This study demonstrates that γ -Tubulin Complex Protein 4 plays a crucial role in γ -tubulin–mediated microtubule nucleation and organization during cell division and morphogenesis in Arabidopsis.
Abstract
Microtubule (MT) nucleation and organization depend on the evolutionarily conserved protein γ -tubulin, which forms a complex with GCP2-GCP6 (GCP for γ -Tubulin Complex Protein). To date, it is still unclear how GCP4-GCP6 (the non-core GCPs) may be involved in acentrosomal MT nucleation in plant cells. We found that GCP4 was associated with γ -tubulin in vivo in Arabidopsis thaliana. When GCP4 expression was repressed by an artificial microRNA, transgenic plants exhibited phenotypes of dwarfism and reduced organ size. In mitotic cells, it was observed that the γ -tubulin signal associated with the mitotic spindle, and the phragmoplast was depleted when GCP4 was downregulated. Consequently, MTs failed to converge at unified spindle poles, and the bipolar phragmoplast MT array frequently had discrete bundles with extended minus ends, resulting in failed cytokinesis as reflected by cell wall stubs in leaf epidermal cells. In addition, cortical MTs in swollen guard cells and pavement cells of the leaf epidermis became hyperparallel and bundled, which was likely caused by frequent MT nucleation with shallow angles on the wall of extant MTs. Therefore, our results support the notion that GCP4 is an indispensable component for the function of γ -tubulin in MT nucleation and organization in plant cells.
INTRODUCTION
γ -Tubulin serves as the most important factor for microtubule (MT) nucleation and organization among eukaryotic cells (Wiese and Zheng, 2006). It forms the γ -tubulin ring complex, or γ TuRC, with five proteins. Collectively, the six proteins are called γ -Tubulin Complex Proteins (GCPs), with γ -tubulin itself being GCP1. The γ TuRC specifically binds to MT minus ends, thereby allowing them to resist depolymerization (Job et al., 2003). In fungal and animal cells, components of the γ TuRC preferentially appear at the MT organizing centers (MTOCs), the spindle pole body, and the centrosome (Raynaud-Messina and Merdes, 2007). Studies in animal systems show that inhibition or depletion of γ -tubulin and other γ TuRC components inhibits centrosome-based MT nucleation, completely abolishing regular MT organization patterns in both interphase and mitotic cells (Raynaud-Messina and Merdes, 2007).
In acentrosomal plant cells, γ -tubulin was initially discovered to have a punctate distribution pattern on all MT arrays during mitosis (Liu et al., 1993). In the mitotic spindle and the phragmoplast, γ -tubulin appears along MTs but preferentially decorates MT minus ends toward spindle poles and the edges of phragmoplast MTs facing daughter nuclei (Liu et al., 1993, 1995). The localization of γ -tubulin along MTs has been verified in spindles and midbodies in animal cells (Lajoie-Mazenc et al., 1994; Lüders and Stearns, 2007). γ -Tubulin also appears at MT nucleation sites in nondividing plant cells. For example, in the stomatal guard cells, γ -tubulin is concentrated at the cortical site interior to the ventral cell wall (McDonald et al., 1993). In early land plants, however, γ -tubulin is highly concentrated in MTOCs like the plastid surface and polar organizers at spindle poles (Shimamura et al., 2004; Brown and Lemmon, 2006, 2007). The difference between γ -tubulin distribution in early and advanced land plants may reflect the evolution of mechanisms underlying MT organization.
There are two paralogous γ -tubulin genes in the genome of Arabidopsis thaliana (Liu et al., 1994). Downregulation or knockout of both genes causes failed organization of cortical MTs as well as spindle and phragmoplast MT arrays (Binarova et al., 2006; Pastuglia et al., 2006). Thus, plant γ -tubulin, as its fungal and animal counterpart, also plays an essential role in MT organization at various stages of cell growth.
The GCPs are found from fungi to mammals (Murphy et al., 1998; Wiese and Zheng, 2006). In fact, GCPs other than γ -tubulin are structurally related to each other (Gunawardane et al., 2000; Murphy et al., 2001). They all contain two conserved domains, namely the Spindle Pole body Component1 (SPC1)/ γ -tubulin Ring Protein1 (grip1) and SPC2/grip2 motifs (Gunawardane et al., 2000). It is believed that the presence of these two motifs is essential for their assembly into γ TuRC. γ -Tubulin, GCP2/Spc97p, and GCP3/Spc98p form the core of the γ TuRC, which is often referred to as the γ -Tubulin Small Complex ( γ TuSC; Wiese and Zheng, 2006). The budding yeast Saccharomyces cerevisiae contains only γ TuSC (Vinh et al., 2002). The fission yeast Schizosaccharomyces pombe, however, contains components corresponding to all animal GCPs (Fujita et al., 2002; Venkatram et al., 2004; Zimmerman and Chang, 2005; Anders et al., 2006). The non-core components of the γ TuRC play essential roles in γ -tubulin–linked activities in animal cells (Gunawardane et al., 2000; Murphy et al., 2001). However, in fission and in the filamentous fungus Aspergillus nidulans, in which γ -tubulin was initially discovered, the counterparts of GCP4, 5, and 6 are not essential for organizing relatively simple MT arrays (Venkatram et al., 2004; Anders et al., 2006; Xiong and Oakley, 2009). Interestingly, when the function of γ -tubulin is compromised to a certain degree, these nonessential GCPs become indispensable (Fujita et al., 2002). Fission yeast cells lacking the nonessential GCP homologs exhibited reduced MT nucleation activity (Samejima et al., 2005; Sawin and Tran, 2006). Thus, these results suggest that the intact γ TuRC may be required for more complex activities of MT nucleation and organization found in metazoans or that it may support robust MT dynamics. This notion was supported by delocalization of the γ -tubulin complex from the cytoplasmic MTOC (other than the spindle pole body) in fission yeast by knocking out genes encoding the GCP5 homolog MT organizer 1 (Mto1p) or Mto2p, a γ -tubulin complex-interacting protein (Venkatram et al., 2004; Zimmerman and Chang, 2005). However, these results leave us questioning whether GCP4, 5, and 6 mediate the interaction between γ TuSC and other cellular factors. Indeed, it was recently found that GCP5 directly interacts with glycogen synthase kinase-3 β, and the interaction is required for keeping the γ TuRC level at the spindle pole balanced (Izumi et al., 2008).
Land plants contain homologs of all GCP proteins (Fava et al., 1999; Murata et al., 2007). In Arabidopsis, the core γ TuRC components GCP2 and GCP3 decorate the nuclear envelope when ectopically expressed in tobacco (Nicotiana tabacum) BY-2 suspension cells (Seltzer et al., 2007). They are required for nuclear envelope–based MT nucleation and consequently for cell division (Erhardt et al., 2002; Nakamura and Hashimoto, 2009). Biochemical evidence suggests that Arabidopsis GCP2 and GCP3 form a soluble complex with γ -tubulin in the cytoplasm (Seltzer et al., 2007). However, it remains unclear whether such a complex contains other plant GCP components. A functional γ TuRC has yet to be demonstrated biochemically in plant cells.
The plant γ -tubulin complex can initiate MT nucleation at ∼ 40 ° after binding to the wall of extant MTs (Murata et al., 2005). When newly polymerized MTs meet other MTs, shallow angle encounters would promote MT stabilization and coalignment (Dixit and Cyr, 2004). Hence, the nucleation angle would directly influence the fate of the new MT. For example, the spiral3 (spr3) mutation at the GCP2 locus affects the nucleation angle and induces the formation of left-handed MT helices and right-handed helical growth without altering MT dynamics or nucleation efficiency (Nakamura and Hashimoto, 2009). However, it is unclear how the spr3 mutation may have affected the organization of the γ -tubulin complex. Moreover, we lack knowledge about mechanisms underlying the interaction between the complex and the wall of MTs. Besides spr3, mutations in genes encoding α - and β -tubulins or MT-interacting factors often affect the dynamic behavior of cortical MTs and consequently cause defects in cell morphogenesis (Buschmann and Lloyd, 2008). To understand the relationship between the γ -tubulin complex and MT organization, we wished to determine how MT arrays would respond to altered activity of the γ -tubulin complex in interphase and dividing cells.
The discrepancy of the requirement of non-core GCP components in MT nucleation in animal and fungal cells has prompted us to test the functions of these GCPs. We initiated our analysis in Arabidopsis by focusing on GCP4. Earlier efforts failed to isolate an inheritable mutation at the genetic locus encoding GCP4 in Arabidopsis. We took an alternative approach of designing an artificial/synthetic microRNA construct (amiR-GCP4) specifically targeted at GCP4 mRNA. Expression of amiR-GCP4 resulted in reduction of the GCP4 mRNA level. The transgenic lines exhibited altered localization of γ -tubulin in the mitotic spindle and the phragmoplast. Consequently, MT arrays were disorganized, and the plants showed drastically retarded growth phenotypes.
