Abstract
Spt4–Spt5, a general transcription elongation factor for RNA polymerase II, also has roles in chromatin regulation. However, the relationships between these functions are not clear. Previously, we isolated suppressors of a Saccharomyces cerevisiae spt5 mutation in genes encoding members of the Paf1 complex, which regulates several cotranscriptional histone modifications, and Chd1, a chromatin remodeling enzyme. Here, we show that this suppression of spt5 can result from loss of histone H3 lysines 4 or 36 methylation, or reduced recruitment of Chd1 or the Rpd3S complex. These spt5 suppressors also rescue the synthetic growth defects observed in spt5 mutants that also lack elongation factor TFIIS. Using a FLO8 reporter gene, we found that a chd1 mutation caused cryptic initiation of transcription. We further observed enhancement of cryptic initiation in chd1 isw1 mutants and increased histone acetylation in a chd1 mutant. We suggest that, as previously proposed for H3 lysine 36 methylation and the Rpd3S complex, H3 lysine 4 methylation and Chd1 function to maintain normal chromatin structures over transcribed genes, and that one function of Spt4–Spt5 is to help RNA polymerase II overcome the repressive effects of these histone modifications and chromatin regulators on transcription.
EUKARYOTES package their genomes into nucleosomes to form chromatin. Although nucleosomes and higher order chromatin structures permit significant compaction of the genome, they also inhibit transcription by blocking access to underlying DNA and by forming a repeating barrier to elongating RNA polymerases. Strategies used to overcome this inhibition and regulate transcription include: post-translational modification of histone tails; remodeling, eviction, or movement of nucleosomes by both ATP-dependent and -independent mechanisms; and incorporation of histone variants into nucleosomes (Saunders et al. 2006; Li et al. 2007a; Williams and Tyler 2007).
In contrast to promoters, which are often persistently nucleosome free, the bodies of actively transcribed genes are typically still nucleosome assembled, even though nucleosomes strongly inhibit elongation by purified RNA polymerase II (Studitsky et al. 2004; Pokholok et al. 2005; Saunders et al. 2006; Rando and Ahmad 2007). These observations imply that eukaryotes must possess activities that transiently alter or remove nucleosomes to permit elongation and then restore them to their prior state. Failure to restore chromatin structure after elongation may reveal cryptic promoters, leading to aberrant transcription initiation from internal positions within a gene (Kaplan et al. 2003; Mason and Struhl 2003; Carrozza et al. 2005). Thus, maintenance of chromatin structure over transcribed sequences presents a unique set of challenges and is critical to appropriate regulation of a cell's transcriptome.
The Spt4–Spt5 complex is an essential, highly conserved regulator of transcription elongation by RNAPII in eukaryotes (Hartzog et al. 2002). It joins elongation complexes soon after initiation (Andrulis et al. 2000; Ping and Rana 2001) and associates with RNAPII along the entire length of the gene (Kim et al. 2004). Although the precise function of Spt4–Spt5 is not known, in vitro studies show that it can repress transcription elongation at promoter proximal locations and can promote elongation under nucleotide limiting conditions (Wada et al. 1998). Furthermore, a wealth of genetic data implicate it in regulation of elongation and RNA processing in vivo (Cui and Denis 2003; Lindstrom et al. 2003; Kim et al. 2004; Bucheli and Buratowski 2005; Burckin et al. 2005; Kaplan et al. 2005; Xiao et al. 2005). In addition, spt4 and spt5 mutations share a number of phenotypes with histone mutations and genetically interact with mutations in genes encoding chromatin remodeling factors, suggesting that the function of Spt4–Spt5 is connected to chromatin (Swanson and Winston 1992; Squazzo et al. 2002; Simic et al. 2003).
We previously identified a mutation in the Saccharomyces cerevisiae SPT5 gene, spt5-242, which confers a cold-sensitive (Cs−) growth defect (Hartzog et al. 1998). We also identified two classes of suppressors of the Cs− phenotype of spt5-242 cells. The first class includes mutations in either of the two large, catalytic subunits of RNAPII (Hartzog et al. 1998). One of these mutations, rpb2-10, displays a decreased elongation rate and lower processivity in vitro (Powell and Reines 1996), and rpb1 suppressors of spt5-242 alter residues implicated in elongation (Hartzog et al. 1998). In addition, spt5-242 is suppressed by 6-azauracil ((Hartzog et al. 1998), which inhibits nucleotide biosynthesis and is believed to impede elongation in vivo by starving the polymerase of substrate nucleotides (Exinger and Lacroute 1992). Thus, it appears that the spt5-242 mutation is suppressed by decreased RNAPII elongation rates. The second class of spt5-242 suppressors is composed of mutations that likely perturb chromatin structure or dynamics. These include mutations in CHD1 (Simic et al. 2003), which encodes an ATP-dependent chromatin remodeling enzyme (Tran et al. 2000; Stockdale et al. 2006), with a pair of conserved N-terminal chromodomains, a central Snf/Swi type helicase domain and a C-terminal domain that resembles Myb-type DNA binding domains (Woodage et al. 1997). In addition, mutations that perturb the Paf1 complex, which regulates the activity of several histone-modifying enzymes, also suppress spt5-242 (Squazzo et al. 2002).
In this work, we investigate the potential roles of this second class of spt5-242 suppressors in transcription elongation. We show that these chromatin-based suppressors have effects on the transcription apparatus that are distinct from elongation rate-based suppression. We show that loss of a specific subset of Paf1 complex functions, methylation of histone H3 lysines 4 and 36, are involved in suppression of spt5-242. We present evidence that recruitment of Chd1 to transcribed genes may depend in part upon H3K4 and H3K36 methylation; we further show that all three conserved domains of Chd1 are required for its recruitment to chromatin and function. Finally, we find that loss of Chd1 contributes to the appearance of cryptic transcripts, suggesting that Chd1 plays a role in maintaining nucleosomes over transcribed regions. We suggest that the Spt4–Spt5 complex promotes transcription elongation across chromatin templates, acting in opposition to Rpd3S, Chd1, as well as histone H3K4 and H3K36 methylation and downstream effectors of these marks.
MATERIALS AND METHODS
Media and genetic methods:
Strain construction and other genetic manipulations were carried out by standard methods (Rose et al. 1990). Yeast media was made as described previously (Rose et al. 1990). All S. cerevisiae strains used in this study (supporting information, Table S1) are isogenic to S288C and are GAL2+ (Winston et al. 1995). The GAL1-FLO8-HIS3 reporter construct was a gift of Fred Winston and was integrated as described by Cheung et al. (2008). For spot dilutions, strains were grown in rich or synthetic media. Cells were counted with a hemocytometer, 1 × 107 cells were pelleted in a microcentrifuge, and resuspended in 1 ml of water. Fivefold serial dilutions of these cells were prepared and 5–10 μl of each dilution was spotted on rich or synthetic media and incubated at the indicated temperature.
Plasmids:
All plasmids used in this study are listed in Table S2. Chromodomain mutations in CHD1 were constructed in a HA3-CHD1 URA3 CEN plasmid, pGH269 (Simic et al. 2003). AgeI sites flanking the chromodomains were generated by PCR mutagenesis. This plasmid was digested with AgeI, purified, and religated to create the complete chromodomain deletion, pTQ5. PCR products flanked by AgeI sites containing the first chromodomain and intervening sequence, or the intervening sequence and the second chromodomain, were synthesized and ligated into pTQ5 to create pTQ4 and pTQ3. The plasmid containing a Y316E point mutation in CHD1 was also derived from pGH269 and was a gift of Patrick Grant (Pray-Grant et al. 2005). rtf1 mutant plasmids in Figure 5A were gifts of Karen Arndt (Warner et al. 2007). For use in ChIP assays, rtf1 mutant plasmids were digested with NdeI to remove the HA3 tag and religated.
Figure 5.—
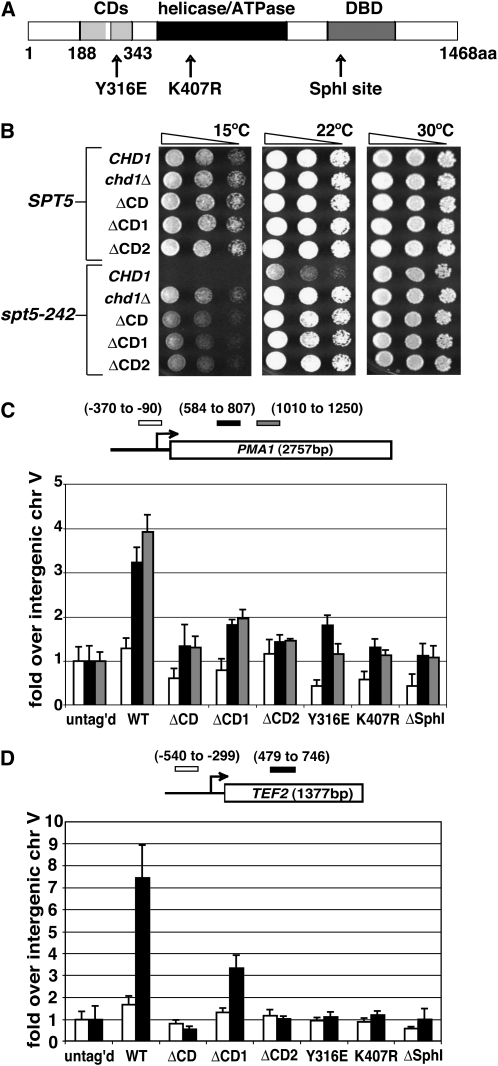
All three conserved domains of Chd1 are required for its function and localization to chromatin. (A) The location of the conserved sequence motifs of Chd1 as well as sites targeted for mutations. (B) Deletion of either or both chromodomains of Chd1 is sufficient for suppression of spt5-242. SPT5 chd1Δ and spt5-242 chd1Δ strains were transformed with URA3 CEN plasmids carrying the indicated chd1 mutations expressed from the normal CHD1 promoter. Serial dilutions of cells with the indicated genotypes were spotted to SC −Ura media and grown at the indicated temperature for 4 days. (C and D) All three conserved domains of Chd1 are required for its association with chromatin. Strains expressing HA3-tagged forms of Chd1 were subjected to anti-HA1 ChIP followed by QPCR analysis using primers directed against the promoters and transcribed regions of PMA1 (C) and TEF2 (D).
