Abstract
Epigenetically inherited aggregates of the yeast prion [PSI+] cause genomewide readthrough translation that sometimes increases evolvability in certain harsh environments. The effects of natural selection on modifiers of [PSI+] appearance have been the subject of much debate. It seems likely that [PSI+] would be at least mildly deleterious in most environments, but this may be counteracted by its evolvability properties on rare occasions. Indirect selection on modifiers of [PSI+] is predicted to depend primarily on the spontaneous [PSI+] appearance rate, but this critical parameter has not previously been adequately measured. Here we measure this epimutation rate accurately and precisely as 5.8 × 10−7 per generation, using a fluctuation test. We also determine that genetic “mimics” of [PSI+] account for up to 80% of all phenotypes involving general nonsense suppression. Using previously developed mathematical models, we can now infer that even in the absence of opportunities for adaptation, modifiers of [PSI+] are only weakly deleterious relative to genetic drift. If we assume that the spontaneous [PSI+] appearance rate is at its evolutionary optimum, then opportunities for adaptation are inferred to be rare, such that the [PSI+] system is favored only very weakly overall. But when we account for the observed increase in the [PSI+] appearance rate in response to stress, we infer much higher overall selection in favor of [PSI+] modifiers, suggesting that [PSI+]-forming ability may be a consequence of selection for evolvability.
THE yeast phenotype [PSI+] is characterized by prion aggregates of the protein Sup35. Cells are in either a [psi−] (normal) or [PSI+] state, depending on the absence or presence of the prion aggregates (Figure 1, a and b). Sup35 prion aggregates replicate in a similar fashion to mammalian prions but are cytoplasmic and, as such, the prion state is cytoplasmically inherited (Wickner et al. 1995).
Figure 1.—
Comparison between the three possible modes ([PSI+], genetic mimic, point mutation revertant) of the expression of 3′-UTR sequences in yeast. (a) The normal [psi−] phenotypic state; (b) the [PSI+] prion causes readthrough and low-level expression of 3′-UTRs across multiple genes, appearing at rate mPSI; (c) a genetic mimic of [PSI+] such as the sal3-4 mutant of Sup35 (Eaglestone et al. 1999) appearing at rate mmimic not reversible by the application of guanidine hydrochloride; (d) a point mutation in a single stop codon at rate μpoint, leading to incorporation of formerly 3′-UTR into a single coding sequence. (e) [PSI+] can act as a “stop-gap” mechanism, buying a lineage more time to acquire one or more adaptive stop codon readthrough point mutations. When this genetic assimilation is complete, [PSI+] can revert to [psi−] (Masel and Bergman 2003; Griswold and Masel 2009).
When not part of an aggregate, Sup35 helps mediate translation termination in yeast (Stansfield et al. 1995b; Zhouravleva et al. 1995). Sup35 molecules that are incorporated into nonfunctional prion aggregates are presumably not available for translation termination, which can lead to the translation of stop codons by near-cognate tRNAs (Figure 1b) (Tuite and Mclaughlin 1982; Pure et al. 1985; Lin et al. 1986). This partial loss of Sup35 function leads to an increased frequency of readthrough translation of 3′-untranslated regions (3′-UTR) across all genes (Figure 1b). This increase is modest in wild-type yeast, from an average readthrough rate of 0.3% in [psi−] cells up to 1% in [PSI+] cells (Firoozan et al. 1991). Some [PSI+] yeast strains grow faster than [psi−] controls in certain harsh environments, suggesting that readthrough translation of some 3′-UTRs may be adaptive in certain conditions (True and Lindquist 2000; Joseph and Kirkpatrick 2008). This directly shows that [PSI+]-mediated capacitance may increase evolvability in the laboratory. [PSI+]-mediated phenotypes have a complex genetic basis, involving multiple loci (True et al. 2004).
As an epigenetically inherited protein aggregate, [PSI+] can easily be lost after some generations (Cox et al. 1980). This returns the lineage to its normal [psi−] state and restores translation fidelity. If a subset of revealed phenotypic variation is adaptive, it may have lost its dependence on [PSI+] by this time (True et al. 2004). This process of genetic assimilation may, for example, involve one or more point mutations in stop codons, increasing readthrough up to 100% (Figure 1e) (Griswold and Masel 2009). This leaves the yeast with a new adaptive trait and with no permanent load of other, deleterious variation.
In general, stop codons can be lost either directly through point mutations or indirectly through upstream indels. This leads to novel coding sequence coming from in-frame and out-of-frame 3′-UTRs, respectively. [PSI+] is expected to facilitate only the former, while mutation bias favors the latter. Yeasts show a much higher ratio of in-frame to out-of-frame 3′-UTR incorporation events than mammals do (Giacomelli et al. 2007), confirming a role for [PSI+] in capacitance-mediated evolvability in natural populations.
