOP activity is also consistent with the observation that driving AS2 transcription in the BOP1 expression domain is sufficient to partially rescue the bop phenotype in some plants. However, the fact that the percentage of rescued plants is low indicates that additional target genes, as yet to be identified, also play biologically relevant roles in the BOP leaf morphogenesis regulatory pathway.
We find that BOP1 protein is specifically recruited to the AS2 promoter where it can function as part of a transcriptional activator complex to induce AS2 expression. The promoter elements with which BOP1 associates are positioned between 2.6 and 3.2 kb upstream of the AS2 start site. This is considerably upstream of a binding site for KAN1, a negative regulator of AS2, which lies 1.4 kb upstream of the ATG (Wu et al., 2008). Because the BOP proteins lack a discernable DNA binding domain, no information is available about the specific cis-element(s) with which BOP1 may associate at the AS2 promoter. However, the genomic region responsible for the BOP1-mediated induction of AS2 expression coincides with the region to which BOP1 protein binds in vivo. This region contains an A-box bZIP transcription factor binding site, the presence of which may indicate involvement of TGA transcription factors in BOP1 DNA binding, as has been proposed for NPR1 (Rochon et al., 2006). BOP proteins physically interact with the TGA transcription factor PERIANTHIA (PAN) in yeast (Hepworth et al., 2005), and the fact that bop1 bop2 and pan plants have overlapping phenotypes indicates that they function in the same genetic pathway. However, neither BOP1 nor BOP2 modulate the transcriptional activity of PAN in transactivation assays nor is AS2 transcription regulated by PAN (see Supplemental Figure 11 online). We therefore predict that the BOP proteins may regulate AS2 expression in concert with multiple bZIP transcription factor family members.
The AS1-AS2 chromatin-remodeling complex plays a key role in class I KNOX gene repression in Arabidopsis lateral organs (Guo et al., 2008); conversely, AS1 is negatively regulated by STM in the SAM to maintain these cells in a proliferative state (Byrne et al., 2000). However, the regulatory mechanism that excludes AS2 from the SAM has not been understood. Our study shows during the early stages of stm embryogenesis, AS2 expression is excluded from the presumptive shoot apex region and is ectopically expressed only at a late stage of embryo development when the stm phenotype is already evident. Thus, AS2 is regulated by a factor functional in the SAM region, but not directly by STM. At the late embryonic stage, AS2 expression adjacent to the shoot apex is dependent on BOP function in both wild-type and stm plants. This finding implicates BOP function in the suppression of meristematic activity during late embryogenesis through the induction of AS2 in the adaxial, proximal region of the cotyledons.
Our results also uncover mutual negative regulation between the BOP genes and the class I KNOX genes. We find that STM directly or indirectly represses BOP1 and BOP2 expression in the SAM and that the ectopic activation of AS2 across the shoot apex region of stm embryos is dependent on BOP activity. Furthermore, the BOP genes (as well as AS1 and AS2) negatively regulate embryonic BP expression at the hypocotyl-cotyledon junction, such that in the absence of BOP activity, BP can substitute for STM to establish a functional vegetative shoot meristem following germination. Interestingly, we observed that in as1, as2, and bop1 bop2 embryos, BP misexpression occurred at the hypocotyl-cotyledon junction in two stripes of cells in the procambium layer that do not normally express AS1, AS2, or the BOP genes. These data suggest that, in addition to their roles in lateral organ determinacy and patterning, these genes act non-cell-autonomously to repress BP transcription. It remains to be seen whether this non-cell-autonomous activity of BOP1 and BOP2 is mediated via their regulation of AS2.
The molecular mechanisms that establish pattern formation along the proximal-distal leaf axis are still poorly defined. Leaf differentiation occurs from the distal blade toward the proximal petiole, reflected by a cell cycle arrest front moving gradually from tip to base (Nath et al., 2003). Studies have shown that growing leaves display a gradient of cell division rates in which the highest rates occur in the proximal regions (Poethig and Sussex, 1985). Class I KNOX gene activity creates an environment that promotes and sustains cell proliferation, and their ectopic expression in developing leaves prolongs leaf cell division and confers indeterminate features such as lobing and ectopic meristem formation (Chuck et al., 1996).