Chromatin immunoprecipitation:
ChIP assays were performed as described previously (Simic et al. 2003). Strains containing hht2 or rtf1 mutations were grown in SC −Trp media and strains containing plasmid-borne chd1 mutations were grown in SC −Ura to maintain plasmid selection. All other strains were grown in YPD media. RNAPII was precipitated with a monoclonal antibody, 8WG16 (Covance), and H3K9/14Ac with antihistone H3 K9/14Ac antibody (Upstate). Total histone H3 precipitated with an antibody directed against the C terminus of histone H3 (Abcam). The anti-HA antibody is described in Simic et al. (2003).
Quantitative PCR analysis was performed as follows on a Bio-Rad iCycler: 40 cycles of 95° for 30 sec, 55° for 30 sec, and 72° for 45 sec. PCR was carried out in 20-μl reactions in 96-well plates using Eurogentec qPCR MasterMix Plus for SYBR Green I Low ROX. Primers directed against TEF2 and PMA1 (Simic et al. 2003), YLR454W (Mason and Struhl 2005), and intergenic chromosome V (Komarnitsky et al. 2000) were described previously. For ChIP of STE11, the promoter primers amplified nucleotides −320 to −563 (relative to the ATG), the 5′ primers amplified nucleotides +1 to +330, and the 3′ primers amplified nucleotides +1641 to +1915. For ChIP of FLO8, the promoter primers amplified nucleotides −65 to −287, the 5′ primers amplified nucleotides +60 to +408, and the 3′ primers amplified nucleotides +1969 to +2349.
In Figure 5 and Figure S1, fold enrichment over a nontranscribed sequence on chromosome V was calculated as described previously (Ausubel et al. 1991). These values were then normalized to values obtained from control immunoprecipitations from an untagged strain. The RNAPII ChIPs in Figure 1 were analyzed as described in Mason and Struhl (2005), using the chromosome V sequence as a control. The ChIPs of H3AcK9/14 in Figure 9 were normalized to precipitations of total histone H3 to account for any strain-to-strain variation in nucleosome density. The H3Ac ChIPs from the set2 mutant in Figure 9 are derived from two independent experiments. All other ChIP data were derived from three or more independent experiments.
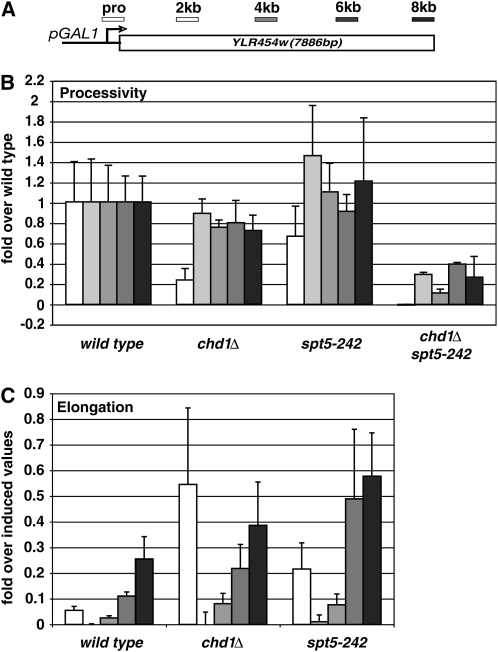
Figure 1.—
Reduced rate of RNA polymerase II elongation in spt5-242 mutant. (A) Location of ChIP probes on hybrid GAL1-YLR454W gene. (B) Measurement of RNAPII processivity. ChIP of RNAPII across the hybrid GAL1-YLR454W gene in wild-type cells and the indicated mutants was performed under inducing conditions. For each mutant, IP/Input values for each ChIP probe were determined and normalized to the corresponding value measured in the wild-type strain. (C) Measurement of elongation rate. Galactose-induced cells were treated with glucose to repress transcription from the GAL1-YLR454W gene and samples were processed for ChIP 5 min later. IP/Input values for each probe are expressed relative to the corresponding value for that probe measure just prior to addition of glucose.
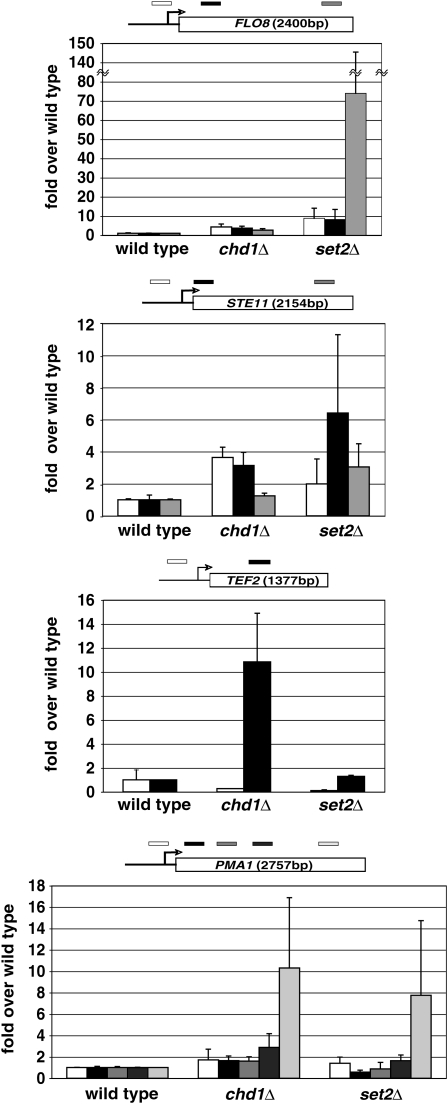
Figure 9.—
Loss of Chd1 leads to increased H3 acetylation. Wild-type, chd1Δ, and set2Δ strains were subjected to ChIP with anti-H3Ac and anti-H3 antisera followed by QPCR analysis using primers directed against the promoters and transcribed regions of FLO8, STE11, PMA1, and TEF2. Bar graphs present H3Ac values relative to total histone H3 and are normalized to wild type.
Northern blots:
Northern blotting was performed as described previously (Ausubel et al. 1991; Swanson et al. 1991; Kaplan et al. 2003). Probes for FLO8, STE11, and RAD18 were PCR amplified from genomic DNA. The FLO8 3′ probe covers nucleotides +1595 to +2349 relative to the ATG. The STE11 3′ probe covers nucleotides +1641 to +2153 relative to the ATG. The RAD18 3′ probe covers nucleotides +472 to +1472 relative to the ATG. Probes were labeled by random priming as described previously (Ausubel et al. 1991).
RESULTS
The spt5-242 mutation reduces the rate of elongation in vivo:
To examine the rate of transcription elongation in vivo, we used a chromatin immunoprecipitation assay. The GAL1 promoter, which is induced in galactose media and repressed in the presence of glucose, was integrated upstream of the ∼7.9 kb YLR454W gene (Figure 1A) and crossed into strains carrying the spt5-242 and chd1Δ mutations. Cells were grown to log phase in media containing raffinose, which neither represses nor induces the GAL1 promoter. The GAL1 promoter was then induced with galactose for 90 min, an aliquot of each culture was formaldehyde crosslinked and frozen, and the remainder of the culture was treated with glucose to repress the GAL1 promoter. Five minutes after addition of glucose an additional sample of cells was crosslinked and frozen. Both samples were then processed for ChIP using an antibody directed against RNA polymerase II, and a set of five PCR primers was used to monitor the distribution of RNAPII across the hybrid GAL1-YLR454W gene.
Previously, Mason and Struhl (2005) used this assay to monitor RNAPII processivity by examining polymerase distribution across the induced GAL1-YLR454W gene, and to monitor elongation rate by examining the decay of the RNAPII ChIP signal across GAL1-YLR454W following glucose repression. Examining a large collection of mutations, they found that deletion of CHD1 did not affect processivity or elongation rate, whereas loss of SPT4 resulted in decreased polymerase processivity, but a normal elongation rate. In addition, they found that rpb2-10, a mutation that suppresses spt5-242 (Hartzog et al. 1998) and reduces elongation and processivity in vitro (Powell and Reines 1996), also reduced processivity and elongation rate across GAL1-YLR454W in vivo.