RESULTS
Arabidopsis GCP4 Is an Integral Component of the γ -Tubulin Complex
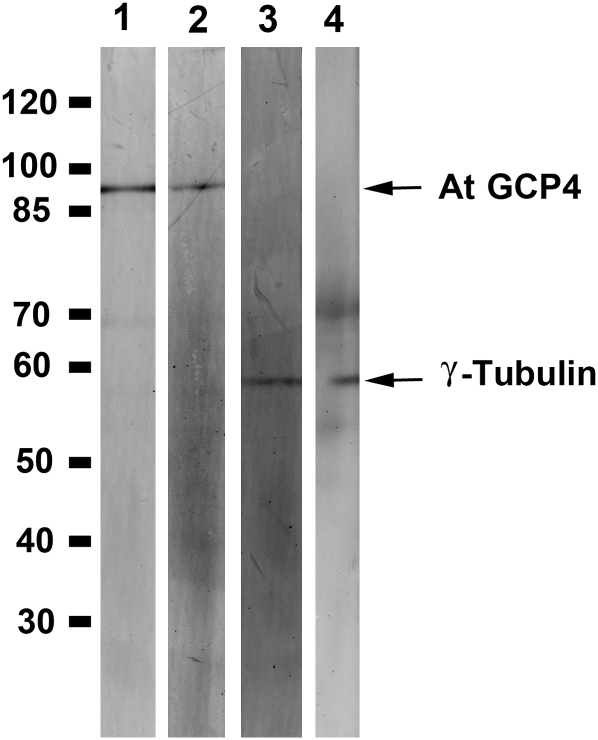
Although At GCP4 shows ∼ 35% sequence identity with its animal counterparts, it has not been determined whether it forms part of the γ -tubulin complex. If it is, we would expect that γ -tubulin would be copurified with GCP4 in vivo or vice versa. An Arabidopsis GCP4-FLAG fusion protein was expressed under the control of the native GCP4 promoter. In an immune-purified fraction enriched with GCP4-FLAG, as probed by both anti-GCP4 and anti-FLAG antibodies, γ -tubulin was also detected using two different antibodies (Figure 1). Therefore, we conclude that At GCP4 and γ -tubulin are associated with each other in vivo.
Figure 1.
Interaction between At GCP4 and γ -Tubulin in Vivo.
Proteins affinity purified from transgenic Arabidopsis lines expressing GCP4-FLAG were probed with anti-AtGCP4 (lane 1) and anti-FLAG (lane 2) to detect isolated GCP4 and with the antibodies GTA (lane 3) and R-70 (lane 4) to detect γ -tubulin. The molecular mass markers (in kD) are shown at the left.
Downregulation of Arabidopsis GCP4 Expression by Artificial MicroRNA Causes Growth Retardation
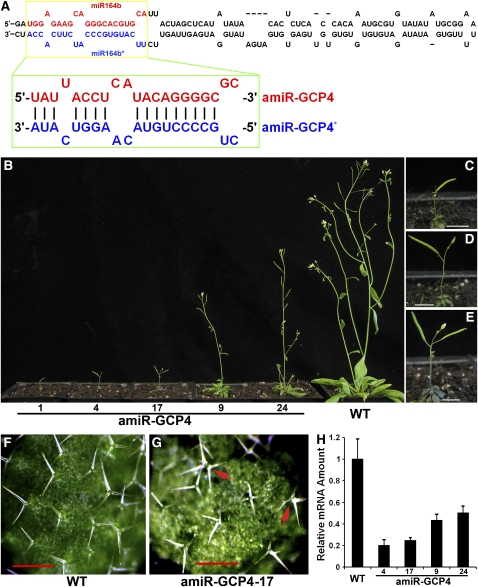
To downregulate GCP4 expression, we designed an artificial microRNA using the backbone of the miR164b gene and replaced the 21-nucleotide miR164b target sequence with a 21-nucleotide region unique to the At GCP4 cDNA sequence that was designed according to the criteria described in previous reports (see Methods) (Alvarez et al., 2006; Schwab et al., 2006). The resulting artificial microRNA (amiR) amiR-GCP4:amiR-GCP4* duplex was expected to silence endogenous At GCP4 expression (Figure 2A). Stable transgenic lines were selected according to their growth phenotypes and brought to the homozygous state (Figure 2B). Consistent phenotypes were observed in four generations. Among the progeny of the transgenic plants exhibiting the most severe growth defects, ∼ 76% of them (n = 97) were very sick and sterile and gradually died. The remaining 24% produced only one or two siliques with a few seeds, as shown for line 1 (Figures 2B and 2C). Most offspring produced by lines #4 (63%, n = 99) and #17 (57%, n = 107) also gradually died, and the surviving plants produced two to five siliques (Figures 2B, 2D, and 2E). Severely dwarfed and small plants derived from these lines produced rosette leaves with smaller sizes than those of the wild type. Other lines, like #9 and #24, also exhibited similar growth retardation phenotypes, but to a lesser extent than observed in the stronger lines of # 1, #4, and #17 (Figure 2B). When monitored in a 6-week period, inhibited growth was exhibited by the mutant plants as early as 2 weeks after seed germination, and the difference compared with the wild-type control became greatly enhanced at later times (see Supplemental Figure 1 online).
Figure 2.
Knocking Down the Expression of Arabidopsis GCP4 by amiR-GCP4.
(A) Predicted folding of miR164b (top) and replacement of the miRNA164b and miRNA164b* sequences with the amiR-GCP4 and amiR-GCP4* sequences, respectively (bottom).
(B) Growth phenotypes of 7-week-old plants of amiR-GCP4 lines (#1, #4, #17, #9, and #24) compared with the wild-type control.
(C) to (E) Close-up views of amiR-GCP4 lines 1, 4, and 17, respectively.
(F) In wild-type leaves, trichomes predominantly bear three branches.
(G) In amiR-GCP4 leaves, trichomes often bear four or five branches (arrows).
(H) Assessment of the expression levels of At GCP4 in the representative amiR-GCP4 lines (#4, 0.20 ± 0.057; #17, 0.24 ± 0.028; #9, 0.43 ± 0.056; and #24, 0.50 ± 0.065) compared with the wild-type control (1.00 ± 0.19) by quantitative RT-PCR.
Bars = 0.5 cm in (C) to (E) and 500 μ m (F) and (G).
These amiR-GCP4 transgenic plants also formed leaf trichomes with increased branches compared with the wild-type control (Figures 2F and 2G). Wild-type leaves formed trichomes with predominantly three branches (Table 1). We found that 29 to 63% of the trichomes of different amiR-GCP4 lines had four branches (Table 1). Because trichome branching is regulated by MTs (Ishida et al., 2008), this phenotype suggested that the abnormal branching phenotype was probably caused by malfunctions of MTs in mutant trichomes.
Table 1.
Trichome Phenotype of amiR-GCP4 Linesa
| Number of Trichome Branches |
||||||
| 1 | 2 | 3 | 4 | 5 | Totalb | |
| Wild type (Columbia-0) | 0 | 0.49% | 88.94% | 11.00% | 0 | 409 |
| amiR-GCP4-4 | 0 | 1.68% | 49.72% | 48.60% | 0 | 179 |
| amiR-GCP4-17 | 0.67% | 2.01% | 30.20% | 63.76% | 3.36% | 149 |
| amiR-GCP4-9 | 0 | 0.89% | 55.02% | 43.79% | 0.30% | 338 |
| amiR-GCP4-24 | 0.46% | 2.06% | 68.42% | 29.06% | 0 | 437 |
The sixth rosette leaves of 3-week-old plants were used for trichome phenotype analysis.
Total number of trichomes examined.
We examined whether there was a correlation between the severity of the growth defects and the degree of downregulation of GCP4 expression. Quantitative RT-PCR data showed that the severe amiR-GCP4 line, #4, had ∼ 20% of the GCP4 expression level found in the wild-type control, and the least severe line, #24, had ∼ 50% (Figure 2H). Hence, we concluded that the lower the GCP4 expression level, the greater the growth defects exhibited by the transgenic seedlings.
To further confirm that the growth phenotypes we observed were due to the downregulation of At GCP4, an amiR-GCP4–resistant version of GCP4 (mGCP4) containing eight silent mutations (see Methods) compared with the amiR-GCP4* strand was transformed into lines #9 and #24. This mGCP4 construct was designed to disrupt amiR-GCP4–mediated regulation without altering the amino acid sequence of GCP4. mGCP4 expression restored growth to a level comparable to the wild-type control (see Supplemental Figure 2 online). Therefore, the phenotypes observed in the amiR-GCP4 mutants were the result of downregulation of GCP4 expression by amiR-GCP4.