The formation of ectopic outgrowths of blade tissue along the leaf petiole accompanied by ectopic KNOX gene expression in bop1-1 and bop1 bop2 mutants indicates that BOP1 and BOP2 are redundantly required for KNOX repression in the proximal region of developing leaves (Ha et al., 2003, 2007). Furthermore, BOP1 and BOP2 are expressed specifically in the proximal, adaxial region of the leaf beginning at the time of primordia initiation. The BOP expression domain overlaps with that of AS2 (Iwakawa et al., 2007), and BOP activity is required to induce AS2 transcription in the proximal, adaxial region of organ primordia. In addition, the BOP genes and AS2 promote adaxial organ identity (Lin et al., 2003; Xu et al., 2003; Ha et al., 2007). as2 and bop mutants have some overlapping phenotypes, although the bop phenotype is more severe than the as2 phenotype in proximal tissues. However, bop as2 plants display a synergistic phenotype (Ha et al., 2003, 2007), indicating that AS2 and the BOP genes are part of overlapping genetic pathways rather than a single linear pathway. Therefore, the BOP genes likely regulate other factors in the proximal leaf domain in addition to AS2, potentially including other ASL/LBD genes. However, ectopic BOP activity does not suppress normal blade development. This suggests that the function of the BOP genes as suppressors of blade formation is executed within a certain developmental context in the proximal region of the leaf.
An additional target of BOP proximal-distal regulation is the C2H2 zinc-finger putative transcription factor gene JAGGED (JAG). Loss-of-function jag mutations result in incomplete formation of distal tissues in developing leaves and floral organs, which is associated with the premature cessation of cell division (Dinneny et al., 2004; Ohno et al., 2004). Ectopic JAG expression in jag-5D mutants causes the formation of ectopic blade tissue along the leaf petiole (Dinneny et al., 2004), similar to the loss-of-function bop phenotypes. The specific role of JAG in promoting leaf growth suggests that it acts downstream of leaf patterning genes; indeed, BOP1 and BOP2 repress the expression of JAG and its close relative NUBBIN (NUB; aka JGL) in the proximal leaf domain (Norberg et al., 2005).
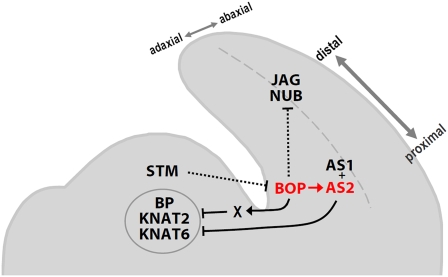
Our data that BOP1 is required to specifically induce AS2 expression in proximal cotyledon and rosette leaf cells begins to uncover a molecular mechanism for Arabidopsis proximal-distal polarity establishment (Figure 8). The BOP and AS2 expression patterns at the base of the leaf primordia are mutually exclusive with the class I KNOX gene expression patterns in the SAM (Pautot et al., 2001; Belles-Boix et al., 2006), and the proximal BOP expression pattern is reciprocal to the distal JAG and NUB expression patterns (Dinneny et al., 2004; Ohno et al., 2004). During organ patterning, BOP1/2 operate in the proximal region of the primordia to induce the transcription of AS2 and other target genes to suppress inappropriate class I KNOX gene activation. They also directly or indirectly repress JAG and NUB expression, restricting their activity to distal leaf cells. The sum of these activities promotes determinacy in proximal leaf cells, ensuring the proper specification of petiole tissue adjacent to the SAM.
Figure 8.
Model for Arabidopsis Vegetative Organ Initiation and Patterning.
The class I KNOX protein STM is restricted to the SAM and represses the expression of organ patterning genes, such as BOP1 and BOP2 (BOP). The BOP genes are transcribed at the adaxial base of vegetative primordia, where they directly activate AS2 expression and directly or indirectly repress JAG and NUB expression in the proximal domain to pattern the proximal-distal axis. AS2 and AS1 form a protein complex that is active throughout the adaxial organ domain and directly represses BP and KNAT2 transcription to restrict their activity to the SAM. The BOP proteins also repress BP, KNAT2, and KNAT6 transcription in the proximal region of organ primordia, likely indirectly through an unknown factor (X).