Consistent with these previous results, we found that RNAPII processivity in a chd1Δ strain closely matched that observed in wild type (Figure 1B). In glucose repressed samples, the chd1 deletion resulted in only a modest increase in the density of RNAPII across GAL1-YLR454W. Polymerase density across the induced GAL1-YLR454W gene in the spt5-242 mutant was similar to that observed in wild-type cells. In contrast, following glucose repression, RNAPII levels at GAL1-YLR45W in spt5-242 cells were higher than in wild-type cells (Figure 1C). This effect was most obvious for ChIP probes at the 3′ end of the gene, indicating a decreased rate of clearance of RNAPII from GAL1-YLR454W. Thus the spt5-242 mutation leads to a decreased transcription elongation rate in vivo.
Two mechanisms for genetic suppression of spt5-242:
We were unable to reproducibly measure RNAPII levels at GAL1-YLR454W in glucose repressed chd1Δ spt5-242 double mutants. This was likely due to the substantial decrease in RNAPII density in this double mutant observed even under inducing conditions (Figure 1A). As an alternative approach to determining the mechanism by which loss of Chd1 leads to suppression of spt5-242, we examined genetic interactions of spt5-242 and chd1 with a null allele of DST1, which encodes transcription elongation factor TFIIS and functions to overcome transcription arrest by RNAPII (Fish and Kane 2002). At each step in RNA chain elongation, RNAPII may add nucleotides to the 3′ end of the nascent RNA, pause, or arrest. Arrest occurs when the polymerase backtracks, leaving its active site misaligned over the DNA:RNA hybrid of the transcription bubble rather than over the 3′ end of the nascent RNA. TFIIS binds arrested RNAPII elongation complexes, stimulates cleavage of the nascent transcript, creating a new 3′ end that is properly aligned with the active site of the enzyme, allowing elongation to resume.
Mutations in RNAPII subunits, including several rpb1 and rpb2 alleles that suppress spt5-242, cause synthetic growth defects or lethality when combined with TFIIS mutations (Hartzog et al. 1998; Lennon et al. 1998). These observations were interpreted to indicate that the rpb mutations lead to an increased frequency of arrest and dependence on TFIIS. In addition, spt5 dst1Δ mutants exhibit strong synthetic growth defects (Figure 2A; Hartzog et al. 1998). Thus, it is likely that spt5 mutations also lead to increased arrest and dependence on TFIIS.
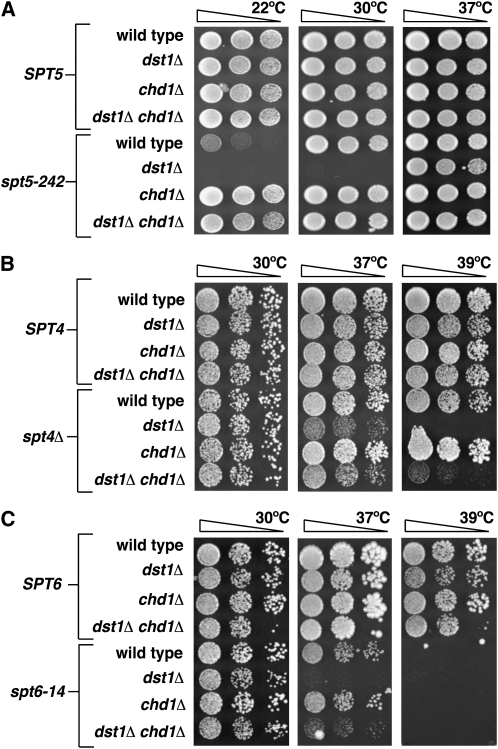
Figure 2.—
chd1 mutations suppress growth defects of spt dst1 mutants. Genetic crosses were performed to create strains carrying combinations of complete deletions of DST1 and CHD1 with the cold-sensitive spt5-242 mutation, a complete deletion of SPT4, and the temperature-sensitive spt6-14 mutation. Serial dilutions of these strains were spotted to YPD media and grown at the indicated temperature. (A) chd1Δ suppresses the growth defect of spt5-242 dst1Δ cells at 22° and 30°. (B) chd1Δ suppresses the growth defect of spt4Δ dst1Δ cells at 39°. (C) chd1Δ suppresses the growth defect of spt6-14 dst1Δ cells at 37°.
To determine whether the chromatin-based suppressors of spt5-242 function by a mechanism similar to or distinct from that of the rpb suppressors of spt5-242, we performed genetic crosses to isolate strains with all possible combinations of spt5-242, dst1Δ, and chd1Δ mutations. Although spt5-242 dst1Δ mutants are inviable at temperatures below 37° (Figure 2A; Hartzog et al. 1998), a spt5-242 dst1Δ chd1Δ mutant was viable at temperatures as low as 22° (Figure 2A). In addition, dst1Δ chd1Δ mutants did not display any other obvious new phenotypes. Thus, loss of Chd1 overcomes the synthetic growth defect of spt5-242 dst1Δ mutants.
In contrast to the results with chd1Δ, when we crossed strains carrying dst1Δ, spt5-242, and either the rpb1-221 or rpb1-244 mutations, which suppress spt5-242, we were unable to obtain viable triple mutants. Thus, although rpb1-221 and rpb1-244 suppress spt5-242, they cannot overcome the synthetic growth defects observed in spt5-242 dst1Δ double mutants. These data support the idea that chromatin defects and elongation defective forms of RNAPII suppress spt5-242 by distinct mechanisms.
In addition to enhancing the phenotype of spt5-242, deletion of DST1 causes a temperature sensitive (Ts−) growth defect when combined with other transcription elongation factor mutations, including spt5-194, spt4Δ, and spt6-14 (Hartzog et al. 1998). We therefore crossed strains carrying chd1Δ, dst1Δ, and these spt mutations. Deletion of CHD1 did not alter growth of the spt5-194 dst1Δ strain (data not shown) but did partially suppress the Ts− phenotype of spt4Δ dst1Δ and spt6-14 dst1Δ (Figure 2, B and C). Thus, Chd1 may have a more general role in elongation since its interactions are not factor specific.
Relationships between chromatin-based suppressors of spt5-242:
The data presented above demonstrate that there are at least two distinct mechanisms for genetic suppression of spt5-242, decreasing the elongation rate of RNA polymerase II or disruption of chromatin. We next explored genetic relationships between previously identified chromatin-based suppressor of spt5-242, the Paf1 complex and Chd1.
Disruption of an Rtf1–Chd1 interaction suppresses spt5-242:
Two prior observations suggest that Chd1 interacts with Rtf1, a component of the Paf1 complex. First, a C-terminal fragment of Chd1 interacted with Rtf1 in a two-hybrid experiment. Second, Chd1 did not associate with transcribed chromatin in an rtf1 mutant (Figure S1; Simic et al. 2003). These data suggested the possibility that in addition to its role in Set1 function, the Paf1 complex may play a more direct role in Chd1's association with chromatin via a direct Rtf1–Chd1 interaction.
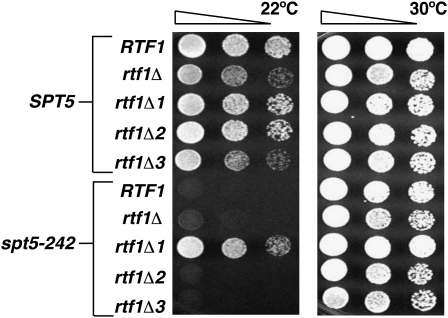
Several recently characterized Rtf1 mutations support this idea (Warner et al. 2007). rtf1Δ1 is an internal deletion that removes amino acids (aa) 3–30 and disrupts the two-hybrid interaction between Chd1 and Rtf1, but does not alter H3K4 methylation. rtf1Δ2, which deletes aa 30–62, does not alter the two-hybrid interaction with Chd1, histone H3 methylation, or H2B ubiquitylation. In contrast, rtf1Δ3 deletes aa 62–109 and causes defects in histone H2B ubiquitylation and H3K4 and H3K79 methylation, but does not alter the Rtf1–Chd1 two-hybrid interaction. Each of these forms of Rtf1 retains the ability to assemble into the Paf1 complex and associate with chromatin. When we combined these rtf1 mutations with spt5-242, we found that rtf1Δ1 suppressed the Cs− phenotype of spt5-242 whereas rtf1Δ2 and rtf1Δ3 did not (Figure 3A). Consistent with this suppression, in ChIP assays, the rtf1Δ1 mutation abolishes Chd1's association with transcribed chromatin (Warner et al. 2007; Figure S1). These data suggest that, in addition to its role in regulating histone H3 modification states, Rtf1 plays an important role in Chd1's recruitment to transcribed chromatin during transcription elongation.
Figure 3.—
The rtf1Δ1 mutation suppresses spt5-242. Serial dilutions of SPT5 rtf1Δ and spt5-242 rtf1Δ strains transformed with the indicated RTF1 plasmids were spotted to SC −Trp media and grown at 22° or 30° for 3 days. Only rtf1Δ1, which disrupts Rtf1–Chd1 interactions, suppressed the cold-sensitive phenotype of spt5-242.