The adaptive evolution both of evolvability in general (Sniegowski and Murphy 2006; Lynch 2007; Pigliucci 2008) and of capacitance in particular (Dickinson and Seger 1999; Wagner et al. 1999; Partridge and Barton 2000; Brookfield 2001; Pal 2001; Meiklejohn and Hartl 2002; Ruden et al. 2003) is highly controversial. In general, any costs of evolvability are borne in the present, while the benefits lie in the future, making it difficult for natural selection to favor an evolvability allele. For example, mutation rates seem to be set according to a trade-off between metabolic cost (favoring higher mutation rates) and the avoidance of deleterious effects (favoring lower mutation rates) (Sniegowski et al. 2000). The fact that mutation creates variation, the ultimate source of evolvability, is merely a fortuitous consequence of the metabolic cost of fidelity.
Previous theoretical population genetic studies have, however, suggested that modifier alleles promoting the formation of [PSI+] might, unlike mutator alleles, be favored for their evolvability properties (King and Masel 2007; Masel et al. 2007; Griswold and Masel 2009; Masel and Griswold 2009). These models depend, however, on a number of parameter estimates. In particular, a number of predictions depend on the spontaneous rate of [PSI+] formation (Masel and Griswold 2009).
[PSI+] appearance rates and the fluctuation test:
The most widely cited spontaneous appearance rate for [PSI+] is mPSI ∼ 10−7–10−5, on the basis of experiments by Lund and Cox (1981). This estimate was calculated as the proportion of colonies scored as [PSI+] after growth over multiple generations from a single founding [psi−] clone. If [PSI+] happens to appear in the first generation of growth, this leads to a “jackpot” event with only one switching event, but many [PSI+] colonies. The proportion of colonies scored as [PSI+] therefore yields a systematic overestimation of the [PSI+] appearance rate.
Various implementations of the fluctuation test (Luria and Delbrück 1943) can address such effects. The mutation rate experiment is replicated many times using independent populations, and a Luria–Delbrück distribution is fitted to the results across all replicates. In a simulation study, Stewart (1994) examined a number of estimators of the underlying Luria–Delbrück distribution and found that the maximum-likelihood estimator performed the best.
Originally developed to study mutation rates, the fluctuation test can also be used for estimating epimutation rates. Fluctuation tests have been used to estimate the rate of gene silencing in Chinese hamster ovary cells (Holliday and Ho 1998) and in the yeast Schizosaccharomyces pombe (Singh and Klar 2002). However, fluctuation tests do not appear to be used routinely for epimutation rate estimates. For example, although the rates of spontaneous appearance and disappearance of [ISP+], a prion-like element in yeast, have been measured using the fluctuation test (Volkov et al. 2002), to the best of our knowledge there are no published estimates of the spontaneous rate of [PSI+] appearance as measured using a fluctuation test. Although results from the fluctuation test can be confounded by reverse epimutation, or back-switching, this is an issue only if the rate of back-switching is very high, e.g., 10−1–10−2 per generation (Saunders et al. 2003). This is not the case for [PSI+], for which the reverse epimutation rate (loss of [PSI+]) is <2 × 10−4 (Tank et al. 2007).
Other [PSI+]-like phenotypes, including genetic mimics:
[PSI+] causes partial loss of Sup35 function, leading to elevated rates of translational readthrough at all stop codons (Figure 1b). There are many other spontaneous changes, presumably mutations, that also lead to elevated translational readthrough (Lund and Cox 1981). Mutations that affect readthrough at all stop codons (Figure 1c) (sometimes called “[PSI+]-like”) can be considered as genetic “mimics” because they produce the same phenotype as the Sup35 aggregate, but are generally not epigenetically inherited. A specific example of such a genetic mimic was characterized by Eaglestone et al. (1999), who identified the sal3-4 point mutation in the SUP35 gene. This leads to a defect in the Sup35 protein structure rendering the termination process less efficient (Eaglestone et al. 1999). The sal3-4 mutant can therefore be considered a partial loss-of-function genetic mimic of [PSI+], since it generates the same readthrough phenotype. Translation termination could also potentially be impaired through other point mutations or deletions, for example, in either the SUP35 or the SUP45 gene (Stansfield et al. 1995a) or in a tRNA that mutates to recognize stop codons at a higher rate. The presence of genetic mimics, whose effects are less reversible than those of [PSI+], can affect the evolution of the evolvability properties of the [PSI+] system such as its epimutation rate (Lancaster and Masel 2009). Note that genetic mimics are quite different from much rarer point mutations that convert stop codons into coding sequence (Figure 1d), resulting in readthrough at a single gene rather than multiple genes.
Here we performed experiments to obtain accurate and precise estimates of the baseline appearance rates of both [PSI+] and [PSI+]-like phenotypes in permissive laboratory conditions, excluding stop codon point mutations that affect only a single gene. Our estimates are superior to previous estimates, since we use the fluctuation test. We consider the consequences of these estimates for the evolution of the [PSI+] system.