[See online article for color version of this figure.]
METHODS
Plant Materials
Arabidopsis thaliana plants were grown as described (Ha et al., 2004). All plants, including bop1-1 (Ha et al., 2003), bop1-4 bop2-11 (Ha et al., 2004), as1-1 (Byrne et al., 2000) stm-11 (Long and Barton, 1998), and bp-1 (Venglat et al., 2002), were in the Ler ecotype, except for pan-1 in the Wassilewskija background (Hepworth et al., 2005) and those plants carrying the as2-1 allele (Semiarti et al., 2001), which were introgressed three times into Columbia (Col-0) prior to analysis. The 35Spro:BOP1 line was previously described (Ha et al., 2007).
Construction of Transgenic Plants
The BOP1-GR fusion construct was created by cloning the BOP1 full-length cDNA into the BamHI and XhoI restriction sites of the GR vector (Huq et al., 2004). The BOP1pro:AS2 construct was generated by cloning 6.0 kb of BOP1 upstream sequence into pCAMBIA1302 (CAMBIA) adjacent to a full-length AS2 cDNA using the EcoRI, PstI, and BstEII restriction sites. To generate the GUS constructs, regions of the AS2 upstream sequence or the 6.0-kb BOP1 upstream sequence were amplified from Col genomic DNA and blunt end cloned into pENTR/D-TOPO (Invitrogen), then recombined into pBGWFS7,0 using Gateway LR recombination (Invitrogen). The full-length 4.8-kb promoter region of AS2 was used to generate the AS2pro:GUS reporter construct. For each construct, the binary vector in the Agrobacterium tumefaciens strain GV3101 was introduced into plants by the floral dip method (Clough and Bent, 1998). Primer sequences are listed in Supplemental Table 3 online.
Transactivation Assays
For the Gal4 DNA BD fusions, the BOP1, BOP2, or bop1-1 coding sequences were cloned into the BamHI and KpnI restriction sites in frame to the BD in the pMN6 plasmid (Huq et al., 2003). Primer sequences are given in Supplemental Table 3 online. pMN6 alone was used as a negative control. A Renilla luciferase (LUC) gene under the control of a cauliflower mosaic virus 35S promoter was used as an internal control, and a firefly LUC gene under the control of four copies of the Gal4 upstream activating sequence fused to a minimal promoter served as a reporter (Huq et al., 2003). The LUC gene in the pGLL reporter plasmid (Huq et al., 2003) was driven by a chimeric promoter containing two copies of the lac operator and four copies of the Gal4 binding site fused upstream of a minimal 35S promoter fragment. Col whole seedlings or leaves were bombarded with the effector constructs along with the 4 × USAGal4: firefly luciferase reporter and the cauliflower mosaic virus 35S:renilla luciferase internal standard vector. For the bop1-1 transactivation assays, the BD-bop1-1 construct was cobombarded with either the BD:BOP1 or the BD-BOP2 construct at two different ratios (2.5:1 and 1:1). Up to 5 μ g of each effector and reporter plasmid, as well as 0.2 μ g of internal control plasmid, were delivered to the tissues by particle bombardment. Tissue was extracted in LUC extraction buffer (Roche). Transcription activity was measured using the dual-Luciferase system (Promega) from three bombardments repeated three times.
Yeast Two-Hybrid Assays
The coding sequences of BOP1, BOP2, or bop1-1 were blunt end cloned into the pENTR/D-TOPO vector (Invitrogen) and then fused into the pDEST 32 BD and pDEST 22 AD vectors (Invitrogen) using Gateway LR recombination (Invitrogen). Primer sequences are given in Supplemental Table 3 online. The bait and prey constructs were transformed into the yeast strain MaV203 (MAT α ). β -Gal activity in transformed yeast cells was measured using o-nitrophenyl- β -d-galactopyranoside as the substrate according to the manufacturer's protocol (Invitrogen).