Evidence that methylation of histone H3 lysines 4 and 36 impact transcription elongation:
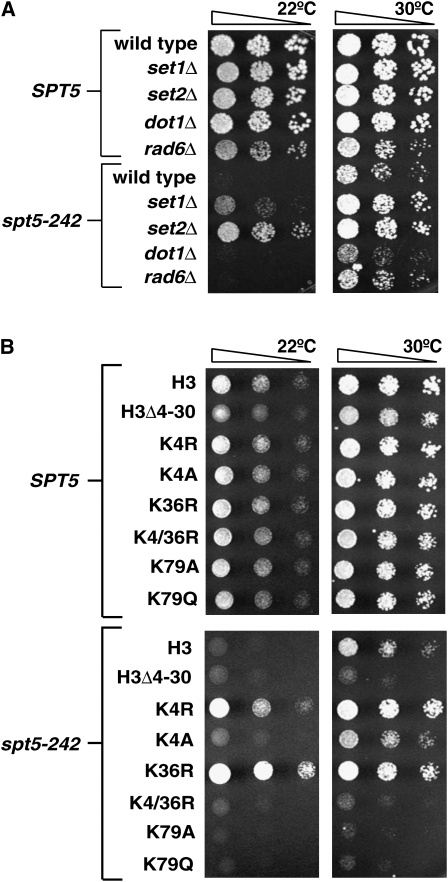
The Paf1 complex plays a role in recruiting and regulating the histone H3 methyltransferases Set1, Set2, and Dot1 and also the Rad6/Bre1 histone H2B ubiquitin ligase (Krogan et al. 2003a,b; Ng et al. 2003; Wood et al. 2003). Suppression of spt5-242 by perturbation of the Paf1 complex could be due to loss of one or more of these enzymes. We therefore crossed an spt5-242 mutant to strains carrying deletions of SET1, SET2, DOT1, and RAD6. We also created spt5-242 strains that carried substitutions of histone H3 lysine 4 (H3K4), H3K36, or H3K79 as their sole source of these histones. The progeny of these crosses were monitored for growth at a variety of temperatures (Figure 4 and data not shown). The set1Δ mutation suppressed the Cs− phenotype of spt5-242 weakly at 15° and more strongly at 22°. A similar level of suppression was observed when H3K4 was substituted with either alanine (H3K4A) or arginine (H3K4R). The set2Δ and H3K36R mutations both strongly suppressed the growth defect of spt5-242 at 15° and 22°. We further observed that spt5-242 set1Δ set2Δ mutants showed the same level of growth as spt5-242 set2Δ mutants. Curiously, spt5-242 H3K4/K36R mutants did not grow at 15° or 22°. In contrast to the results with SET1 and SET2, deletion of DOT1 or mutation of its target, H3K79, failed to suppress spt5-242. In sum, these data suggest a role for Set1, Set2, and methylation of histone H3 lysines 4 and 36 in Spt5 function.
Figure 4.—
spt5-242 is suppressed by loss of H3K4 or H3K36 methylation. (A) Spt5-242 is suppressed by loss of the H3K4 or H3K36 methyltransferases Set1 and Set2 but not by loss of the H3K79 methyltrasferase Dot1 nor the ubiquitin conjugating enzyme Rad6. Strains with the indicated genotypes were spotted to YPD and incubated at 30° for 2 days or 22° for 4 days. (B) Mutations altering H3K4 or H3K36 suppress spt5-242. SPT5 and spt5-242 strains carrying deletions of both histone H3–H4 loci and a CEN URA3 HHT1-HHF1 plasmid were transformed with plasmids carrying the indicated histone H3 allele. Trp+ transformants were spotted directly to 5FOA and incubated at 30° for 2 days or 22° for 3 days.
Normal methylation of H3K4 and H3K79 depends upon Rtf1, Rad6, and H2BK123 ubiquitylation (Dover et al. 2002; Ng et al. 2002; Sun and Allis 2002; Krogan et al. 2003a). Curiously, mutations that abolished H2BK123 ubiquitylation, rtf1Δ, rtf1Δ3, and rad6Δ, all failed to suppress spt5-242 (Figures 3 and 4; Squazzo et al. 2002). Thus, loss of H2BK123 ubiquitylation appears to impact elongation in a manner that is distinct from and epistatic to the loss of H3K4 or H3K36 methylation. One potential explanation for these observations is that, in addition to its role in histone methylation, H2BK123 ubiquitylation may directly facilitate elongation (Pavri et al. 2006; Fleming et al. 2008).
Chd1's conserved domains are required for association with transcribed chromatin:
The results described above raised the possibility that methylation of H3K4 or K36 might mediate recruitment of Chd1 to chromatin. We therefore sought to identify the domain(s) of Chd1 that mediate its association with chromatin. We focused on Chd1's three conserved domains: its pair of N-terminal chromodomains (CDs), its Swi/Snf-like helicase domain, and its C-terminal domain with homology to Myb-type DNA binding domains (Figure 5A). We previously constructed mutations altering each of these domains in a triple HA epitope-tagged form of Chd1 (HA3-Chd1), and found that the altered proteins were expressed at normal levels and that the mutations suppressed spt5-242, although not always to the same extent as a complete chd1Δ (Simic et al. 2003).
To examine the contribution of the individual CDs, we generated individual deletions of CDs 1 and 2 (ΔCD1 and ΔCD2) as well as a mutation altering a single residue of CD2, Y316E, which was previously shown to disrupt binding to dimethylated H3K4 peptides in vitro (Pray-Grant et al. 2005). Western blot analyses indicated that these altered forms of HA3-Chd1 were expressed at wild-type levels (data not shown). Like the double chromodomain deletion (ΔCD), alterations of individual Chd1 chromodomains suppressed the growth defect of the spt5-242 mutation moderately at 15° and as well as chd1Δ at 22° (Figure 5B and data not shown), implying that deletion of either CD is genetically equivalent to deletion of both and that the CDs play an important role in Chd1 function.
To ask whether these genetic interactions reflect a failure of Chd1 to associate with chromatin, we performed ChIP assays with the chromodomain deletion forms of HA3-Chd1. Deletion of both CDs, deletion of CD2 alone, and the Y316E mutation all decreased the HA3-Chd1 ChIP signal over PMA1 and TEF2 to background levels (Figure 5, C and D). Deletion of CD1 resulted in a similar decrease at PMA1, but only a partial decrease over TEF2. These data suggest that CD2 is essential and CD1 is important for the association of Chd1 with actively transcribed chromatin.
We next examined the helicase and C-terminal domains of Chd1. To determine whether Chd1's ATPase activity is required for its association with chromatin, we performed ChIP assays with the HA3-K407R form of Chd1, which is expressed at wild-type levels and suppresses spt5-242. Lysine 407 falls in the adenine nucleotide-binding motif of Chd1, and similar substitutions in other chromatin remodeling enzymes abolishes their ATP-binding and remodeling activities (Corona et al. 1999). Like the CD deletion mutants, we found this mutation to also cause a dramatic decrease in Chd1 levels over the PMA1 and TEF2 ORFs (Figure 5, C and D). Consistent with our data, mutations that alter the helicase or chromodomains of mouse Chd1 alter its nuclear distribution in cultured cells (Kelley et al. 1999). Finally, we examined a truncated form of Chd1 lacking most of the putative C-terminal DNA-binding domain (HA3-ΔSphI, Δaa 1083–1468) and found that it also showed a decreased association of Chd1 with PMA1 and TEF2 (Figure 5, C and D).
In each of the experiments above, mutations in the chromodomains, ATP-binding site and C terminus of Chd1 gave a similar set of loss-of-function phenotypes. To further examine the effects of these chd1 mutations on gene expression, we performed DNA microarray analysis of each of them. We compared the expression profiles of these mutants to a set of profiles from 80 mutants defective for a wide variety of functions in gene expression. Hierarchical clustering of the resulting expression data showed no striking differences between these chd1 mutants in overall gene expression or pre-mRNA splicing (data not shown). In summary, both genetic and DNA microarray analyses suggest that mutation of any one of Chd1's three conserved domains either leads to a nonfunctional protein or prevents Chd1 from reaching its site of action.
Chd1 recruitment does not generally depend upon H3K4 or H3K36 methylation:
We next considered the hypothesis that suppression of spt5-242 by set1, set2, H3K4, and H3K36 mutations stems from a failure to recruit Chd1 to transcribed chromatin. This hypothesis was suggested by previous observations of chromodomains that bind methylated histone tails (Bannister et al. 2001; Lachner et al. 2001; Cao et al. 2002). Furthermore, one report suggests that yeast Chd1 may bind to H3 tails methylated at lysine 4 (Pray-Grant et al. 2005), but others argue that although human Chd1 binds H3K4 methylated tails, yeast Chd1 lacks this activity (Flanagan et al. 2005, 2007; Sims et al. 2005). We therefore performed ChIP of HA3-Chd1 from strains carrying the set1Δ or set2Δ mutations, or from strains that expressed the H3K4R or H3K36R as their only source of histone H3. However, the resulting data did not show consistent changes in the association of HA3-Chd1 with transcribed chromatin (data not shown).