MATERIALS AND METHODS
Experiments measuring conversion from [psi−] to [PSI+]:
We assessed conversion from [psi−] to [PSI+] in strain 74-D694. 74-D694 carries the ade1-14 allele that contains a premature stop codon that is suppressible by [PSI+]. A [psi−] strain of 74-D694 was generated by overexpression of Hsp104p. This [psi−] strain was transformed with a plasmid expressing either the [PSI+] suppressible marker ura3-14 (Manogaran et al. 2006) or ura3-197 (Kurahashi and Nakamura 2007), both of which also contain premature stop codons suppressible by [PSI+]. In addition, a transformation was performed with a plasmid containing wild-type URA3 as a control.
The transformed strains were grown to OD600 ∼ 1.6 and 150 μl of the resulting cell culture was placed onto plates of synthetic defined medium (SD) lacking adenine and uracil (SD −Ura −Ade). After 4 weeks of growth at 21° on the selective medium the number of colonies was counted. These colonies derive from cells that acquired a readthrough phenotype during the growth of the cell culture. Each colony was spotted and scored as true [PSI+] if it underwent a color change from white to red following growth on guanidine hydrochloride (GdnHCl). GdnHCl cures [PSI+] permanently but only transiently affects readthrough from genetic mimics of nonsense suppression (Bradley et al. 2003).
The total number of cells at the end of growth was determined by plating 200 μl of a 1:10,000 dilution onto medium (SD −Leu) that selects for the presence of the transformed plasmids. The total number of cells on the SD −Ura −Ade plates is the product of the number of colonies that grew on the SD −Leu medium after 3–4 days and the dilution factor between the amount of culture plated for total cell counting and double-readthrough selective cell counting. We performed a total of 18 replicate experiments, 7 of which had a dilution factor of 22,500 and 11 of which had a dilution factor of 15,000. Twelve of these replicates were double-marker experiments where colonies were simultaneously selected for readthrough at both Ade and Ura loci, while 6 of them were single-marker experiments using a URA3 wild-type control plasmid.
Fluctuation test analysis:
We performed the fluctuation test on the replicate colony counts. The code originally developed by Shaver and Sniegowski (2005) was modified by Maughan et al. (2006) to fix some minor issues. We modified the last version from Maughan et al. (2006) to fit a maximum-likelihood function for the Luria–Delbrück distribution for counts with two different dilution factors but the same underlying mutation rate. Ninety-five percent confidence intervals were estimated using bootstrap sampling with replacement within each of the two dilution factor subsets of the data. When bootstrapping confidence intervals on the ratio between [PSI+] and [PSI+]-mimic appearance rates, [PSI+] and [PSI+]-mimic colony counts from the same replicate experiment were resampled as a pair, as their errors may not be independent. Independent Luria–Delbrück distributions were then fitted to the sets of [PSI+] and [PSI+]-mimic colony counts.
RESULTS
Estimating [PSI+] absolute appearance rate using a fluctuation test:
We selected for readthrough phenotypes and scored the number of resulting colonies. Those colonies that grew on GdnHCl were determined to be true [PSI+], and the remainder were assumed to be genetic mimics of [PSI+] (Table 1). Using all 18 replicates we performed the fluctuation test described previously to compute the maximum-likelihood estimate (MLE) and 95% confidence intervals (C.I.'s) for the spontaneous [PSI+] appearance rate mPSI and mimic appearance rate mmimic. The MLE for mPSI is 5.79 × 10−7 with a C.I. of (4.59–7.46) × 10−7; the MLE for mmimic is 2.34 × 10−6, with a C.I. of (2.04–2.77) × 10−6.
TABLE 1.