Aborted Cytokinesis and Cell Enlargement Caused by amiR-GCP4
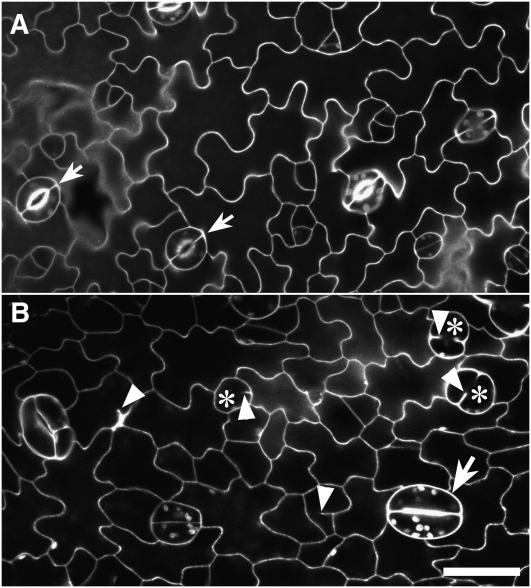
Further still, we questioned the cause of the amiR-GCP4 plants bearing leaves that were significantly smaller than those of the wild type. The morphology of epidermal cells was examined using the propidium iodine staining, which outlined cells after diffusion into the cell wall. In the wild type, the epidermis had paired kidney-shaped guard cells of consistent sizes, which were scattered among the lobed pavement cells (arrows, Figure 3A). In leaves of amiR-GCP4 lines, however, guard cells exhibited irregular forms (Figure 3B). A few pairs of guard cells exhibited kidney shapes (arrow), but they were considerably larger than their counterparts in wild-type leaves. Guard mother cells frequently failed to develop into functional guard cells, and they contained autofluorescent chloroplasts (asterisks, Figure 3B). Single spherical cells were frequently observed instead of paired guard cells. When shown by serial optical sections, these defective spherical cells were found to have cell wall invaginations on one side of the cells and abnormal accumulation of cell wall materials (see Supplemental ). Guard cells of the wild-type control exhibited typical cell wall thickening on the ventral sides, which is required for stomatal pore opening and closure (see Supplemental ). Defects were also observed in pavement cells of the amiR-GCP4 plants. The pavement cells and aborted guard mother cells often had cell wall stubs, likely resulting from incomplete cell plates caused by failed cytokinesis (arrowheads, Figure 3B). Thus, the mutant leaf epidermal cells showed defects in both cell division and anisotropic cell enlargement.
Figure 3.
Outlines of Leaf Epidermal Cells in Wild-Type and amiR-GCP4 Plants.
(A) Propidium iodide staining highlights the outlines of lobed pavement cells and pairs of guard cells (arrow) in a wild-type leaf.
(B) In the leaf of amiR-GCP4 plants, some differentiated guard cells are abnormally swollen (arrow). Frequently, no paired guard cells were formed between pavement cells due to failed cell division (asterisks). Cell wall stubs were found in both pavement cells and aborted guard cells (arrowheads).
Bar = 20 μ m.
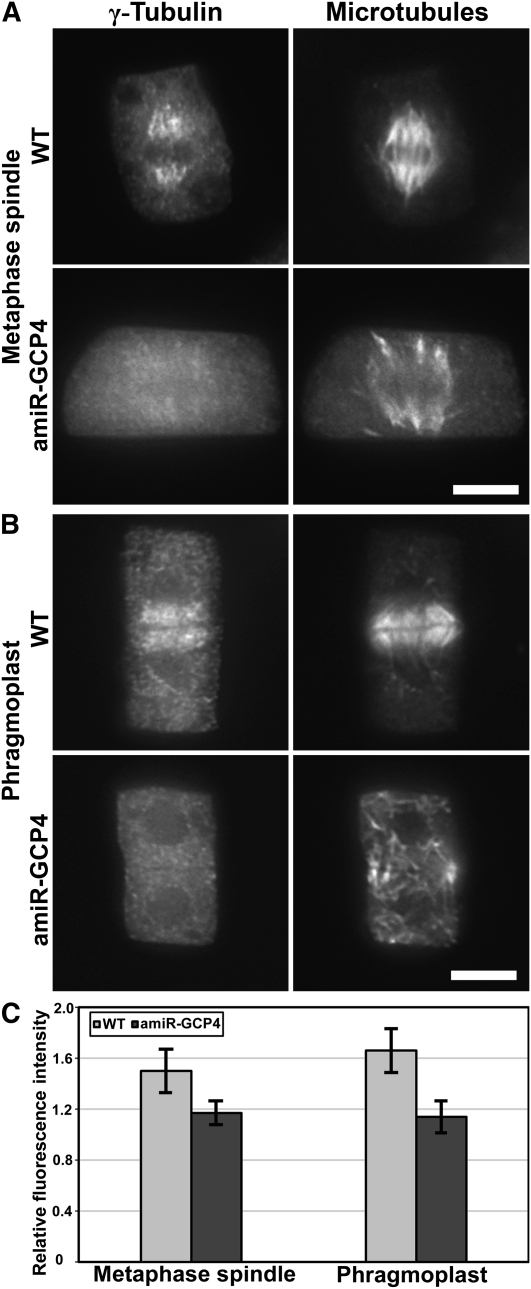
Reduction of γ -Tubulin Association with Spindles and the Phragmoplast in amiR-GCP4 Cells
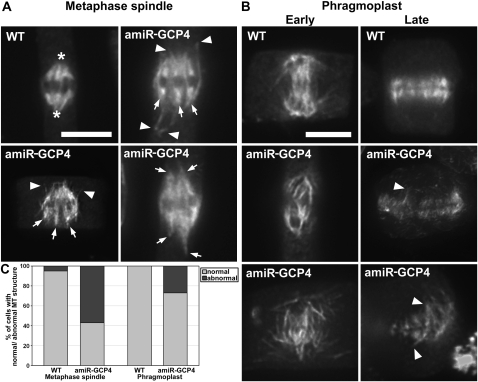
We questioned whether the cytokinetic defect observed in mutant cells was caused by disorganized MT arrays in dividing cells. Although lacking the centrosome, wild-type spindle MTs converged toward spindle poles in root meristematic cells in Arabidopsis (asterisks, Figure 4A). In the amiR-GCP4 cells, metaphase spindles exhibited at least three abnormal appearances (Figure 4A). First, MTs of the kinetochore fibers often failed to converge toward common poles (arrows). Second, these MTs often extended to reach discrete sites. Third, there were nonkinetochore MT bundles initiated from regions near the mitotic spindles (arrowheads, Figure 4A). Quantitative assessment of the spindles of the wild-type control and the mutant revealed that 57% (n = 54) of the amiR-GCP4 spindles were abnormal, compared with 5% (n = 20) of the control spindles (Figure 4C).
Figure 4.
Abnormal MT Organization in the Mitotic Spindle and the Phragmoplast of amiR-GCP4 Cells.
(A) Wild-type spindles have MTs converging toward spindle poles (asterisks). In the mutant spindles, MTs often converge in several discrete sites (arrows) and have prominent nonspindle MTs (arrowheads).
(B) MTs in wild-type phragmoplasts are organized in a bipolar fashion. In amiR-GCP4 cells, some early phragmoplasts have disorganized MTs that extend beyond the region normally occupied by the phragmoplast. In mature phragmoplasts of wild-type cells, MTs have the most prominent signals toward the periphery. In amiR-GCP4 cells, MTs often appear in discrete, thin bundles or extend beyond normal positions (arrowheads).
(C) Quantitative assessment of abnormal spindles and phragmoplasts in amiR-GCP4 plants compared with the control ones.
Bars = 5 μ m.
Abnormal MT organization was also observed in the mutant phragmoplasts. Upon the completion of anaphase, interzonal MTs coalesced and were later rearranged into a bipolar array with the dark line lacking the antitubulin fluorescence appearing in the phragmoplast midline (Figure 4B). In the mutant cells, MT bundles from the two sides often failed to engage with each other, although the antiparallel pattern was still established. In the mature phragmoplast of the control cells, MTs form striking bundles and appeared more prominent at the phragmoplast periphery than at its center (Figure 4B). In the amiR-GCP4 cells, however, parallel MT bundles appeared across the phragmoplast, and MTs at the distal edges often reached out to different sites showing irregular lengths (arrowheads, Figure 4B). Later, the mutant phragmoplast MTs failed to exhibit coalesced bundles as seen in the control. Nearly 27% (n = 117) of the mutant phragmoplasts exhibited abnormal MT organization patterns, which were not (n = 45) observed in the wild-type control cells (Figure 4C).
We investigated whether the defects in MT organization in the mutant cells were due to altered activities of the γ -tubulin complex. Using the G9 antibody (Horio et al., 1999), γ -tubulin was detected in the spindle and phragmoplast of control cells, exhibiting a preference for MT minus ends toward spindle poles and phragmoplast edges (WT in Figures 5A and 5B). This striking pattern of γ -tubulin distribution was replaced by a faint and diffuse signal in the spindle and phragmoplast in the amiR-GCP4 mutant cells (amiR-GCP4 in Figures 5A and 5B). We measured the anti- γ -tubulin fluorescence intensity and found that, in wild-type cells, the ratio of the signal on metaphase spindle MTs to the diffuse cytoplasmic signal was 1.5 ± 0.17 (n = 35) (Figure 5C). At identical stages, the mutant cells had an average ratio of 1.17 ± 0.09 (n = 43). Similarly, in the phragmoplast, wild-type cells had a phragmoplast/cytoplasm fluorescence intensity ratio of 1.66 ± 0.17 (n = 32), but the mutant cells had a ratio of only 1.2 ± 0.12 (n = 47) (Figure 5C). The results suggested that a significant portion of γ -tubulin was no longer associated with spindle and phragmoplast MTs in the mutant cells. Instead, it might have become soluble in the cytoplasm.