Genetic evidence that the Rpd3S complex opposes Spt4–Spt5 function:
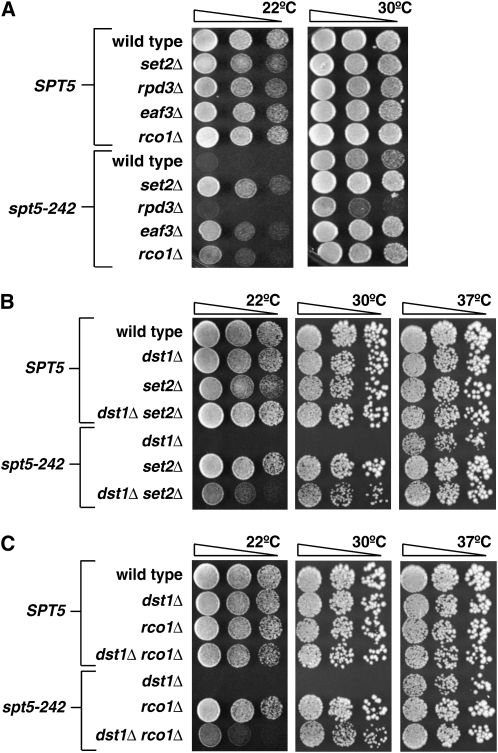
If altered H3K4 or K36 methylation does not result in decreased recruitment of Chd1 to chromatin, then what mechanism might explain suppression of spt5-242 when H3K4 or K36 methylation is perturbed? In the case of H3K36, a potential explanation is provided by the observation that nucleosomes methylated at H3K36 are targeted by the Rpd3S histone deacetylase complex (Carrozza et al. 2005; Keogh et al. 2005). This raises the possibility that the genetic interaction between SET2 and SPT5 reflects a role for the Rpd3S complex in transcription elongation and Spt5 function. To test this genetically, we crossed the spt5-242 strain to strains lacking Rpd3, the catalytic subunit of the Rpd3S complex (Figure 6A). The resulting double mutant did not grow at 22° and grew more poorly than spt5-242 alone at 30°. This enhancement of the spt5-242 growth defect could be due to Rpd3's participation in multiple histone deacetylase complexes (Carrozza et al. 2005; Keogh et al. 2005). Thus, we examined mutations of two other genes. One, EAF3 encodes a chromodomain protein that binds H3K36 methylated nucleosomes and is found in the Rpd3S histone deacetylase and NuA4 histone acetyltransferase complexes (Eisen et al. 2001; Carrozza et al. 2005; Joshi and Struhl 2005; Keogh et al. 2005; Li et al. 2007b). The other, RCO1, encodes a protein that interacts with Eaf3 and is required for proper Rpd3S complex assembly (Carrozza et al. 2005). Both eaf3Δ and rco1Δ mutations suppressed spt5-242's Cs− growth defect (Figure 6A). Because Rco1 is found exclusively in the Rpd3S complex (Carrozza et al. 2005; Keogh et al. 2005), these data suggest a role for the Rpd3S complex in Spt5 function.
Figure 6.—
Mutations that disrupt Rpd3S function suppress spt5-242. (A) To determine whether loss or reduced recruitment of the Rpd3S complex leads to suppression of spt5-242, strains carrying set2Δ, rpd3Δ, eaf3Δ, or rco1Δ mutations in combination with wild-type SPT5 or spt5-242 were spotted on YPD and incubated at 22° or 30° for 2 days. (B) Loss of Set2 suppresses the growth defect of spt5-242 dst1Δ double mutants. Strains carrying the indicated combinations of set2Δ, dst1Δ, and spt5-242 mutations were serially diluted on YPD media and incubated at 22°, 30°, or 37° for 3 days. (C) Loss of Rco1, a subunit of Rpd3S, suppresses the growth defect of spt5-242 dst1Δ double mutants. Strains carrying the indicated combinations of rco1Δ, dst1Δ, and spt5-242 mutations were serially diluted on YPD media and incubated at 22°, 30°, or 37° for 3 days.
We next performed genetic crosses to create spt5-242 dst1Δ set2Δ and spt5-242 dst1Δ rco1Δ mutants. As with chd1Δ, we found that the set2Δ and rco1Δ mutations suppressed the growth defects of the spt5-242 dst1Δ mutant (Figure 6, B and C). Thus, like chd1Δ, suppression of spt5-242 by loss of Set2 or the Rpd3S complex occurs by a pathway that is distinct from that used by elongation defective forms of RNAPII.
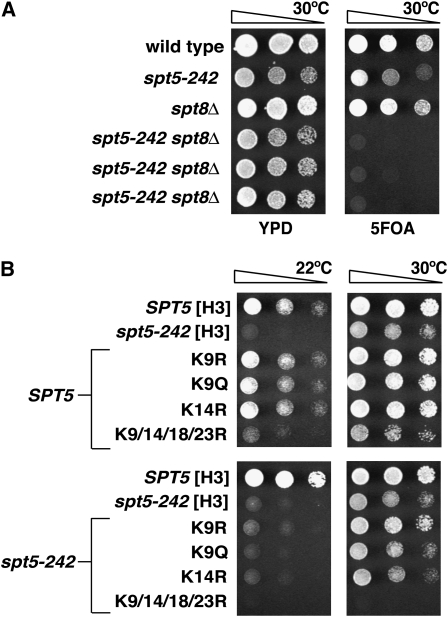
If loss of the Rpd3S complex, which leads to hyperacetylation of histones (Carrozza et al. 2005; Joshi and Struhl 2005; Keogh et al. 2005), suppresses spt5-242, we would expect that decreased histone acetylation should enhance spt5-242 phenotypes. To test this idea, we crossed spt5-242 to deletions of GCN5, SPT3, SPT7, and SPT8, genes encoding subunits of the SAGA histone acetyltransferase (Grant et al. 1997). In each case, the double mutants derived from these crosses failed to grow at 22° and grew very poorly at 30° (Figure 7A and data not shown). In addition, we found that spt5-242 cells grew poorly when their only source of histone H3 was H3K9/14R or H3K9/14/18/23R (Figure 7B). These observations suggest that histone acetylation plays an important role in transcription elongation, particularly when Spt5 function is compromised.
Figure 7.—
Mutations that interfere with histone acetylation do not suppress spt5-242. (A) Loss of the SAGA subunit Spt8 decreases the viability of spt5-242 mutants. A strain containing spt5-242 was crossed to a strain lacking SPT8. The resulting diploid was transformed with a URA3 SPT5 plasmid and then followed through sporulation. The parental strains, a wild-type control, and three representative double mutants were selected and spotted to YPD and 5FOA and incubated at 30° for 2 days. (B) Mutation of H3K9 or H3K14 fails to suppress spt5-242, but mutation of H3K9/14/18/23 leads to enhanced spt5-242 phenotypes. SPT5 and spt5-242 strains carrying deletions of both histone H3–H4 loci and a URA3 HHT1-HHF1 plasmid were transformed with plasmids carrying the indicated histone H3 allele. Trp+ transformants were spotted directly to 5FOA and incubated at 30° for 2 days or 22° for 3 days.
A role for Chd1 in prevention of cryptic transcription initiation:
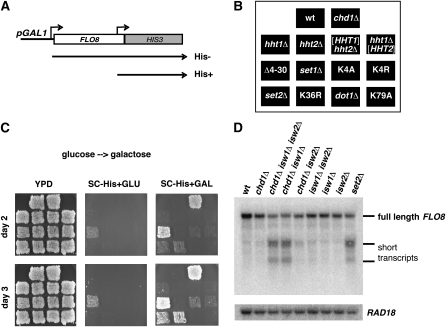
The Rpd3S complex is implicated in maintaining normal chromatin structure over transcribed regions, and loss of Rpd3S function leads to transcription initiation from cryptic internal promoters (Carrozza et al. 2005; Li et al. 2007b; Cheung et al. 2008). To assay for this defect in chd1Δ cells, we used a genetic reporter of cryptic initiation over the FLO8 gene, which has been extensively characterized for cryptic initiation (Kaplan et al. 2003; Carrozza et al. 2005; Cheung et al. 2008). In the reporter, FLO8 transcription is driven by the strong GAL1 promoter and the 3′ end of FLO8 is replaced by the HIS3 gene (Figure 8A; Cheung et al. 2008). Transcription initiation at the normal FLO8 start site produces a transcript in which the HIS3 open reading frame is out of frame and not translated. Internal initiation at FLO8, however, allows translation of a functional His3 protein, complementing the His− phenotype of the reporter strain. Consistent with previous reports, we found that set2 deletion and H3K36R substitution mutations gave strong His+ phenotypes in the reporter strain. In contrast, a set1 null mutation gave a His− phenotype and H3K4 substitutions gave weak His+ phenotypes that were only apparent after 4–5 days incubation. Interestingly, deletion of the N-terminal tail of histone H3 gave a strong His+ phenotype. Finally, we observed that a chd1 null mutation gave a strong His+ phenotype (Figure 8C).