Population and mutant counts to estimate rate of appearance of readthrough phenotype in yeast
| Mutant counts |
|||||||
|---|---|---|---|---|---|---|---|
| Replicate | URA3 allele | No. of plates | Dilution factor | GdnHCl curable ([PSI+]) | Grown in SD −Ura −Ade (total) | Non-[PSI+] (total − [PSI+]) | Population counts (grown in SD −Leu) |
| 1 | ura3-14 | 3 | 22,500 | 8 | 45 | 37 | 207 |
| 2 | ura3-14 | 2 | 15,000 | 4 | 36 | 32 | 214 |
| 3 | ura3-14 | 3 | 22,500 | 1 | 44 | 43 | 188 |
| 4 | ura3-14 | 2 | 15,000 | 4 | 17 | 13 | 285 |
| 5 | ura3-14 | 2 | 15,000 | 5 | 22 | 17 | 228 |
| 6 | ura3-14 | 2 | 15,000 | 4 | 79 | 75 | 281 |
| 7 | ura3-197 | 3 | 22,500 | 4 | 44 | 40 | 250 |
| 8 | ura3-197 | 3 | 22,500 | 16 | 61 | 45 | 232 |
| 9 | ura3-197 | 2 | 15,000 | 37 | 76 | 39 | 210 |
| 10 | ura3-197 | 2 | 15,000 | 3 | 14 | 11 | 210 |
| 11 | ura3-197 | 2 | 15,000 | 2 | 22 | 20 | 160 |
| 12 | ura3-197 | 2 | 15,000 | 9 | 40 | 31 | 314 |
| 13 | URA3 | 3 | 22,500 | 7 | 42 | 35 | 222 |
| 14 | URA3 | 3 | 22,500 | 1 | 74 | 73 | 158 |
| 15 | URA3 | 3 | 22,500 | 2 | 35 | 33 | 181 |
| 16 | URA3 | 2 | 15,000 | 5 | 23 | 18 | 234 |
| 17 | URA3 | 2 | 15,000 | 2 | 14 | 12 | 261 |
| 18 | URA3 | 2 | 15,000 | 5 | 23 | 18 | 413 |
We expect a single-locus stop codon point mutation reversion rate of only ∼10−9, which is expected to make an insignificant contribution to the total. In agreement with this, using only the 12 double-selection replicates, which should almost completely eliminate the possibility of including single-locus revertants, we obtained mmimic = 2.23 × 10−6 [C.I.: 2.1–3.2 × 10−6], while the 6 single-marker replicates gave us mmimic = 1.93 × 10−6 [C.I.: 1.68–2.17 × 10−6]. Since these confidence intervals overlap, and the estimates are in fact in the opposite direction to what would be expected if single-locus revertants made a significant contribution, we used all replicates (both the single- and the double-marker experiments) interchangeably in subsequent analyses. Note that there may be weak [PSI+] variants in the population that are unable to overcome the auxotrophic selection: this would make our spontaneous appearance rate an underestimate.
A large fraction of readthrough phenotypes are mimics of [PSI+]:
On the basis of the MLEs for mPSI and mmimic from these experiments, we can now estimate the quantity
 |
Using the bootstrap analysis described in materials and methods, we obtained a 95% confidence interval for R of 0.166–0.246.
This agrees with a meta-analysis of previous data. Lund and Cox (1981) scored the proportion of spontaneous ADE2 readthrough colonies that were GdnHCl curable. In our meta-analysis, we include only 6 replicates treated by KCl and 4 replicates treated by dimethyl sulfoxide as the other treatments were mutagens, which could overestimate the number of genetic mimics. We also include similar spontaneous ADE2 readthrough experiments of Shewmaker and Wickner (2006) (2 replicates in each of two yeast strains). For this meta-analysis, we estimate R independently for each replicate experiment, using raw colony counts, and average this estimate across replicates. Systematic overestimation due to jackpots affects both the numerator and the denominator and so may be relatively unimportant for measuring relative rather than absolute rates. When including only the previous 14 replicates from Lund and Cox (1981) and Shewmaker and Wickner (2006), we obtain a mean, median, and standard error of the mean of R of 0.194, 0.186, and 0.034, respectively.
Using instead the 18 replicates from our new experiments in the same way for the purposes of comparison, we estimate the mean, median, and standard error of the mean of R as 0.170, 0.186, and 0.023, respectively. Assuming the estimator of R is normally distributed, an approximate 95% C.I. for R can be obtained from the mean plus or minus twice the standard error of the mean, yielding a C.I. of [0.124, 0.216]. This corresponds fairly well to the more reliable 95% C.I. of [0.166, 0.246] estimated using the bootstrap method. Our new data are in good agreement with our meta-analysis of literature data, which yield a C.I. of [0.126, 0.262]. Since we do not have the raw data necessary to do a more accurate and precise fluctuation test on these literature data, here we use the ML estimate for data in this article only. We do note, however, the consistency between previous literature and our data, giving broad support to the conclusion that ∼80% of all spontaneous [PSI+]-like phenotypes are actually genetic mimics.
Implications for natural selection on modifiers of the [PSI+] system:
Evolvability will be favored by natural selection so long as NeΩ > 1 (King and Masel 2007; Griswold and Masel 2009), where Ω is the probability per generation that [PSI+] becomes adaptive. We estimate the yeast effective population size Ne as ∼3 × 106–6 × 106, computed from Ne = (Θ(1 + F))/(4μ) using average pairwise divergence of sequences Θ ∼ 0.0032–0.0038 (Tsai et al. 2008), inbreeding coefficient F = 0.98 (Tsai et al. 2008), and a genomewide per-base pair point mutation rate ∼μ = 3.3 × 10−10 (Lynch et al. 2008) to μ = 5 × 10−10 (Lang and Murray 2008). At first sight, however, Ω seems an unknowable quantity.