Figure 5.
Reduction of the γ -Tubulin Signal on Spindle and Phragmoplast MTs in amiR-GCP4 Cells.
(A) In wild-type cells, γ -tubulin appears prominently on spindle MTs. Greatly reduced signals are associated with abnormal amiR-GCP4 spindle MTs.
(B) The wild-type phragmoplast also contains a prominent γ -tubulin signal. In amiR-GCP4 mutant cells, the γ -tubulin signal is greatly diminished in phragmoplast MTs.
(C) Quantitative assessment of relative signal intensity in the control cells and amiR-GCP4 cells.
Bars = 5 μ m.
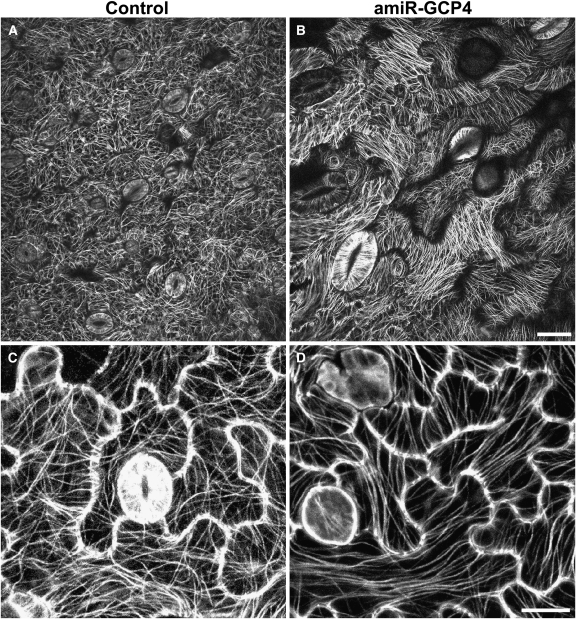
Cortical MT Organization Depends on GCP4 Activities
Prompted by the abnormal cell morphology exhibited by the amiR-GCP4 lines, we set out to examine whether the phenotype was caused by abnormal behavior of cortical MTs. Using a green fluorescent protein (GFP)-TUB6 fusion that does not cause any noticeable difference in MT organization or seedling growth (Nakamura et al., 2004), MT organization was observed in live cells by confocal microscopy. When amiR-GCP4 was expressed in plants bearing the GFP-TUB6 fusion, identical phenotypes were observed as when it was expressed in the absence of the fusion. In the control GFP-TUB6 plants, pavement cells of the epidermis exhibited a fine and complex MT network with nonuniform orientations (Figure 6A). Locally, some cortical MTs were aligned in parallel with each other, but beyond the localized area other MTs assumed new orientations (Figure 6C). Such a phenomenon was largely absent in the amiR-GCP4 cells. Cortical MTs became highly ordered, while the MTs of some pavement cells were oriented in only one parallel direction (Figure 6B). Long parallel MTs were also observed along the long axis of other pavement cells (Figure 6D).
Figure 6.
MT Organization in Epidermal Cells of the Wild-Type Control and amiR-GCP4 Leaves Expressing GFP-TUB6.
(A) In the control leaf epidermal pavement cells, cortical MTs exhibit a complex fine network.
(B) In the amiR-GCP4 mutant, cortical MTs exhibit highly ordered parallel bundles in most pavement cells and enlarged guard cells.
(C) In a control leaf, all pavement cells exhibit fine cortical MTs with nonuniform orientations within individual cells.
(D) In an amiR-GCP4 mutant leaf, highly bundled MTs exhibit parallel configurations.
Bars = 20 μ m in (A) and (B) and 10 μ m in (C) and (D).
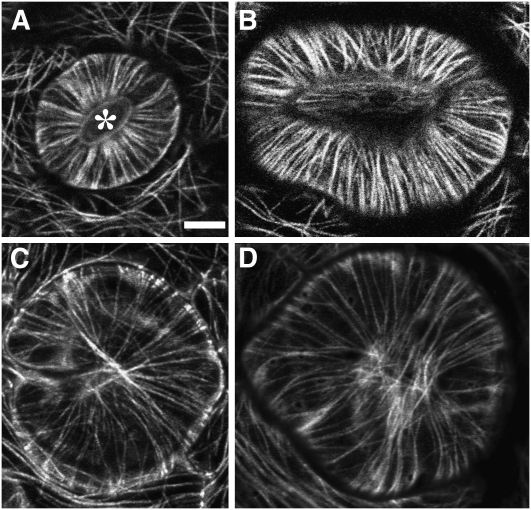
We noticed that the guard cells of the amiR-GCP4 mutants exhibited phenotypes of abnormal swelling and wondered whether the morphological changes were associated with altered MT behaviors. In control cells, MTs exhibited the characteristic radial array (Figure 7A). These MTs radiated from the cortex next to the ventral wall of the guard cells facing the stomatal pore (asterisk, Figure 7A). This result is consistent with the centrally located MT-organizing zone in this region (Marc et al., 1989). In highly enlarged guard cells of the mutant, MTs did not exhibit a radiating pattern. Instead, they often formed nearly parallel arrays that were composed of prominent bundles (Figure 7B). A great proportion of guard mother cells failed to complete cell division, leaving behind round- or horseshoe-shaped swollen cells (Figures 7C and 7D). In these cells, cortical MTs encircled the entire cells without any centralized nucleating site. The abnormal MT organization pattern exhibited by these cells could be the result of failed MT nucleation at a defined site like that next to the ventral cell wall following aborted divisions of guard mother cells.
Figure 7.
Abnormal MT Organization in Swollen Guard Cells.
(A) In the control guard cells, radial MTs decorated by GFP-TUB6 are nucleated from the central region facing the ventral cell wall surrounding the stomatal pore (asterisk).
(B) to (D) In the amiR-GCP4 line expressing GFP-TUB6, abnormally swollen guard cells contain highly bundled parallel cortical MTs (B). Other cells derived from failed division of the guard mother cell. Cortical MTs are aligned uniformly in all directions, resulting in enlarged spherical cells ([C] and [D]).
Bar = 10 μ m.
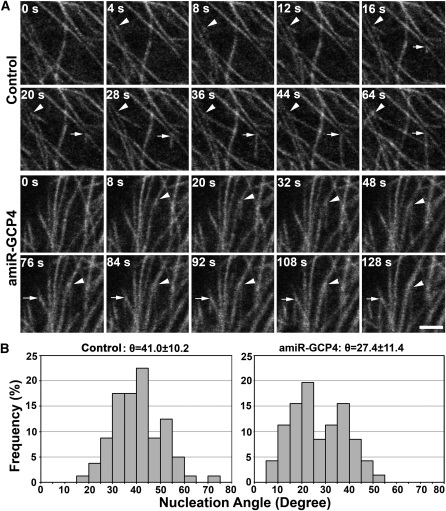
MT Nucleation at Shallow Angles on Extant MTs in the Mutant Cells
We examined whether the abnormal MT organization was attributed to altered nucleation patterns in the mutant cells. At the cell cortex, newly polymerized MTs on the wall of extant MTs were monitored in pavement cells of the control and amiR-GCP4 plants expressing GFP-TUB6 in time-lapsed movies (see Supplemental Movies 1 and 2 online). Published data show that such new MTs would assume orientations with an average angle of 40 ° relative to the old ones (Murata et al., 2005). Frequent MT nucleation events were detected on extant MT walls in both the control and mutant cells (Figure 8A). In the two events shown in the control cell in Figure 8A, the branch angles were 39.7 ° and 41.6 ° (see Supplemental Movie 1 online). The two events shown in the amiR-GCP4 cell, however, had branch angles of 14.0 ° and 30.6 ° (Figure 8A; see Supplemental Movie 2 online). In the control cells, the branch angles had an average of 41.0 ° ± 10.2 ° (n = 80) (Figure 8B). We never detected a branch angle smaller than 15 °. In amiR-GCP4 cells, the average branch angle was 27.4 ° ± 11.4 ° (n = 71) (Figure 8B). Over 15% of the angles were smaller than 15 °. Therefore, downregulation of GCP4 expression caused a significant reduction in branch angles of MTs nucleated on the extant MT wall.
Figure 8.
MT Nucleation on Extant MTs in Control and amiR-GCP4 Cells.
(A) Two representative nucleation events are shown in both control cells and amiR-GCP4 cells expressing GFP-TUB6. In the control panels, two nucleation sites are indicated by an arrowhead and an arrow with respective MT-branching angles of 39.7 ° and 41.6 °. In the amiR-GCP4 panels, the two MT-branching angles are 14.0 ° (arrowhead) and 30.6 ° (arrow). Bar = 2.5 μ m.