Figure 8.—
Cryptic, internal initiation of transcription in a chd1 mutant. Two approaches were used to determine whether loss of Chd1 leads to the appearance of cryptic, internally initiated transcripts over transcribed sequences. (A) Diagram of the pGAL1-FLO8-HIS3 reporter gene. Transcription initiation from the normal FLO8 start site produces a transcript in which HIS3 is out of frame and not translated. Internal initiation of this transcript produces in frame transcripts and a His+ phenotype. (B) Diagram showing the pattern of cells carrying the pGAL1-FLO8-HIS3 reporter and indicated genotypes that were patched onto YPD media. The strains labeled hht1Δ or hht2Δ lack one HHT-HHF locus. [HHT1] and [HHT2] refer to a strain with deletions of both HHT–HHF loci complemented by a plasmid-borne copy of one of these two loci; in the patch labeled Δ4-30, this strain carries an hht2–HHF2 plasmid, carrying a deletion of codons 4–30 of histone H3. Patches K4A and K4R indicate similar strains with plasmids carrying H3K4 mutations. (C) The YPD plate described in B was replica plated onto SC −His media utilizing either glucose or galactose as the carbon source and incubated at 30° for the indicated number of days. (D) RNA was isolated from the indicated strains and subjected to Northern blot analysis to detect short transcripts from STE11 (top) and FLO8 (middle). Hybridization to a RAD18 probe (bottom) was used as a loading control.
These observations suggest that the chd1 mutation causes internal initiation of transcription from the FLO8-HIS3 reporter. To test for internal initiation defects in the chromosomal FLO8 gene, we assayed Northern blots of wild-type and mutant strains with probes derived from the 3′ end of FLO8, but did not observe clear evidence of internal initiation (Figure 8D). This observation mirrors recent results of Winston and colleagues, who observed a strong effect of a chd1 deletion on the FLO8-HIS3 reporter but very weak effects on internal initiation of normal chromosomal genes as measured by Northern blots (Cheung et al. 2008). We reasoned that the internal initiation defect caused by a chd1Δ mutation might be weak on its own, but enhanced in the presence of other mutations. This phenomenon has been observed for several mutations that activate the FLO8-HIS3 reporter, but only show clear molecular evidence of internal initiation when combined with other mutations (Prather et al. 2005; Nourani et al. 2006; Chu et al. 2007). Because previous reports have shown synthetic growth defects and chromatin disruption in an isw1Δ isw2Δ chd1Δ mutant (Tsukiyama et al. 1999; Xella et al. 2006), we analyzed RNA from strains carrying all combinations of chd1Δ, isw1Δ, and isw2Δ mutations (Figure 8D). We observed that the chd1 mutation showed a strong synthetic cryptic initiation defect when combined with isw1Δ. Furthermore, we obtained similar results when we probed Northern blots for STE11; neither the isw1Δ nor chd1Δ single mutants displayed internal initiation, but the isw1Δ chd1Δ mutant produced a strong internally initiated STE11 transcript (data not shown).
Increased histone acetylation in chd1Δ strains:
We next asked whether the cryptic initiation defect of a chd1Δ mutation reflected altered histone methylation. Western blot analysis of crude extracts revealed no obvious changes in modifications H3K4me3, H3K36me3, H3K79me2, and H3K9/14Ac (data not shown). We performed ChIP with H3K4me3 and H3K36me3 antibodies and did not observe alterations of these methyl marks in a chd1Δ strain. In contrast, when we performed ChIP with antibodies directed against histone H3 acetylated at lysines 9 and 14, we observed increased acetylation in chd1 and set2 mutants (Figure 9). Consistent with the relatively weak cryptic initiation phenotype of chd1Δ, the increase in acetylation observed at FLO8 and STE11 in the chd1Δ strain was less than that observed for a set2Δ strain.
DISCUSSION
Spt5 is an essential and highly conserved transcription elongation factor that is a core component of transcribing RNAPII. In this work, we have used a unique mutation in the yeast SPT5 gene to genetically probe the RNAPII transcription apparatus. This mutation, spt5-242, causes transcription defects and a cold-sensitive growth defect that can be suppressed by conditions that decrease the rate of transcription elongation. The data presented here show that there is a second mechanism for suppression of spt5-242 that involves particular perturbations of chromatin. Thus, there appear to be two distinct paths to suppression of spt5-242, via decreased elongation rate or alteration of chromatin.
What mechanisms might explain these two pathways to suppression of spt5-242? Several groups have previously argued that Set2, Chd1, and Rpd3S oppose transcription (Carrozza et al. 2005; Joshi and Struhl 2005; Keogh et al. 2005; Kizer et al. 2005; Biswas et al. 2006, 2007). Evidence for this idea includes the observations that mutations in SET2, CHD1, and genes encoding subunits of Rpd3S display partial resistance to the elongation inhibitor 6AU (Woodage et al. 1997; Keogh et al. 2005; Kizer et al. 2005; Biswas et al. 2006). In addition, similar to the results presented here for suppression of spt5-242, set2, rpd3S, and chd1 mutations genetically suppress mutations affecting FACT and the Bur1 kinase (Keogh et al. 2005; Biswas et al. 2006, 2007; Chu et al. 2006). Thus, the genetic relationships between Spt4–Spt5 and chromatin modifiers presented here likely reflect a general set of antagonistic relationships between chromatin and transcription elongation factors (Figure 8).
One clue to a potential biochemical explanation for these relationships is provided by our observations that chromatin-based suppressors of spt5-242 also alleviated the growth defect of an spt5-242 dst1Δ mutant. In contrast, the rpb1 suppressors of spt5-242 display synthetic sickness or lethality when combined with dst1Δ and do not overcome spt5-242 dst1Δ growth defects (Figure 1; Hartzog et al. 1998). These observations are consistent with a model in which our chromatin suppressor mutations decrease the probability of transcription arrest and the need for TFIIS in vivo. Although TFIIS has been recently implicated in transcription initiation as well as elongation (reviewed in Sikorski and Buratowski 2009), several observations show that Spt4–Spt5's functions are restricted to postinitiation events. First, Spt4–Spt5 does not join transcription complexes until after promoter clearance (Ping and Rana 2001; Bourgeois et al. 2002) and only exerts its negative effect on transcription after the nascent transcript has been extended 50–60 nucleotides (Yamaguchi et al. 1999; Bourgeois et al. 2002). Second, Spt4–Spt5 does not alter initiation in vitro (Guo et al. 2000; Bourgeois et al. 2002; Yamada et al. 2006; Zhu et al. 2007). Third, although proteomic studies have revealed a myriad of interactions between Spt4–Spt5 and RNAPII, elongation factors and RNA processing factors, Spt4–Spt5 has not been observed to physically associate with initiation-specific factors such as the mediator complex (Krogan et al. 2002, 2006; Lindstrom et al. 2003; Gavin et al. 2006). Finally, using ChIP of RNAPII, two groups have observed decreased processivity of RNAPII in spt4 mutants (Morillon et al. 2003; Mason and Struhl 2005). Thus, the increased dependence of spt4 and spt5 mutants on TFIIS function is consistent with the idea that transcription arrest occurs more frequently in the absence of normal Spt4–Spt5 function. Furthermore, barriers to RNAPII elongation, such as nucleosomes, are known to provoke transcription arrest (Fish and Kane 2002; Kireeva et al. 2005). Thus we believe that the chromatin-based suppressors disrupt, fail to reassemble, or alter the stability, spacing or location of nucleosomes, which would otherwise lead to increased dependence upon TFIIS, perhaps due to transcription arrest in spt5-242 cells.
How is Chd1 recruited to chromatin? Given the shared phenotypes of chd1, set1, and set2 mutants, and that Chd1 contains chromodomains, an attractive model is that the H3K4me or H3K36me marks are used to recruit Chd1 to chromatin. Chromatin immunoprecipitation experiments and staining of Drosophila polytene chromosomes indicate that Chd1 associates with actively transcribed genes (Stokes et al. 1996; Simic et al. 2003; Srinivasan et al. 2005; McDaniel et al. 2008). However, the mechanism by which Chd1 is recruited to these loci is not clear. In humans, in vitro data show that the chromodomains recognize H3K4 methylated nucleosomes (Flanagan et al. 2005). However, there are conflicting data regarding the ability of yeast Chd1 to bind H3K4 methylated nucleosomes (Santos-Rosa et al. 2003; Pray-Grant et al. 2005; Sims et al. 2005; Flanagan et al. 2007; McDaniel et al. 2008). Furthermore, genetic data indicate that H3K4 methylation is unlikely to be the sole determinant of Chd1's localization; in Drosophila, mutations in the Kismet gene prevent association of Chd1 with polytene chromosomes (Srinivasan et al. 2005) but have no apparent affect on H3K4me levels (Srinivasan et al. 2008), and mutations of the H3K4 demethylase, Lid, result in increased H3K4me without a clear increase in Chd1 levels on polytene chromosomes (Eissenberg et al. 2007). Consistent with these observations, Stillman and colleagues have observed that set1 and H3K4 mutations enhance phenotypes of spt16 mutations whereas chd1 mutations suppress spt16 phenotypes, suggesting distinct functions for Chd1 and H3K4 methylation (Biswas et al. 2006, 2007, 2008). Our data suggest that methylation of histone H3 does not have a strong effect on Chd1's localization. In contrast, the rtf1Δ1 mutation, which abolishes Chd1–Rtf1 interactions, was a strong suppressor of spt5-242. These data suggest that Chd1 is initially recruited to transcription complexes via an interaction with Rtf1. It remains possible that H3K4 or H3K36 methylation may affect some other aspect of Chd1 function, such as the persistence of its association with a gene, its ability to maintain contact with histone H3 during remodeling, or its rate of action. The exact relationships between Set1 and H3K4 methylation, Set2 and H3K36 methylation, and Chd1 are difficult to determine. The strong suppression of spt5-242 by set2, chd1, and H3K36 mutations prevented clear determination of whether or not set1 mutations could act additively with these other suppressors of spt5-242.