If, however, we assume optimality, then the rate of spontaneous [PSI+] appearance mPSI = 4.59 × 10−7–7.46 × 10−7 that we have measured here can be used to infer Ω. Without genetic mimics, [PSI+] is expected to evolve toward an optimal rate of [PSI+] appearance equal to the probability Ω that [PSI+] appearance will be adaptive (Lachmann and Jablonka 1996; Kussell and Leibler 2005; Kussell et al. 2005; Wolf et al. 2005; King and Masel 2007). When genetic mimics are common, a higher rate of [PSI+] appearance is expected (Lancaster and Masel 2009).
The modifier model of Lancaster and Masel (2009) allows the calculation of the evolved rate of [PSI+] appearance mPSI as a function of the rate of genetic mimic mutations mmimic, the effective population size Ne, and the rate Ω at which environmental change makes [PSI+]-like phenotypes adaptive.
The final parameter of this model is the amount of population structure Nemk, where mk is the proportion of individuals that migrate between demes. Recent genomewide polymorphism surveys of yeast suggest that there is substantial population structure in both the wild yeast S. paradoxus (Liti et al. 2009) and S. cerevisiae (Liti et al. 2009; Schacherer et al. 2009). Almost all the strains of S. paradoxus surveyed appear to fall into one of three subpopulations (Liti et al. 2009). S. cerevisiae population structure appears to be more complex, with the first survey finding five subpopulations, Malaysian, “sake,” North African, West African, and wine/European (Liti et al. 2009), and the second finding three subpopulations, sake, wine/European, and laboratory strains (Schacherer et al. 2009). Subpopulations appear to group by ecological niche rather than strict geographical location (Schacherer et al. 2009), consistent with another S. cerevisiae study (Aa et al. 2006) where significant population structure was found between oak tree strains and vineyard strains, independent of location. Taken together these data show that wild yeast populations are structured, although this structure is possibly more driven by ecological specialization than by geographical proximity. Structured populations mean that gene flow between subpopulations is in the range 0 < Nemk < 1. We confirmed this by estimating Nemk on the basis of FST values from Aa et al. (2006) for a limited number of genes (data not shown).
Using the model of Lancaster and Masel (2009), we calculated optimal mPSI as a function of Ne, Ω, Nemk, and mmimic. We explored the 95% confidence interval for mmimic of (2.04–2.77) × 10−6 and an effective population size Ne in the range (3–6) × 106. By assuming that observed mPSI is equal to the calculated optimal, we were able to infer NeΩ (Figure 2). Most of the uncertainty in the inferred range of NeΩ estimates is due to the uncertainty in Ne rather than in mmimic or Nemk. Assuming optimality implies extremely low values of Ω ≈ (3.3–5) × 10−7, leading to very weak selection for the ability to form [PSI+], with marginal values of NeΩ ≈ 1–3. Evolvability properties will drive the evolution of the [PSI+] system only if NeΩ > 1 (King and Masel 2007; Griswold and Masel 2009). This undermines the assumption of optimality and seems to suggest that [PSI+] has not evolved because of its evolvability.
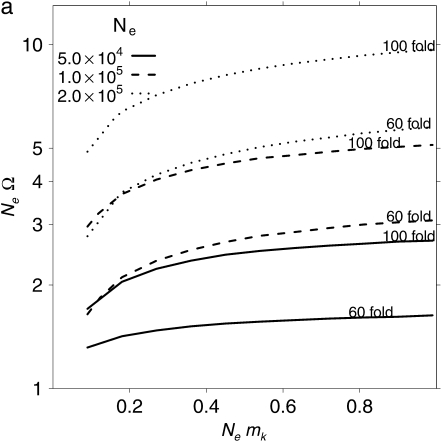
Figure 2.—
The model of Lancaster and Masel (2009) predicts the parameter range (shaded) for which mPSI (expressed as a function of the elevation from the unstressed baseline observed value of 5.8 × 10−7) is compatible with both a 95% C.I. of mmimic of (2.04–2.77) × 10−6 and Ne = (3–6) × 106. The upper bound is given by the combination {mmimic = 2.04 × 10−6, Ne = 6 × 106} and the lower bound by {mmimic = 2.77 × 10−6, Ne = 3 × 106}.
However, [PSI+] appearance rates are not constant, but instead are strongly dependent on environmental conditions. Stressful conditions that affect the [PSI+] appearance rate include low temperatures (up to 100-fold increase) (Chernoff et al. 1995; Derkatch et al. 2000) and high salt concentrations, oxidative stress, and high temperatures (up to 60-fold increase) (Tyedmers et al. 2008). Stressful conditions may induce a higher [PSI+] appearance rate at times when an opportunity for [PSI+]-mediated adaptation exists. The model of Lancaster and Masel (2009) considers only a single [PSI+] appearance rate, rather than one with and one without stress. Optimal switching is a trade-off between the benefits of switching when needed and the costs of switching when not. However, the optimal switching rate is determined primarily by the benefits rather than the costs (King and Masel 2007; Lancaster and Masel 2009). The model is therefore a predictor of optimal induced rather than optimal baseline [PSI+] appearance rate. In Figure 2 we therefore explore a range of values of induced mPSI from our measured baseline “unstressed” rate, up to a 100-fold elevation of the baseline. If we consider induced values of mPSI up to 60-fold higher at times when [PSI+] is most likely to be adaptive, we infer much higher values of NeΩ ∼ 3–50 (Figure 2). These values are compatible with evolvability properties driving the evolution of the [PSI+] system.