(B) Quantitative analysis of MT branching angles in both the control (n = 80) and amiR-GCP4 cells expressing GFP-TUB6 (n = 71). Statistically significant difference was shown between amiR-GCP4 cells and the control cells (Student's t test; P < 0.01).
DISCUSSION
Our results show that GCP4 is indispensable for γ -tubulin function in MT nucleation and organization during cell division and anisotropic cell expansion in Arabidopsis. Reduction of GCP4 activity altered normal activities of γ -tubulin in all MT arrays in dividing cells and rapidly growing cells. Therefore, GCP4 plays a critical role in the interaction between γ -tubulin and MTs and consequently in organizing complex MT arrays.
Viable Mutants with Compromised Functions of the γ -Tubulin Complex
In plant cells, the essential function of γ -tubulin in MT nucleation and organization depends on GCP2 and the γ TuRC-interacting protein NEDD1 (for neural precursor cell-expressed developmentally downregulated1) (Nakamura and Hashimoto, 2009; Zeng et al., 2009). Null mutations in the GCP2 and NEDD1 genes must be maintained in heterozygous states, which prevents us from studying their functions in vegetative cells (Nakamura and Hashimoto, 2009; Zeng et al., 2009). Earlier studies using the RNA interference approach have downregulated the expression of γ -tubulin in Arabidopsis seedlings and caused devastating cell division phenotypes (Binarova et al., 2006).
We successfully generated mutants in which GCP4 expression was suppressed at different levels. Although a null mutation at the GCP4 locus would likely cause a lethal phenotype, as does that of γ -tubulin or GCP2, our mutant plants exhibited stable growth phenotypes when the amiR-GCP4 transgene was brought to the homozygous state. The phenotypes exhibited by these mutants were truly due to the reduced function of GCP4 as the amiR-GCP4 resistant form, mGCP4, was able to complement the loss of endogenous GCP4 activity. This approach also has the advantage over the traditional RNA interference approach because the target of amiR-GCP4 is a very small region. More importantly, we generated a series of mutants with different levels of GCP4 expression, which corresponded to the severity of growth phenotypes of the whole plants. These mutants would be very useful for us to further assess the requirement of GCP4 for γ -tubulin function. Besides being used for analyzing the growth phenotypes caused by amiR-GCP4, the mutants also allowed us to conclude the requirement of GCP4 for γ -tubulin association with MT arrays, such as spindles and phragmoplasts. Because in the past such stable mutations in genes encoding γ -tubulin or its interacting factors could not be kept in a homozygous fertile plant, the use of these mutants was limited. The amiR-GCP4 mutants reported here can be used for testing many speculations and hypotheses regarding the function of the γ -tubulin complex.
Redistribution of Active γ -Tubulin and MT Organization in the Spindle and Phragmoplast
Our results indicated that γ -tubulin accumulated on the nuclear envelope and capped the poles of prophase prespindles in the mutant cells, as observed in control cells. The disturbance of γ -tubulin localization in spindle and phragmoplast MTs suggests that the γ -tubulin complex may function in generating new MTs in the mitotic spindle and the phragmoplast in dividing cells. Unfortunately, the density of MTs and the resolution limit of fluorescence microscopy prevent us from resolving individual MTs in these arrays. Although it has been suggested that mechanisms underlying MT organization in mitotic cells can be different from those in interphase cells (Wasteneys, 2002), it remains to be tested whether new MTs can branch off from extant MTs in the spindle and phragmoplast.
In higher plant cells, γ -tubulin is not concentrated at sites like the centrosome in animal cells. Instead, its signal appears as a gradient with more prominent signal toward the spindle poles and the edges of the phragmoplast (Liu et al., 1993, 1994, 1995). Such a distribution pattern was later confirmed in animal cells (Lajoie-Mazenc et al., 1994; Lüders and Stearns, 2007). Recently, it has been found that γ -tubulin tightly interacts with the centrosome to nucleate MTs, while it binds to spindle MTs weakly and transiently (Hallen et al., 2008). This conclusion would support the notion that MT-dependent MT nucleation may require intact γ TuRC.
It has been proposed that the assembly of the γ TuSC precedes that of the γ TuRC (Jeng and Stearns, 1999). Therefore, the assembly of the γ TuSC with other GCP components to form the γ TuRC could be required for the interaction with MTs. Similarly to what has been suggested in the case of animal spindles, the γ -tubulin complex may bind weakly to both spindle and phragmoplast MTs, and this binding requires noncore GCPs. When such an association is downregulated, less MT nucleation takes place on existing MTs. Consequently, interactions among MT bundles would not happen robustly, and the bundles would become discrete. This would explain why the mutant spindle often contained MT bundles that were not fully converged, and the mutant phragmoplasts had discrete, thin MT bundles. It appears that cytokinesis can be seriously harmed when this takes place. Surprisingly, mitosis did not seem to be affected to a noticeable level, but this is consistent with our earlier findings showing that the nedd1 mutation causes a serious defect in cytokinesis but not in sister chromatid segregation (Zeng et al., 2009).
Published studies indicate that the minus-end–directed class 14 kinesin KATA (for kinesin-like protein in Arabidopsis thaliana A)/ATK1 (for Arabidopsis thaliana Kinesin 1) plays a critical role in organizing converged spindle poles, as the atk1-1 mutation renders wide spindle poles (Chen et al., 2002; Ambrose et al., 2005). However, KATA/ATK1 acts at the MT plus end and is thus unlikely to interact with the γ -tubulin complex directly (Ambrose et al., 2005). Interestingly, in fission yeast, a similar kinesin motor acts synergistically with γ -tubulin to regulate MT organization during mitosis (Paluh et al., 2000). Whether a similar relationship between γ -tubulin and KATA/ATK1 exists in plant cells awaits further experiments.
GCP4-Dependent Function of the γ -Tubulin Complex in Organizing Cortical MTs
The presence of GCP4 in the γ -tubulin complex in Arabidopsis supports the possibility that the γ -tubulin complex adopts a ring formation, as reported in its animal counterparts (Zheng et al., 1995; Murphy et al., 2001). In plant cells, cytoplasmic γ -tubulin has been detected in two protein complexes of 750 and 1500 kD, respectively (Stoppin-Mellet et al., 2000). To date, the molecular composition of these two complexes is unclear. It would be interesting to test the functionality of the two complexes in MT nucleation. In most fungi, the small complex is sufficient for basic MT nucleation function, although they have a complete set of GCP proteins as in animals (Fujita et al., 2002; Xiong and Oakley, 2009). In animal cells, GCP4 plays a critical role in astral MT nucleation (Fava et al., 1999). In a separate report, it has been found that γ TuSC can use an alternative pathway apart from assembling the γ TuRC to target γ -tubulin at the centrosome for MT nucleation (Vérollet et al., 2006). It has been suggested that the γ TuRC is only essential for noncentrosomal localization of γ -tubulin (Vérollet et al., 2006). Because of the absence of a structurally defined MTOC in plant cells, our findings further support the notion that GCP4-dependent assembly of a complex like γ TuRC is essential for centrosome-independent function of γ -tubulin in MT nucleation.
The appearance of highly parallel MT bundles in the amiR-GCP4 plants is likely linked to the reduced branch angles during MT nucleation on extant MTs. Such reduced angles may result from the assembly of an incomplete γ -tubulin complex. It has been shown that a putative GCP6 ortholog was able to localize to MTOC in the absence of GCP4 in A. nidulans (Xiong and Oakley, 2009). Therefore, GCP6 might interact with γ -tubulin to form a complex to nucleate MTs. MTs nucleated by such a complex instead of a γ TuRC-like complex would tend to have smaller branch angles from extant MTs.
Our results demonstrate that the formation of the complex cortical MT array during interphase in cells like pavement cells depends on a γ TuRC-like complex. When a polymerizing MT encounters a preexisting one, their encounter angle governs the fate of this polymerizing MT (Dixit and Cyr, 2004). Smaller branch angles detected in the amiR-GCP4 cells would favor shallow-angle encounters. Therefore, the outcome would be bundling of the interacting MTs. Besides recapitulating MT branching, a recent report also showed that a significant portion of newly polymerized MTs follow the axes of extant MTs (Chan et al., 2009). Whether such a parallel MT polymerization event depends on γ TuRC remains to be addressed. However, a combination of MT polymerization along existing ones and shallow-angle MT–MT interaction would probably account for the formation of hyperparallel bundles seen in mutant cells. Here, we hypothesize that, in the absence of a complete γ TuRC due to the lack of GCP4, the balance between MT branching and MT bundling might have been disturbed. Many microtubule-associated proteins like Tobacco Microtubule-Associated Protein 200/Microtubule Organization1 and End Binding protein-1 would favor MT bundle formation by stabilizing MTs (Hamada, 2007; Guo et al., 2009).
It should be noted that the phenotype of cortical MTs exhibited by the amiR-GCP4 mutants differed from that by downregulation or knockout of γ -tubulin itself. When the function of γ -tubulin is compromised, cortical MTs become randomized and eventually disappear (Binarova et al., 2006; Pastuglia et al., 2006). Our observations suggest that the phenotype exhibited by the amiR-GCP4 lines was not caused by a complete loss of function of γ -tubulin. Instead, it was caused by altered activities of γ -tubulin.