Expression Analysis
GUS staining, tissue embedding, and sectioning were performed as described (Sieburth and Meyerowitz, 1997). Plant fixation and in situ hybridization were performed as described (Jackson, 1992). Probes were generated with the primers given in Supplemental Table 3 online, except for BP, which was directly linearized from a plasmid provided by Sarah Hake (University of California at Berkeley). For the chemical induction treatments, whole seedlings were transferred into and immersed in Murashige and Skoog media containing 10 μ M Dex and/or 50 μ M CHX.
Total RNA extraction and RT-PCR was performed as described (Ha et al., 2007). Quantitative real-time PCR was performed using the SYBR Green PCR master mix (Applied Biosystems with the primers shown in Supplemental Table 3 online). PCR reactions were run and analyzed using a MyiQTM Single-Color real-time PCR detection system (Bio-Rad). Quantification of all real-time PCR experiments was performed using three biological replicates.
Bimolecular Fluorescence Complementation Assays
The coding sequences of BOP1, BOP2, or bop1-1 were blunt end cloned into the pENTR/D-TOPO vector (Invitrogen) and fused to the dissected YFP at the N or C terminus in pE-SPYNE-GW (N-terminal) or pE-SPYCE-GW (C-terminal) (Weltmeier et al., 2006) using Gateway LR recombination (Invitrogen). Five micrograms of each YFP fusion construct plasmid and 200 ng of a pRecA-red fluorescent protein construct (Thompson et al., 2009) were used. For transient expression in onion cells, particle bombardment was performed using a Biollistic PDS-1000/He unit (Bio-Rad). For fluorescence visualization, epidermal peels were examined 24 h after bombardment using a Zeiss Axiophot microscope.
ChIP
ChIP was performed as described (Lawrence et al., 2004) using an anti-GR antibody (Santa Cruz Biotechnology) on seedlings mock or Dex treated for 4 h. Quantification of immunoprecipitated DNA was performed by real-time PCR. Comparison of the abundance of promoter fragments in +Dex versus –Dex treated 35Spro:BOP1-GR bop1-1 plants was performed by first normalizing each amplification value to the unrelated TUB4 sequence and then dividing by the value for input samples to obtain input-normalized values. The relative abundance of fragments in +Dex versus –Dex treated samples was calculated by determining the ratio of these input-normalized values. Primer sequences are listed in Supplemental Table 3 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: BOP1 (AT3G57130), BOP2 (AT2G41370), AS2 (AT1G65620), ASL4/LOB (AT5G63090), LBD36 (AT5G66870), STM (AT1G62360), BP (AT4G08150), KNAT2 (AT1G70510), KNAT6 (AT1G23380), PHB (AT2G34710), FIL (AT2G45190), JAG (AT1G68480), NUB (AT1G13400), and PAN (AT1G68640).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Subcellular Localization of BOP1 Protein.
Supplemental Figure 2. Dex-Dependent Enrichment of BOP1-GR in the Nuclear Fraction.
Supplemental Figure 3. Phenotypes of Ler and bop1-1 Plants after Dex Treatment.
Supplemental Figure 4. Phenotypes of 35Spro:BOP1-GR bop1-4 bop2-11 Plants.
Supplemental Figure 5. Direct Regulation of AS2 Expression by BOP1.
Supplemental Figure 6. Polar Regulation of AS2 Expression by BOP in Seedlings.
Supplemental Figure 7. Comparison of AS2, BOP1, and BOP2 Expression Patterns.
Supplemental Figure 8. Ectopic Shoot Meristem Formation in Rescued stm-11 Plants.
Supplemental Figure 9. Regulation of Embryonic BP Expression by BOP1 and BOP2.
Supplemental Figure 10. Multiple Sequence Alignment of the BOP1, BOP2, and NPR1 Proteins.
Supplemental Figure 11. Analysis of the Relationship between BOP, AS2, and PAN.
Supplemental Table 1. Rescue of Leaf Phenotypes by the BOP1pro:AS2 Construct.
Supplemental Table 2. AS2pro:GUS Expression in Wild-Type, stm, and stm bop1 bop2 Embryos.
Supplemental Table 3. Oligonucleotides Used in This Study.