What is the function of Chd1? Chd1 mutants display few growth defects and only modest defects in gene expression (Tran et al. 2000), suggesting that Chd1 does not play a direct or critical role in transcription. Several reports implicate Chd1 in ATP-dependent nucleosome assembly and spacing (Tran et al. 2000; Robinson and Schultz 2003; Lusser et al. 2005; Stockdale et al. 2006), suggesting a role in establishment or maintenance of chromatin structure over transcribed genes. However, chd1 mutants do not display obvious defects in bulk or transcribed chromatin structure (Xella et al. 2006), although it is possible that Chd1 plays a redundant or transient role in establishing or maintaining chromatin structures during transcription.
Another possibility is that Chd1 helps maintain epigenetic information, i.e., histones with specific post-translational marks, over transcribed sequences. We did not observe any clear alterations in levels of H3K4me3 or H3K36me3 in Western blots of chd1Δ cell extracts or by ChIP with antisera directed against these modifications, suggesting that Chd1 does not regulate these histone modifications (T. K. Quan and G. A. Hartzog, unpublished data). However, the observation that disruption of H3K36 methylation suppresses spt5-242 suggested a link to histone acetylation over transcribed genes via the Rpd3S complex. In support of this idea, we found that disruption of Rpd3S is sufficient to suppress spt5-242. In contrast, disruption of the SAGA histone acetyltransferase, or H3 residues targeted by SAGA, enhances the phenotypes of an spt5-242 mutant. Furthermore, just as disruption of the Rpd3S complex leads to inappropriate transcription initiation events from the middle of genes, we found that a chd1 mutation strongly activates a genetic reporter of cryptic initiation and in Northern blot analysis, causes internal initiation when combined with an isw1 mutation. Although it is intriguing to note that Isw1 apparently binds to di- and trimethylated H3K4 and depends upon Set1 for its association with chromatin (Santos-Rosa et al. 2003), we do not observe suppression of spt5-242 by an isw1 mutation (T. K. Quan and G. A. Hartzog, unpublished data). Internal transcription initiation per se is unlikely to explain suppression of spt5-242 by chd1Δ as a number of other mutations that give internal initiation phenotypes, spt6, H2BK123R, spt16, do not suppress spt5-242 (T. K. Quan and G. A. Hartzog, unpublished data). The observation that H3K9/14Ac is increased over transcribed sequences in a chd1 mutant may indicate a role for Chd1 in recruitment of Rpd3S to chromatin, or in the dynamics of acetylated histones. Our working model is that Rpd3S, Chd1, and methylation of H3K4 and H3K36 play important roles in maintaining normal chromatin structures over transcribed regions. Loss of these factors or modifications reduces the dependence of elongating RNAPII on Spt4–Spt5 and other positively acting elongation factors at the price of an increased probability of transcription initiation from cryptic promoters.
Acknowledgments
We gratefully acknowledge Ines Pinto, Fred Winston, Patrick Grant, Kevin Struhl, Sharon Dent, Mary Ann Osley, Mary Bryk, LeAnne Howe, and Karen Arndt for their gifts of plasmids and strains, Craig Kaplan for many helpful suggestions, and Karen Arndt for her comments on the manuscript. This work was supported by a National Institutes of Health (NIH) grant GM060479 (to G.A.H.). T.K.Q. was supported in part by NIH grant T32 GM08646.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.111526/DC1.
References
- Andrulis, E. D., E. Guzman, P. Doring, J. Werner and J. T. Lis, 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14 2635–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al., 1991. Current Protocols in Molecular Biology. Greene Publishing Associates and Wiley-Interscience, New York.
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas et al., 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410 120–124. [DOI] [PubMed] [Google Scholar]
- Biswas, D., R. Dutta-Biswas, D. Mitra, Y. Shibata, B. D. Strahl et al., 2006. Opposing roles for Set2 and yFACT in regulating TBP binding at promoters. EMBO J. 25 4479–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, D., R. Dutta-Biswas and D. J. Stillman, 2007. Chd1 and yFACT act in opposition in regulating transcription. Mol. Cell. Biol. 27 6279–6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, D., S. Takahata, H. Xin, R. Dutta-Biswas, Y. Yu et al., 2008. A role for Chd1 and Set2 in negatively regulating DNA replication in Saccharomyces cerevisiae. Genetics 178 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois, C. F., Y. K. Kim, M. J. Churcher, M. J. West and J. Karn, 2002. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol. Cell. Biol. 22 1079–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheli, M. E., and S. Buratowski, 2005. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. EMBO J. 24 2150–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckin, T., R. Nagel, Y. Mandel-Gutfreund, L. Shiue, T. A. Clark et al., 2005. Exploring functional relationships between components of the gene expression machinery. Nat. Struct. Mol. Biol. 12 175–182. [DOI] [PubMed] [Google Scholar]
- Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage et al., 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298 1039–1043. [DOI] [PubMed] [Google Scholar]
- Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson et al., 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123 581–592. [DOI] [PubMed] [Google Scholar]
- Cheung, V., G. Chua, N. N. Batada, C. R. Landry, S. W. Michnick et al., 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6 e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, Y., R. Simic, M. H. Warner, K. M. Arndt and G. Prelich, 2007. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 26 4646–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, Y., A. Sutton, R. Sternglanz and G. Prelich, 2006. The BUR1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by SET2. Mol. Cell. Biol. 26 3029–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona, D. F., G. Langst, C. R. Clapier, E. J. Bonte, S. Ferrari et al., 1999. ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell 3 239–245. [DOI] [PubMed] [Google Scholar]
- Cui, Y., and C. L. Denis, 2003. In vivo evidence that defects in the transcriptional elongation factors RPB2, TFIIS, and SPT5 enhance upstream poly(A) site utilization. Mol. Cell. Biol. 23 7887–7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover, J., J. Schneider, M. A. Tawiah-Boateng, A. Wood, K. Dean et al., 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277 28368–28371. [DOI] [PubMed] [Google Scholar]
- Eisen, A., R. T. Utley, A. Nourani, S. Allard, P. Schmidt et al., 2001. The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J. Biol. Chem. 276 3484–3491. [DOI] [PubMed] [Google Scholar]
- Eissenberg, J. C., M. G. Lee, J. Schneider, A. Ilvarsonn, R. Shiekhattar et al., 2007. The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nat. Struct. Mol. Biol. 14 344–346. [DOI] [PubMed] [Google Scholar]
- Exinger, F., and F. Lacroute, 1992. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 22 9–11. [DOI] [PubMed] [Google Scholar]
- Fish, R. N., and C. M. Kane, 2002. Promoting elongation with transcript cleavage stimulatory factors. Biochim. Biophys. Acta 1577 287–307. [DOI] [PubMed] [Google Scholar]
- Flanagan, J. F., L. Z. Mi, M. Chruszcz, M. Cymborowski, K. L. Clines et al., 2005. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438 1181–1185. [DOI] [PubMed] [Google Scholar]
- Flanagan, J. F., B. J. Blus, D. Kim, K. L. Clines, F. Rastinejad et al., 2007. Molecular implications of evolutionary differences in CHD double chromodomains. J. Mol. Biol. 369 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming, A. B., C. F. Kao, C. Hillyer, M. Pikaart and M. A. Osley, 2008. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell 31 57–66. [DOI] [PubMed] [Google Scholar]
- Gavin, A. C., P. Aloy, P. Grandi, R. Krause, M. Boesche et al., 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440 631–636. [DOI] [PubMed] [Google Scholar]
- Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell et al., 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11 1640–1650. [DOI] [PubMed] [Google Scholar]
- Guo, S., Y. Yamaguchi, S. Schilbach, T. Wada, J. Lee et al., 2000. A regulator of transcriptional elongation controls vertebrate neuronal development. Nature 408 366–369. [DOI] [PubMed] [Google Scholar]
- Hartzog, G. A., T. Wada, H. Handa and F. Winston, 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog, G. A., J. L. Speer and D. L. Lindstrom, 2002. Transcript elongation on a nucleoprotein template. Biochim. Biophys. Acta 1577 276–286. [DOI] [PubMed] [Google Scholar]
- Joshi, A. A., and K. Struhl, 2005. Eaf3 chromodomain interaction with methylated H3–K36 links histone deacetylation to Pol II elongation. Mol. Cell 20 971–978. [DOI] [PubMed] [Google Scholar]
- Kaplan, C. D., L. Laprade and F. Winston, 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301 1096–1099. [DOI] [PubMed] [Google Scholar]
- Kaplan, C. D., M. J. Holland and F. Winston, 2005. Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10–GAL7 locus. J. Biol. Chem. 280 913–922. [DOI] [PubMed] [Google Scholar]
- Kelley, D. E., D. G. Stokes and R. P. Perry, 1999. CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma 108 10–25. [DOI] [PubMed] [Google Scholar]
- Keogh, M. C., S. K. Kurdistani, S. A. Morris, S. H. Ahn, V. Podolny et al., 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123 593–605. [DOI] [PubMed] [Google Scholar]