Activation of GCP4 for the Formation of a MT Nucleation-Competent γ -Tubulin Complex
Because γ -tubulin was copurified with GCP4 from seedling extracts, indicating that they are in the same complex, it was expected that GCP4 would colocalize with γ -tubulin. However, our polyclonal anti-GCP4 antibodies detected abundant signal in the cytoplasm. Similar results were obtained with the anti-FLAG antibody in cells expressing GCP4-FLAG under the control of the native promoter (see Supplemental Figure 4 online). We suspect that GCP4 is largely present in an inactive form in the cytoplasm. Upon activation via an unknown mechanism, it may form a complex with γ -tubulin that is able to associate with the wall of MTs and to nucleate new MTs.
In summary, our results indicate that the γ -TuRC protein, GCP4, plays an essential role in MT organization in plant cells. When its function is compromised, plant growth is severely retarded due to the collective effect of cell division failures, abnormal cell enlargement, and the loss of functional stomata, as observed in our mutants.
METHODS
Plant Materials, Growth Conditions, and Transformation
Wild-type Arabidopsis thaliana (Columbia-0) plants and transgenic lines harboring 35S:GFP-TUB6 (Nakamura et al., 2004) were used in this study. Plants were grown under a 16-h-light and 8-h-dark cycle with 70% relative humidity at 22 ° C.
The standard floral dipping method was used for plant transformation as described before (Clough and Bent, 1998). The Agrobacterium tumefaciens strain GV3101 was used for transformation. Homozygous T3 seedlings were used for phenotype analysis, confocal microscopy observations, and immunoblotting tests.
Construction of the amiR-GCP4 Plasmid
An artificial miRNA construct was designed according to published protocols (Alvarez et al., 2006; Schwab et al., 2006) (http://wmd2.weigelworld.org/cgi-bin/mirnatools.pl) with modifications. Briefly, the amiR-GCP4 construct had miR164b precursor as the backbone, and its 21-nucleotide miRNA and miRNA* regions were replaced by 21-nucleotide sequences based on the At GCP4 cDNA sequence (Figure 2A). The 282-bp amiR-GCP4 precursor sequence was as follows: 5 ′ -GGTACCGAGAATGATGAAGGTGTGTGATGAGCAAGATATTACCTCATACAGGGGCGCTTACTAGCTCATATATACACTCTCACCACAAATGCGTGTATATATGCGGAATTTTGTGATATAGATGTGTGTGTGTGTTGAGTGTGATGATATGGATGAGTTAGTTCCTGCCCCTGTACAAGGTCATATCATGACCACTCCACCTTGGTGACGATGACGACGAGGGTTCAAGTGTTACGCACGTGGGAATATACTTATATCGATAAACACACACGTGCGGGATCC-3 ′. Note that the amiR-GCP4 and amiR-GCP4* are highlighted in bold and in italics plus bold, respectively, and the cloning sites KpnI at the 5 ′ end and BamHI sites at the 3 ′ end are underlined. This construct was synthesized commercially (GeneScript). After digestion by KpnI and BamHI, this 282-bp amiR-GCP4 precursor was cloned into the pART7 vector at the corresponding sites. The resulting 35S(p):amiR-GCP4 fragment, which was produced by NotI digestion of the pART7-amiR-GCP4 plasmid, was then cloned into the binary vector pML-BART at the NotI site. The resulting plasmid was confirmed by sequencing. This amiR-GCP4 construct was introduced into Arabidopsis (Columbia-0) wild-type plants and the 35S:GFP-TUB6 marker plants. Transgenic T1 plants were selected by spraying Finale (AgrEvo) for Basta resistance.
Construction of Arabidopsis GCP4-FLAG and amiR-GCP4-Resistant GCP4-FLAG Plasmids
The PGCP4:GCP4-FLAG construct contained a 5146-bp fragment, including a 962-bp fragment with noncoding sequence between the start codon of the preceding gene (At3g53770) and the At GCP4 coding region. The entire genomic sequence was amplified by PCR using primers GCP4_5.1K-F, 5 ′ -CACCTAATCCTCTCAAATCTCCTATGAAGAAGAT-3 ′, and GCP4_5.1K-R, 5 ′ -TGCGCCTGCGCCCAATGGAAGCGCTGGGCGTTGTCGGAC-3 ′. The underlined CACC sequence in GCP4_5.1K-F was designed for directional cloning, and the 12-bp underlined sequence in GCP4_5.1K-R was aimed for encoding a Gly-Ala duplicate linker between At GCP4 and the affinity tag FLAG. After amplification with the Phusion High-Fidelity DNA polymerase (New England Biolabs), this genomic fragment was cloned into the Gateway pENTR/D-TOPO vector using the cloning kits provided by the manufacturer (Invitrogen). An LR recombination reaction was performed between the entry clone containing the PGCP4:GCP4 fragment and the destination vector pEarleyGate-302 containing a FLAG tag (Earley et al., 2006). The resulting plasmid was confirmed by sequencing and transformed into wild-type plants.
To make a construct that contains an amiR-GCP4–resistant version of GCP4 (mGCP4), eight silent mutations were introduced into the abovementioned entry clone using the Change-IT multiple mutation site-directed mutagenesis kit (USB). Specifically, the amiR-GCP4 complementary site 5 ′ -CCGCCCCTGTATGAGGTAATT-3 ′ was replaced by 5 ′ -CCtCCgtTaTAcGAaGTcATc-3 ′, with base changes indicated by lowercase letters. This entry clone of PGCP4:mGCP4 was introduced into a binary vector pGWB10 for generating a FLAG fusion (Nakagawa et al., 2007) by the LR reaction as described above. The resulting PGCP4:mGCP4-FLAG construct was confirmed by sequencing and transformed into the weaker amiR-GCP4 lines 9 and 24.
RNA Extraction and Real-Time Quantitative RT-PCR
RNA samples were prepared from young rosette leaves of 4-week-old amiR-GCP4 lines and wild-type control plants. Protocols for RNA preparation and real-time RT-PCR were adopted from a previous study (Kong et al., 2006), with modifications. Briefly, the cDNA samples were diluted to 20 and 5 ng/ μ L. Triplicate quantitative assays were performed with 1 μ L of each cDNA dilution in a 25- μ L reaction containing IQ SYBR Green Supermix using an iCycler IQ5 real-time PCR system (Bio-Rad). The relative quantification method Delta-Delta CT was used to evaluate quantitative variations between replicates examined. Three biological replicates were performed. The amplification of PP2A, encoding protein phosphatase 2A, was used as a reference gene to normalize the data as reported (Czechowski et al., 2005). Gene-specific primers for GCP4 were Q53760-543F, 5 ′ -GCTACGGTTACAGAGGGGCTC-3 ′, and Q53760-745R, 5 ′ -GCTGATTATACATGACCTGATGTCCATTC-3 ′, which span the predicted amiR-GCP4 cleavage site.
Preparation of GCP4 Antibodies
Using the cDNA clone U65252 (ABRC) as the template, an GCP4 cDNA fragment encoding amino acids 238 to 494 was amplified by PCR with primers 53760P-F, 5 ′ -CACCTCTAGATGGTGATTTAGATC-3 ′ (italicized CACC designed for directional cloning; the XbaI site underlined), and 53760P-R, 5 ′ -CCCAAGCTTCCCGGTGAGAGATAC-3 ′ (HindIII site underlined). The fragment was then cloned into gateway pENTR/D-TOPO vector using the pENTR/D-TOPO Kit (Invitrogen). After digestion with XbaI and HindIII, the fragment was cloned into the glutathione S-transferase (GST) vector pGEX-KG (Guan and Dixon, 1991). The GST-GCP4 fusion protein was expressed and purified before being used as the antigen for antibody production as described previously (Lu et al., 2005). Monospecific anti-At GCP4 antibodies were purified using a blot-based purification protocol (Olmsted, 1981).6
Immunopurification and Protein Gel Blotting Analyses
Sterilized Arabidopsis seeds were stratified for 2 d before being germinated at room temperature in liquid medium containing half-strength Murashige and Skoog salts (Invitrogen) by shaking at 100 rpm for 60 h in dark. Approximately 1 g of seedlings was grinded in liquid nitrogen with a mortar and pestle. The powder was mixed with 6 mL of K-HEPES extraction buffer containing 20 mM HEPES, 110 mM KAc, 2 mM MgCl2, 0.1% Tween 20, 0.2% Triton X-100, and EDTA-free protease inhibitor cocktail (Roche) and incubated at 4 ° C for 1 h. After centrifugation at 18,000g for 20 min at 4 ° C, the supernatant was loaded onto an affinity column containing 1 mL M2 anti-FLAG gel (Sigma-Aldrich). The resin was then washed with 10 mL extraction buffer. Proteins bound to the affinity matrix were eluted under native conditions with the elution solution containing 100 ng/ μ L 3 × FLAG peptide (Sigma-Aldrich) in five fractions. Eluted proteins were separated in a 10% SDS-PAGE and processed for immunoblotting analysis as described previously (Lee et al., 2001). The anti-FLAG M2 monoclonal antibody (1:1000) and anti-GCP4 rabbit polyclonal antibody (1:1000) were used to detect At GCP4 protein, and anti- γ -tubulin antibodies GTA (1:1000) (Silflow et al., 1999) and R70 (1:2000) (Julian et al., 1993) were used for γ -tubulin detection.