Supplemental Methods. Protein Detection, Construction of Transgenic Plants, Expression Analysis, and Histological Analysis.
Supplemental References.
Supplementary Material
Acknowledgments
We acknowledge the ABRC for supplying seeds and thank Sarah Hake, Hirokazu Tsukaya, and members of the lab for helpful comments. This work was supported by a USDA Current Research Information System grant to J.C.F.
References
- Abel S., Nguyen M.D., Theologis A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251: 533–549 [DOI] [PubMed] [Google Scholar]
- Belles-Boix E., Hamant O., Witiak S.M., Morin H., Traas J., Pautot V. (2006). KNAT6: An Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 18: 1900–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M.E., Barley R., Curtis M., Arroyo J.M., Dunham M., Hudson A., Martienssen R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971 [DOI] [PubMed] [Google Scholar]
- Byrne M.E., Simorowski J., Martienssen R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129: 1957–1965 [DOI] [PubMed] [Google Scholar]
- Cao H., Bowling S.A., Gordon S., Dong X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Glazebrook J., Clarke J.D., Volko S., Dong X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Carles C.C., Lertpiriyapong K., Reville K., Fletcher J.C. (2004). The ULTRAPETALA1 gene functions early in Arabidopsis development to restrict shoot apical meristem activity, and acts through WUSCHEL to regulate floral meristem determinacy. Genetics 167: 1893–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Lincoln C., Hake S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8: 1277–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Despres C., Chubak C., Rochon A., Clark R., Bethune T., Desveaux D., Fobert P.R. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15: 2181–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres C., DeLong C., Glaze S., Liu E., Fobert P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12: 279–290 [PMC free article] [PubMed] [Google Scholar]
- Dinneny J.R., Yadegari R., Fischer R.L., Yanofsky M.F., Weigel D. (2004). The role of JAGGED in shaping lateral organs. Development 131: 1101–1110 [DOI] [PubMed] [Google Scholar]
- Dockx J., Quaedvlieg N., Keultjes G., Kock P., Weisbeek P., Smeekens S. (1995). The homeobox gene ATK1 of Arabidopsis thaliana is expressed in the shoot apex of the seedling and in flowers and inflorescence stems of mature plants. Plant Mol. Biol. 28: 723–737 [DOI] [PubMed] [Google Scholar]
- Guo M., Thomas J., Collins G., Timmermans M.C. (2008). Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20: 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha C.M., Jun J.H., Nam H.G., Fletcher J.C. (2004). BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 45: 1361–1370 [DOI] [PubMed] [Google Scholar]
- Ha C.M., Jun J.H., Nam H.G., Fletcher J.C. (2007). BLADE-ON-PETIOLE1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. Plant Cell 19: 1809–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha C.M., Kim G.-T., Kim B.C., Jun J.H., Soh M.S., Ueno Y., Machida Y., Tsukaya H., Nam H.G. (2003). The BLADE-ON-PETIOLE1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development 130: 161–172 [DOI] [PubMed] [Google Scholar]
- Hepworth S.R., Zhang Y., McKim S., Li X., Haughn G. (2005). BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17: 1434–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C., Apel K., Quail P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Quail P.H. (2003). Nuclear translocation of the photoreceptor phytochrome B is necessary for its biological function in seedling photomorphogenesis. Plant J. 35: 660–664 [DOI] [PubMed] [Google Scholar]
- Iwakawa H., Iwasaki M., Kojima S., Ueno Y., Soma T., Tanaka H., Semiarti E., Machida Y., Machida C. (2007). Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 51: 173–184 [DOI] [PubMed] [Google Scholar]
- Iwakawa H., Ueno Y., Semiarti E., Onouchi H., Kojima S., Tsukaya H., Hasebe M., Soma T., Ikezaki M., Machida C., Machida Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43: 467–478 [DOI] [PubMed] [Google Scholar]
- Jackson D. (1992). In situ hybridization in plants. In Molecular Plant Pathology: A Practical Approach, Bowles D.J., Gurr S.J., McPherson R., (Oxford, UK: Oxford University Press; ), 163–174 [Google Scholar]
- Jackson D., Veit B., Hake S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413 [Google Scholar]
- Lawrence R.J., Earley K., Pontes O., Silva M., Chen Z.J., Neves N., Viegas W., Pikaard C.S. (2004). A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 13: 599–609 [DOI] [PubMed] [Google Scholar]