Most of the remaining uncertainty in the shaded regions of Figure 2 derives from uncertainty in the measurement of Ne in Saccharomyces. Further uncertainty comes from the fact that Ne may not have been constant over the evolutionary history of yeast. We therefore explore sensitivity to the parameter Ne more fully in Figure 3. Evolvability is still inferred so long as Ne > ∼105.
Figure 3.—
Sensitivity of our inference to the effective population size, Ne. For Ne ∼ 5 × 106, as estimated for Saccharomyces, we infer a role for natural selection in favoring [PSI+]-mediated evolvability if and only if mPSI is elevated by stress, as has been observed. (a) For Ne ≲ 105 we would not infer evolvability, even with realistically elevated switching rates. (b) Selection for evolvability would be inferred even in the absence of stress-mediated induction for Ne ≳ 4 × 107. Higher values of Ne are not shown because their computation requires excessive memory, but the result is still clear.
DISCUSSION
A previous study estimated the spontaneous appearance rate of [PSI+] (mPSI) as 10−7–10−5 (Lund and Cox 1981). Here we provide a more accurate and precise measurement in unstressed conditions of baseline mPSI as 5.8 × 10−7 with a 95% confidence interval of 4.6 × 10−7–7.5 × 10−7, correcting for jackpot effects of the Luria–Delbrück distribution. The same previous study found many non-[PSI+] colonies with elevated rates of stop codon readthrough at a single locus (Lund and Cox 1981). We have confirmed that these colonies typically show readthrough at multiple loci and are best thought of as genetic mimics of [PSI+] rather than as single-locus revertants. Only 20% of phenotypically [PSI+]-like colonies represent true [PSI+]. Both of these new quantitative estimates have implications for understanding the selective forces operating on [PSI+].
Deleterious selection against [PSI+] modifiers under usual conditions is very weak:
[PSI+] is rare in natural yeast populations, despite the fact that outcrossed sex should cause it to spread, suggesting that it is deleterious (Nakayashiki et al. 2005). If so, modifier alleles that permit [PSI+] formation will also be subject to indirect negative selection with strength mPSINe under usual environmental circumstances during which [PSI+] is deleterious (Masel and Griswold 2009). With a yeast effective population size Ne between 3 × 106 and 6 × 106 (Tsai et al. 2008) and the confidence intervals for mPSI measured here, we compute mPSINe as lying between 1.38 and 4.48; i.e., mPSINe > 1. This means that selection against [PSI+] modifiers at times when [PSI+] is deleterious is only weakly effective relative to genetic drift.
Overall positive selection for [PSI+] modifiers is only strong enough if [PSI+] is induced by stress:
The [PSI+] system will be favored due to its evolvability properties if NeΩ > 1 (King and Masel 2007; Griswold and Masel 2009). By assuming that observed [PSI+] appearance rates are optimal, we have inferred the frequency Ω with which [PSI+] is adaptive. On the basis of our measured [PSI+] appearance rate of 5.8 × 10−7, we infer only a marginal parameter range of NeΩ ∼ 1–3. Higher rates of [PSI+] appearance consistent with stress-mediated induction give NeΩ ∼ 3–50. Stress induction is a necessary and sufficient condition for the inference that [PSI+]-forming ability has been favored by natural selection for evolvability.
Mutational degradation:
Masel et al. (2007) found that for the [PSI+] system to avoid mutational degradation during long intervals when [PSI+] is not adaptive, the condition  > 1.3/Ω must be met. Calculating
> 1.3/Ω must be met. Calculating  as a function of mPSI = 5.8 × 10−7 and Ne = 5 × 106 according to the formulas of Masel et al. (2007), we calculate an upper bound m on the mutation rate with which the ability to form [PSI+] is lost. NeΩ = 3 requires m < 10−6 while NeΩ = 50 requires m < 4.6 × 10−5 to avoid the mutational degradation of switching ability. It seems unlikely that the mutational degradation rate would exceed these thresholds.
as a function of mPSI = 5.8 × 10−7 and Ne = 5 × 106 according to the formulas of Masel et al. (2007), we calculate an upper bound m on the mutation rate with which the ability to form [PSI+] is lost. NeΩ = 3 requires m < 10−6 while NeΩ = 50 requires m < 4.6 × 10−5 to avoid the mutational degradation of switching ability. It seems unlikely that the mutational degradation rate would exceed these thresholds.