Fluorescence and Confocal Microscopy
Root tip cells were processed for indirect immunofluorescence staining as described previously (Lee and Liu, 2000). Sheep polyclonal anti-tubulin antibody (Cytoskeleton) and the G9 mouse monoclonal anti- γ -tubulin antibody (Horio et al., 1999) were used for dual localization of MTs and γ -tubulin. Secondary antibodies are as follows: Texas Red–conjugated donkey anti-goat IgG (Rockland Immunochemicals) and fluorescein isothiocyanate–conjugated donkey anti-mouse IgG (Sigma-Aldrich).
Samples were observed with an Eclipse E600 microscope equipped with epifluorescence optics (Nikon), and images were captured by an Orca CCD camera (Hamamatsu Photonics) using the MetaMorph software package (Molecular Devices).
To quantify the γ -tubulin signal intensity in both amiR-GCP4 cells and wild-type cells, intensities of anti- γ -tubulin fluorescent signal were measured in 16 × 16-pixel regions within metaphase spindles and phragmoplasts. The fluorescent signal in the cytoplasm was used as a reference. The relative fluorescence intensity for γ -tubulin was derived from the ratio of the anti- γ -tubulin signal on MT arrays to the diffuse cytoplasmic signal. Student's t test was used for the significance analysis.
The middle regions of the third true leaves were dissected from 2-week-old seedlings for observations under an LSM700 (Carl Zeiss) or an FV1000 (Olympus) confocal laser scanning microscope. To visualize the epidermal cell outlines, freshly dissected leaves were stained in 1 mg/mL propidium iodide for 2 min. Following rinses with 0.3 M mannitol, leaf segments were mounted in the same solution with a cover slip. The abaxial side of the leaves was viewed with a C-Apochromat × 63/numerical aperture 1.20 water objective, and the samples were excited by a 555-nm helium neon laser. Images were captured using the Zen2009 or FlowView software and processed in Adobe Photoshop.
Similar conditions were applied for visualizing MTs in live epidermal cells expressing GFP-TUB6. The 488-nm argon laser was used for GFP excitation. To minimize the background autofluorescence signal, the GFP signal was collected between 500- and 550-nm emission wavelengths.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers At3g53760 (At GCP4) and At1g13320 (phosphatase 2A). The cDNA clone U65252 contains the full-length coding sequence of At GCP4.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Time Course of Growth Phenotype Exhibited by the amiR-GCP4 Lines.
Supplemental Figure 2. Complementation of the amiR-GCP4 Lines 9 and 24 by the PGCP4:mGCP4 Construct.
Supplemental Figure 3. Failed Stomatogenesis in amiR-GCP4 Lines.
Supplemental Figure 4. Localization of GCP4-FLAG in a Cytokinetic Cell.
Supplemental Movie 1. MT Nucleation Behavior in the Control Cell Expressing GFP-TUB6.
Supplemental Movie 2. MT Nucleation Behavior in the amiR-GCP4 Cell Expressing GFP-TUB6.
Supplemental Movie Legends.
Supplementary Material
Acknowledgments
We thank Michel Wright at Institut de Sciences et Technologies du Médicament de Toulouse of Centre National de la Recherche Scientifique in Toulouse, France for providing the R70 anti- γ -tubulin antiserum and Masayoshi Nakamura and Takashi Hashimoto for the GFP-TUB6 line. We thank Geoffrey Lambright at Carl Zeiss and John Jordan of Olympus USA for their expert help on confocal microscopy and Lindsay Kiyama for critical reading the manuscript. This work was supported by the Chemical Sciences, Geosciences, and Biosciences Program Division of the Office of Basic Energy Sciences of the U.S. Department of Energy under Contract DE-FG02-04ER15554 and the National Science Foundation Grant MCB-0920454. T.H. was a Katherine Esau postdoctoral fellow.
References
- Alvarez J.P., Pekker I., Goldshmidt A., Blum E., Amsellem Z., Eshed Y. (2006). Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose J.C., Li W., Marcus A., Ma H., Cyr R. (2005). A minus-end-directed kinesin with plus-end tracking protein activity is involved in spindle morphogenesis. Mol. Biol. Cell 16: 1584–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders A., Lourenco P.C.C., Sawin K.E. (2006). Noncore components of the fission yeast γ -tubulin complex. Mol. Biol. Cell 17: 5075–5093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binarova P., Cenklova V., Prochazkova J., Doskocilova A., Volc J., Vrlik M., Bogre L. (2006). γ -Tubulin is essential for acentrosomal microtubule nucleation and coordination of late mitotic events in Arabidopsis. Plant Cell 18: 1199–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R.C., Lemmon B.E. (2006). Polar organizers and girdling bands of microtubules are associated with gamma-tubulin and act in establishment of meiotic quadripolarity in the hepatic Aneura pinguis (Bryophyta). Protoplasma 227: 77–85 [DOI] [PubMed] [Google Scholar]
- Brown R.C., Lemmon B.E. (2007). The pleiotrophic plant MTOC: an evolutionary perspective. J. Integr. Plant Biol. 49: 1142–1153 [Google Scholar]
- Buschmann H., Lloyd C.W. (2008). Arabidopsis mutants and the network of microtubule-associated functions. Mol. Plant 1: 888–898 [DOI] [PubMed] [Google Scholar]
- Chan J., Sambade A., Calder G., Lloyd C. (2009). Arabidopsis cortical microtubules are initiated along, as well as branching from, existing microtubules. Plant Cell 21: 2298–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.B., Marcus A., Li W.X., Hu Y., Calzada J.P.V., Grossniklaus U., Cyr R.J., Ma H. (2002). The Arabidopsis ATK1 gene is required for spindle morphogenesis in male meiosis. Development 129: 2401–2409 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R., Cyr R. (2004). Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell 16: 3274–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K.M., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Erhardt M., Stoppin-Mellet V., Campagne S., Canaday J., Mutterer J., Fabian T., Sauter M., Muller T., Peter C., Lambert A.M., Schmit A.C. (2002). The plant Spc98p homologue colocalizes with γ -tubulin at microtubule nucleation sites and is required for microtubule nucleation. J. Cell Sci. 115: 2423–2431 [DOI] [PubMed] [Google Scholar]
- Fava F., Raynaud-Messina B., Leung-Tack J., Mazzolini L., Li M., Guillemot J.C., Cachot D., Tollon Y., Ferrara P., Wright M. (1999). Human 76p: A new member of the gamma-tubulin-associated protein family. J. Cell Biol. 147: 857–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita A., Vardy L., Garcia M.A., Toda T. (2002). A fourth component of the fission yeast γ -tubulin complex, Alp16, is required for cytoplasmic microtubule integrity and becomes indispensable when γ -tubulin function is compromised. Mol. Biol. Cell 13: 2360–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K.L., Dixon J.E. (1991). Eukaryotic proteins expressed in Escherichia coli: An improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192: 262–267 [DOI] [PubMed] [Google Scholar]
- Gunawardane R.N., Martin O.C., Cao K., Zhang L.J., Dej K., Iwamatsu A., Zheng Y.X. (2000). Characterization and reconstitution of Drosophila γ -tubulin ring complex subunits. J. Cell Biol. 151: 1513–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Ho C.-M.K., Kong Z., Lee Y.-R.J., Qian Q., Liu B. (2009). Evaluating the microtubule cytoskeleton and its interacting proteins in monocots by mining the rice genome. Ann. Bot. (Lond.) 103: 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallen M.A., Ho J., Yankel C.D., Endow S.A. (2008). Fluorescence recovery kinetic analysis of γ -tubulin binding to the mitotic spindle. Biophys. J. 95: 3048–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T. (2007). Microtubule-associated proteins in higher plants. J. Plant Res. 120: 79–98 [DOI] [PubMed] [Google Scholar]
- Horio T., Basaki A., Takeoka A., Yamato M. (1999). Lethal level overexpression of γ -tubulin in fission yeast causes mitotic arrest. Cell Motil. Cytoskeleton 44: 284–295 [DOI] [PubMed] [Google Scholar]
- Ishida T., Kurata T., Okada K., Wada T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Izumi N., Fumoto K., Izumi S., Kikuchi A. (2008). GSK-3 β regulates proper mitotic spindle formation in cooperation with a component of the γ -tubulin ring complex, GCP5. J. Biol. Chem. 283: 12981–12991 [DOI] [PubMed] [Google Scholar]
- Jeng R., Stearns T. (1999). γ -Tubulin complexes: Size does matter. Trends Cell Biol. 9: 339–342 [DOI] [PubMed] [Google Scholar]