- Li H., Xu L., Wang H., Yuan Z., Cao X., Yang Z., Zhang D., Huang H. (2005). The putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and microRNA165/166 in Arabidopsis leaf development. Plant Cell 17: 2157–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Shuai B., Springer P.S. (2003). The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and adaxial-abaxial patterning. Plant Cell 15: 2241–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C., Long J., Yamaguchi J., Serikawa K., Hake S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6: 1859–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.A., Barton M.K. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125: 3027–3035 [DOI] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Matsumura Y., Iwakawa H., Machida Y., Machida C. (2009). Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana and functional and molecular comparisons between AS2 and other family members. Plant J. 58: 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z., Fan W., Dong X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944 [DOI] [PubMed] [Google Scholar]
- Nath U., Crawford B.C.W., Carpenter R., Coen E. (2003). Genetic control of surface curvature. Science 299: 1401–1407 [DOI] [PubMed] [Google Scholar]
- Norberg M., Holmlund M., Nilsson O. (2005). The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 132: 2203–2213 [DOI] [PubMed] [Google Scholar]
- Ohno C., Reddy G.V., Heisler M.G.B., Meyerowitz E.M. (2004). The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131: 1111–1122 [DOI] [PubMed] [Google Scholar]
- Ori N., Eshed Y., Chuck G., Bowman J.L., Hake S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127: 5523–5532 [DOI] [PubMed] [Google Scholar]
- Pautot V., Dockx J., Hamant O., Kronenberger J., Grandjean O., Jublot D., Traas J. (2001). KNAT2: Evidence for a link between Knotted-like genes and carpel development. Plant Cell 13: 1719–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps-Durr T.L., Thomas J., Vahab P., Timmermans M.C. (2005). Maize rough sheath2 and its Arabidopsis ortholog ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17: 2886–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R.S., Sussex I.M. (1985). The cellular parameters of leaf development in tobacco: A clonal analysis. Planta 165: 170–184 [DOI] [PubMed] [Google Scholar]
- Rochon A., Boyle P., Wignes T., Fobert P.R., Despres C. (2006). The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal residues. Plant Cell 18: 3670–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiarti E., Ueno Y., Tsukaya H., Iwakawa H., Machida C., Machida Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128: 1771–1783 [DOI] [PubMed] [Google Scholar]
- Shuai B., Reynaga-Pena C.G., Springer P.S. (2002). The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 129: 747–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth L.E., Meyerowitz E.M. (1997). Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y., Spoel S.H., Pajerowska-Mukhtar K., Mou Z., Song J., Wang C., Zuo J., Dong X. (2008). Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B.E., Bartling L., Whipple C., Hall D.H., Sakai H., Schmidt R., Hake S. (2009). Bearded-ear encodes a MADS box transcription factor critical for maize floral development. Plant Cell 21: 2578–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno Y., Ishikawa T., Watanabe K., Terakura S., Iwakawa H., Okada K., Machida C., Machida Y. (2007). Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in Arabidopsis leaves. Plant Cell 19: 445–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venglat S.P., Dumonceaux T., Rozwadowski K., Parnell L., Babic V., Keller W., Martienssen R., Selvaraj G., Datla R. (2002). The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltmeier F., Ehlert A., Mayer C.S., Dietrich K., Wang X., Schütze K., Alonso R., Harter K., Vicente-Carbajosa J., Dröge-Laser W. (2006). Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J. 25: 3133–3343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Lin W., Huang T., Poethig R.S., Springer P., Kerstetter R. (2008). KANADI1 regulates adaxial-abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2. Proc. Natl. Acad. Sci. USA 105: 16392–16397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Xu Y., Dong A., Sun Y., Pi L., Xu Y., Huang H. (2003). Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying adaxial identity. Development 130: 4097–4107 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Fan W., Kinkema M., Li X., Dong X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96: 6523–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.