Acknowledgments
We thank Oliver D. King for C code; Cortland Griswold, John Novembre, Michael Hickerson, Naoki Takebayashi, Grant Peterson, and Jessica Garb for helpful discussions; and the National Institutes of Health for funding (R01 GM076041 to J.M. and R01 GM072228 to H.L.T.). J.M. is a Pew Scholar in the Biomedical Sciences and an Alfred P. Sloan Research Fellow.
References
- Aa, E., J. P. Townsend, R. I. Adams, K. M. Nielsen and J. W. Taylor, 2006. Population structure and gene evolution in Saccharomyces cerevisiae. FEMS Yeast Res. 6 702–715. [DOI] [PubMed] [Google Scholar]
- Bradley, M. E., S. Bagriantsev, N. Vishveshwara and S. W. Liebman, 2003. Guanidine reduces stop codon read-through caused by missense mutations in SUP35 or SUP45. Yeast 20 625–632. [DOI] [PubMed] [Google Scholar]
- Brookfield, J. F., 2001. Evolution: the evolvability enigma. Curr. Biol. 11 R106–R108. [DOI] [PubMed] [Google Scholar]
- Chernoff, Y. O., S. L. Lindquist, B. Ono, S. G. Inge-Vechtomov and S. W. Liebman, 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268 880–884. [DOI] [PubMed] [Google Scholar]
- Cox, B. S., M. F. Tuite and C. J. Mundy, 1980. Reversion from suppression to nonsuppression in SUQ5 [psi+] strains of yeast: the classification of mutations. Genetics 95 589–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch, I. L., M. E. Bradley, S. V. Masse, S. P. Zadorsky, G. V. Polozkov et al., 2000. Dependence and independence of [PSI(+)] and [PIN(+)]: A two-prion system in yeast? EMBO J. 19 1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, W. J., and J. Seger, 1999. Cause and effect in evolution. Nature 399 30. [DOI] [PubMed] [Google Scholar]
- Eaglestone, S. S., B. S. Cox and M. F. Tuite, 1999. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J. 18 1974–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoozan, M., C. M. Grant, J. A. Duarte and M. F. Tuite, 1991. Quantitation of readthrough of termination codons in yeast using a novel gene fusion assay. Yeast 7 173–183. [DOI] [PubMed] [Google Scholar]
- Giacomelli, M. G., A. S. Hancock and J. Masel, 2007. The conversion of 3′ UTRs into coding regions. Mol. Biol. Evol. 24 457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold, C. K., and J. Masel, 2009. Complex adaptations can drive the evolution of the capacitor [PSI], even with realistic rates of yeast sex. PLoS Genet. 5 e1000517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday, R., and T. Ho, 1998. Evidence for gene silencing by endogenous DNA methylation. Proc. Natl. Acad. Sci. USA 95 8727–8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, S. B., and M. Kirkpatrick, 2008. Effects of the [PSI+] prion on rates of adaptation in yeast. J. Evol. Biol. 21 773–780. [DOI] [PubMed] [Google Scholar]
- King, O. D., and J. Masel, 2007. The evolution of bet-hedging adaptations to rare scenarios. Theor. Popul. Biol. 72 560–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi, H., and Y. Nakamura, 2007. Channel mutations in Hsp104 hexamer distinctively affect thermotolerance and prion-specific propagation. Mol. Microbiol. 63 1669–1683. [DOI] [PubMed] [Google Scholar]
- Kussell, E., and S. Leibler, 2005. Phenotypic diversity, population growth, and information in fluctuating environments. Science 309 2075–2078. [DOI] [PubMed] [Google Scholar]
- Kussell, E., R. Kishony, N. Q. Balaban and S. Leibler, 2005. Bacterial persistence: a model of survival in changing environments. Genetics 169 1807–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann, M., and E. Jablonka, 1996. The inheritance of phenotypes: an adaptation to fluctuating environments. J. Theor. Biol. 181 1–9. [DOI] [PubMed] [Google Scholar]
- Lancaster, A. K., and J. Masel, 2009. The evolution of reversible switches in the presence of irreversible mimics. Evolution 63 2350–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, G. I., and A. W. Murray, 2008. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics 178 67–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J. P., M. Aker, K. C. Sitney and R. K. Mortimer, 1986. First position wobble in codon-anticodon pairing: amber suppression by a yeast glutamine tRNA. Gene 49 383–388. [DOI] [PubMed] [Google Scholar]
- Liti, G., D. M. Carter, A. M. Moses, J. Warringer, L. Parts et al., 2009. Population genomics of domestic and wild yeasts. Nature 458 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, P. M., and B. S. Cox, 1981. Reversion analysis of [psi−] mutations in Saccharomyces cerevisiae. Genet. Res. 37 173–182. [DOI] [PubMed] [Google Scholar]