- Job D., Valiron O., Oakley B. (2003). Microtubule nucleation. Curr. Opin. Cell Biol. 15: 111–117 [DOI] [PubMed] [Google Scholar]
- Julian M., Tollon Y., Lajoie-Mazenc I., Moisand A., Mazarguil H., Puget A., Wright A. (1993). γ -Tubulin participates in the formation of the midbody during cytokinesis in mammalian cells. J. Cell Sci. 105: 145–156 [DOI] [PubMed] [Google Scholar]
- Kong Z., Li M., Yang W., Xu W., Xue Y. (2006). A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 141: 1376–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie-Mazenc I., Tollon Y., Detraves C., Julian M., Moisand A., Gueth-Hallonet C., Debec A., Salles-Passador I., Puget A., Mazarguil H., Raynaud-Messina B., Wright M. (1994). Recruitment of antigenic γ -tubulin during mitosis in animal cells: presence of γ -tubulin in the mitotic spindle. J. Cell Sci. 107: 2825–2837 [DOI] [PubMed] [Google Scholar]
- Lee Y.-R.J., Giang H.M., Liu B. (2001). A novel plant kinesin-related protein specifically associates with the phragmoplast organelles. Plant Cell 13: 2427–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.R.J., Liu B. (2000). Identification of a phragmoplast-associated kinesin-related protein in higher plants. Curr. Biol. 10: 797–800 [DOI] [PubMed] [Google Scholar]
- Liu B., Joshi H.C., Palevitz B.A. (1995). Experimental manipulation of γ -tubulin distribution in Arabidopsis using anti-microtubule drugs. Cell Motil. Cytoskeleton 31: 113–129 [DOI] [PubMed] [Google Scholar]
- Liu B., Joshi H.C., Wilson T.J., Silflow C.D., Palevitz B.A., Snustad D.P. (1994). γ -Tubulin in Arabidopsis-gene sequence, immunoblot, and immunofluorescence studies. Plant Cell 6: 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Marc J., Joshi H.C., Palevitz B.A. (1993). A γ -tubulin-related protein associated with the microtubule arrays of higher plants in a cell cycle-dependent manner. J. Cell Sci. 104: 1217–1228 [DOI] [PubMed] [Google Scholar]
- Lu L., Lee Y.-R.J., Pan R., Liu B. (2005). An internal motor kinesin is associated with the Golgi apparatus and plays a role in trichome morphogenesis in Arabidopsis. Mol. Biol. Cell 16: 811–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J., Stearns T. (2007). Microtubule-organizing centres: A re-evaluation. Nat. Rev. Mol. Cell Biol. 8: 161–167 [DOI] [PubMed] [Google Scholar]
- Marc J., Mineyuki Y., Palevitz B.A. (1989). A planar microtubule-organizing zone in guard-cells of Allium - Experimental depolymerization and reassembly of microtubules. Planta 179: 530–540 [DOI] [PubMed] [Google Scholar]
- McDonald A.R., Liu B., Joshi H.C., Palevitz B.A. (1993). γ -Tubulin is associated with a cortical-microtubule-organizing zone in the developing guard cells of Allium cepa L. Planta 191: 357–361 [DOI] [PubMed] [Google Scholar]
- Murata T., Sonobe S., Baskin T.I., Hyodo S., Hasezawa S., Nagata T., Horio T., Hasebe M. (2005). Microtubule-dependent microtubule nucleation based on recruitment of γ -tubulin in higher plants. Nat. Cell Biol. 7: 961–968 [DOI] [PubMed] [Google Scholar]
- Murata T., Tanahashi T., Nishiyama T., Yamaguchi K., Hasebe M. (2007). How do plants organize microtubules without a centrosome?. J. Integr. Plant Biol. 49: 1154–1163 [Google Scholar]
- Murphy S.M., Preble A.M., Patel U.K., O'Connell K.L., Dias D.P., Moritz M., Agard D., Stults J.T., Stearns T. (2001). GCP5 and GCP6: Two new members of the human γ -tubulin complex. Mol. Biol. Cell 12: 3340–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S.M., Urbani L., Stearns T. (1998). The mammalian γ -tubulin complex contains homologues of the yeast spindle pole body components spc97p and spc98p. J. Cell Biol. 141: 663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Hashimoto T. (2009). A mutation in the Arabidopsis γ -tubulin-containing complex causes helical growth and abnormal microtubule branching. J. Cell Sci. 122: 2208–2217 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Naoi K., Shoji T., Hashimoto T. (2004). Low concentrations of propyzamide and oryzalin alter microtubule dynamics in Arabidopsis epidermal cells. Plant Cell Physiol. 45: 1330–1334 [DOI] [PubMed] [Google Scholar]
- Olmsted J.B. (1981). Affinity purification of antibodies from diazotized paper blots of heterogeneous protein samples. J. Biol. Chem. 256: 11955–11957 [PubMed] [Google Scholar]
- Paluh J.L., Nogales E., Oakley B.R., McDonald K., Pidoux A.L., Cande W.Z. (2000). A mutation in γ -tubulin alters microtubule dynamics and organization and is synthetically lethal with the kinesin-like protein pkl1p. Mol. Biol. Cell 11: 1225–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuglia M., Azimzadeh J., Goussot M., Camilleri C., Belcram K., Evrard J.L., Schmit A.C., Guerche P., Bouchez D. (2006). γ -Tubulin is essential for microtubule organization and development in Arabidopsis. Plant Cell 18: 1412–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud-Messina B., Merdes A. (2007). γ -Tubulin complexes and microtubule organization. Curr. Opin. Cell Biol. 19: 24–30 [DOI] [PubMed] [Google Scholar]
- Samejima I., Lourenco P.C.C., Snaith H.A., Sawin K.E. (2005). Fission yeast mto2p regulates microtubule nucleation by the centrosomin-related protein mto1p. Mol. Biol. Cell 16: 3040–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K.E., Tran P.T. (2006). Cytoplasmic microtubule organization in fission yeast. Yeast 23: 1001–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltzer V., Janski N., Canaday J., Herzog E., Erhardt M., Evrard J.L., Schmit A.C. (2007). Arabidopsis GCP2 and GCP3 are part of a soluble gamma-tubulin complex and have nuclear envelope targeting domains. Plant J. 52: 322–331 [DOI] [PubMed] [Google Scholar]
- Shimamura M., Brown R.C., Lemmon B.E., Akashi T., Mizuno K., Nishihara N., Tomizawa K., Yoshimoto K., Deguchi H., Hosoya H., Horio T., Mineyuki Y. (2004). γ -Tubulin in basal land plants: Characterization, localization, and implication in the evolution of acentriolar microtubule organizing centers. Plant Cell 16: 45–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow C.D., Liu B., LaVoie M., Richardson E.A., Palevitz B.A. (1999). γ -Tubulin in Chlamydomonas: Characterization of the gene and localization of the gene product in cells. Cell Motil. Cytoskeleton 42: 285–297 [DOI] [PubMed] [Google Scholar]
- Stoppin-Mellet V., Peter C., Lambert A.M. (2000). Distribution of γ -tubulin in higher plant cells: Cytosolic γ -tubulin is part of higher molecular weight complexes. Plant Biol. 2: 290–296 [Google Scholar]
- Venkatram S., Tasto J.J., Feoktistova A., Jennings J.L., Link A.J., Gould K.L. (2004). Identification and characterization of two novel proteins affecting fission yeast γ -tubulin complex function. Mol. Biol. Cell 15: 2287–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vérollet C., Colombié N., Daubon T., Bourbon H.M., Wright M., Raynaud-Messina B. (2006). Drosophila melanogaster γ -TuRC is dispensable for targeting γ -tubulin to the centrosome and microtubule nucleation. J. Cell Biol. 172: 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinh D.B., Kern J.W., Hancock W.O., Howard J., Davis T.N. (2002). Reconstitution and characterization of budding yeast γ -tubulin complex. Mol. Biol. Cell 13: 1144–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys G. (2002). Microtubule organization in the green kingdom: Chaos or self-order?. J. Cell Sci. 115: 1345–1354 [DOI] [PubMed] [Google Scholar]
- Wiese C., Zheng Y. (2006). Microtubule nucleation: γ -Tubulin and beyond. J. Cell Sci. 119: 4143–4153 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Oakley B.R. (2009). In vivo analysis of the functions of γ -tubulin complex proteins. J. Cell Sci. 122: 4218–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C.J., Lee Y.R., Liu B. (2009). The WD40 repeat protein NEDD1 functions in microtubule organization during cell division in Arabidopsis thaliana. Plant Cell 21: 1129–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Wong M.L., Alberts B., Mitchison T. (1995). Nucleation of microtubule assembly by a γ -tubulin-containing ring complex. Nature 378: 578–583 [DOI] [PubMed] [Google Scholar]
- Zimmerman S., Chang F. (2005). Effects of γ -tubulin complex proteins on microtubule nucleation and catastrophe in fission yeast. Mol. Biol. Cell 16: 2719–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.