- Kim, M., S. H. Ahn, N. J. Krogan, J. F. Greenblatt and S. Buratowski, 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23 354–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva, M. L., B. Hancock, G. H. Cremona, W. Walter, V. M. Studitsky et al., 2005. Nature of the nucleosomal barrier to RNA polymerase II. Mol. Cell 18 97–108. [DOI] [PubMed] [Google Scholar]
- Kizer, K. O., H. P. Phatnani, Y. Shibata, H. Hall, A. L. Greenleaf et al., 2005. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 25 3305–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky, P., E. J. Cho and S. Buratowski, 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor et al., 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22 6979–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt et al., 2003. a The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11 721–729. [DOI] [PubMed] [Google Scholar]
- Krogan, N. J., M. Kim, A. Tong, A. Golshani, G. Cagney et al., 2003. b Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23 4207–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo et al., 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440 637–643. [DOI] [PubMed] [Google Scholar]
- Lachner, M., D. O'Carroll, S. Rea, K. Mechtler and T. Jenuwein, 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410 116–120. [DOI] [PubMed] [Google Scholar]
- Lennon, 3rd, J. C., M. Wind, L. Saunders, M. B. Hock and D. Reines, 1998. Mutations in RNA polymerase II and elongation factor SII severely reduce mRNA levels in Saccharomyces cerevisiae. Mol. Cell. Biol. 18 5771–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., M. Carey and J. L. Workman, 2007. a The role of chromatin during transcription. Cell 128 707–719. [DOI] [PubMed] [Google Scholar]
- Li, B., M. Gogol, M. Carey, D. Lee, C. Seidel et al., 2007. b Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 316 1050–1054. [DOI] [PubMed] [Google Scholar]
- Lindstrom, D. L., S. L. Squazzo, N. Muster, T. A. Burckin, K. C. Wachter et al., 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23 1368–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser, A., D. L. Urwin and J. T. Kadonaga, 2005. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struct. Mol. Biol. 12 160–166. [DOI] [PubMed] [Google Scholar]
- Mason, P. B., and K. Struhl, 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 23 8323–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, P. B., and K. Struhl, 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell 17 831–840. [DOI] [PubMed] [Google Scholar]
- McDaniel, I. E., J. M. Lee, M. S. Berger, C. K. Hanagami and J. A. Armstrong, 2008. Investigations of CHD1 function in transcription and development of Drosophila melanogaster. Genetics 178 583–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon, A., N. Karabetsou, J. O'Sullivan, N. Kent, N. Proudfoot et al., 2003. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell 115 425–435. [DOI] [PubMed] [Google Scholar]
- Ng, H. H., R. M. Xu, Y. Zhang and K. Struhl, 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277 34655–34657. [DOI] [PubMed] [Google Scholar]
- Ng, H. H., F. Robert, R. A. Young and K. Struhl, 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11 709–719. [DOI] [PubMed] [Google Scholar]
- Nourani, A., F. Robert and F. Winston, 2006. Evidence that Spt2/Sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure, and genome stability in Saccharomyces cerevisiae. Mol. Cell. Biol. 26 1496–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri, R., B. Zhu, G. Li, P. Trojer, S. Mandal et al., 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125 703–717. [DOI] [PubMed] [Google Scholar]
- Ping, Y. H., and T. M. Rana, 2001. DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J. Biol. Chem. 276 12951–12958. [DOI] [PubMed] [Google Scholar]
- Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett et al., 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122 517–527. [DOI] [PubMed] [Google Scholar]
- Powell, W., and D. Reines, 1996. Mutations in the second largest subunit of RNA polymerase II cause 6-azauracil sensitivity in yeast and increased transcriptional arrest in vitro. J. Biol. Chem. 271 6866–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather, D., N. J. Krogan, A. Emili, J. F. Greenblatt and F. Winston, 2005. Identification and characterization of Elf1, a conserved transcription elongation factor in Saccharomyces cerevisiae. Mol. Cell. Biol. 25 10122–10135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant, M. G., J. A. Daniel, D. Schieltz, J. R. Yates, 3rd and P. A. Grant, 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433 434–438. [DOI] [PubMed] [Google Scholar]
- Rando, O. J., and K. Ahmad, 2007. Rules and regulation in the primary structure of chromatin. Curr. Opin. Cell Biol. 19 250–256. [DOI] [PubMed] [Google Scholar]
- Robinson, K. M., and M. C. Schultz, 2003. Replication-independent assembly of nucleosome arrays in a novel yeast chromatin reconstitution system involves antisilencing factor Asf1p and chromodomain protein Chd1p. Mol. Cell. Biol. 23 7937–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M. D., F. Winston and P. Hieter, 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Santos-Rosa, H., R. Schneider, B. E. Bernstein, N. Karabetsou, A. Morillon et al., 2003. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell 12 1325–1332. [DOI] [PubMed] [Google Scholar]
- Saunders, A., L. J. Core and J. T. Lis, 2006. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell. Biol. 7 557–567. [DOI] [PubMed] [Google Scholar]
- Sikorski, T. W., and S. Buratowski, 2009. The basal initiation machinery: beyond the general transcription factors. Curr. Opin. Cell Biol. 21 344–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic, R., D. L. Lindstrom, H. G. Tran, K. L. Roinick, P. J. Costa et al., 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22 1846–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims, 3rd, R. J., C. F. Chen, H. Santos-Rosa, T. Kouzarides, S. S. Patel et al., 2005. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 280 41789–41792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic et al., 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21 1764–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, S., J. A. Armstrong, R. Deuring, I. K. Dahlsveen, H. McNeill et al., 2005. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development 132 1623–1635. [DOI] [PubMed] [Google Scholar]
- Srinivasan, S., K. M. Dorighi and J. W. Tamkun, 2008. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet. 4 e1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockdale, C., A. Flaus, H. Ferreira and T. Owen-Hughes, 2006. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J. Biol. Chem. 281 16279–16288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes, D. G., K. D. Tartof and R. P. Perry, 1996. CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc. Natl. Acad. Sci. USA 93 7137–7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studitsky, V. M., W. Walter, M. Kireeva, M. Kashlev and G. Felsenfeld, 2004. Chromatin remodeling by RNA polymerases. Trends Biochem. Sci. 29 127–135. [DOI] [PubMed] [Google Scholar]
- Sun, Z. W., and C. D. Allis, 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418 104–108. [DOI] [PubMed] [Google Scholar]
- Swanson, M. S., E. A. Malone and F. Winston, 1991. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol. Cell. Biol. 11 3009–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, M. S., and F. Winston, 1992. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics 132 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, H. G., D. J. Steger, V. R. Iyer and A. D. Johnson, 2000. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J. 19 2323–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama, T., J. Palmer, C. C. Landel, J. Shiloach and C. Wu, 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai et al., 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, M. H., K. L. Roinick and K. M. Arndt, 2007. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol. Cell. Biol. 27 6103–6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, S. K., and J. K. Tyler, 2007. Transcriptional regulation by chromatin disassembly and reassembly. Curr. Opin. Genet. Dev. 17 88–93. [DOI] [PubMed] [Google Scholar]
- Winston, F., C. Dollard and S. L. Ricupero-Hovasse, 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11 53–55. [DOI] [PubMed] [Google Scholar]
- Wood, A., J. Schneider, J. Dover, M. Johnston and A. Shilatifard, 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278 34739–34742. [DOI] [PubMed] [Google Scholar]
- Woodage, T., M. A. Basrai, A. D. Baxevanis, P. Hieter and F. S. Collins, 1997. Characterization of the CHD family of proteins. Proc. Natl. Acad. Sci. USA 94 11472–11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xella, B., C. Goding, E. Agricola, E. Di Mauro and M. Caserta, 2006. The ISWI and CHD1 chromatin remodelling activities influence ADH2 expression and chromatin organization. Mol. Microbiol. 59 1531–1541. [DOI] [PubMed] [Google Scholar]
- Xiao, Y., Y. H. Yang, T. A. Burckin, L. Shiue, G. A. Hartzog et al., 2005. Analysis of a splice array experiment elucidates roles of chromatin elongation factor Spt4–5 in splicing. PLoS Comput. Biol. 1 e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, T., Y. Yamaguchi, N. Inukai, S. Okamoto, T. Mura et al., 2006. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 21 227–237. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya et al., 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97 41–51. [DOI] [PubMed] [Google Scholar]
- Zhu, W., T. Wada, S. Okabe, T. Taneda, Y. Yamaguchi et al., 2007. DSIF contributes to transcriptional activation by DNA-binding activators by preventing pausing during transcription elongation. Nucleic Acids Res. 35 4064–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]