- Luria, S., and M. Delbrück, 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., 2007. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc. Natl. Acad. Sci. USA 104(Suppl. 1): 8597–8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., W. Sung, K. Morris, N. Coffey, C. R. Landry et al., 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. USA 105 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manogaran, A. L., K. T. Kirkland and S. W. Liebman, 2006. An engineered nonsense URA3 allele provides a versatile system to detect the presence, absence and appearance of the [PSI+] prion in Saccharomyces cerevisiae. Yeast 23 141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel, J., and A. Bergman, 2003. The evolution of the evolvability properties of the yeast prion [PSI+]. Evolution 57 1498–1512. [DOI] [PubMed] [Google Scholar]
- Masel, J., and C. K. Griswold, 2009. The strength of selection against the yeast prion [PSI+]. Genetics 181 1057–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel, J., O. D. King and H. Maughan, 2007. The loss of adaptive plasticity during long periods of environmental stasis. Am. Nat. 169 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maughan, H., V. Callicotte, A. Hancock, C. W. Birky, Jr., W. L. Nicholson et al., 2006. The population genetics of phenotypic deterioration in experimental populations of Bacillus subtilis. Evolution 60 686–695. [PubMed] [Google Scholar]
- Meiklejohn, C., and D. Hartl, 2002. A single mode of canalization. Trends Ecol. Evol. 17 468–473. [Google Scholar]
- Nakayashiki, T., C. P. Kurtzman, H. K. Edskes and R. B. Wickner, 2005. Yeast prions [URE3] and [PSI+] are diseases. Proc. Natl. Acad. Sci. USA 102 10575–10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal, C., 2001. Yeast prions and evolvability. Trends Genet. 17 167–169. [DOI] [PubMed] [Google Scholar]
- Partridge, L., and N. H. Barton, 2000. Evolving evolvability. Nature 407 457–458. [DOI] [PubMed] [Google Scholar]
- Pigliucci, M., 2008. Is evolvability evolvable? Nat. Rev. Genet. 9 75–82. [DOI] [PubMed] [Google Scholar]
- Pure, G. A., G. W. Robinson, L. Naumovski and E. C. Friedberg, 1985. Partial suppression of an ochre mutation in Saccharomyces cerevisiae by multicopy plasmids containing a normal yeast tRNAGln gene. J. Mol. Biol. 183 31–42. [DOI] [PubMed] [Google Scholar]
- Ruden, D. M., M. D. Garfinkel, V. E. Sollars and X. Lu, 2003. Waddington's widget: Hsp90 and the inheritance of acquired characters. Semin. Cell Dev. Biol. 14 301–310. [DOI] [PubMed] [Google Scholar]
- Saunders, N. J., E. R. Moxon and M. B. Gravenor, 2003. Mutation rates: estimating phase variation rates when fitness differences are present and their impact on population structure. Microbiology 149 485–495. [DOI] [PubMed] [Google Scholar]
- Schacherer, J., J. A. Shapiro, D. M. Ruderfer and L. Kruglyak, 2009. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458 342–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaver, A. C., and P. D. Sniegowski, 2005. Fluctuation test analysis programs and source code. University of Pennsylvania, Philadelphia, PA (http://www.bio.upenn.edu/faculty/sniegowski/#software).
- Shewmaker, F., and R. B. Wickner, 2006. Ageing in yeast does not enhance prion generation. Yeast 23 1123–1128. [DOI] [PubMed] [Google Scholar]
- Singh, G., and A. J. Klar, 2002. The 2.1-kb inverted repeat DNA sequences flank the mat2,3 silent region in two species of Schizosaccharomyces and are involved in epigenetic silencing in Schizosaccharomyces pombe. Genetics 162 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski, P. D., and H. A. Murphy, 2006. Evolvability. Curr. Biol. 16 R831–R834. [DOI] [PubMed] [Google Scholar]
- Sniegowski, P. D., P. J. Gerrish, T. Johnson and A. Shaver, 2000. The evolution of mutation rates: separating causes from consequences. BioEssays 22 1057–1066. [DOI] [PubMed] [Google Scholar]
- Stansfield, I., Akhmaloka and M. F. Tuite, 1995. a A mutant allele of the SUP45 (SAL4) gene of Saccharomyces cerevisiae shows temperature-dependent allosuppressor and omnipotent suppressor phenotypes. Curr. Genet. 27 417–426. [DOI] [PubMed] [Google Scholar]
- Stansfield, I., K. M. Jones, V. V. Kushnirov, A. R. Dagkesamanskaya, A. I. Poznyakovski et al., 1995. b The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 14 4365–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, F. M., 1994. Fluctuation tests: How reliable are the estimates of mutation rates? Genetics 137 1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank, E. M., D. A. Harris, A. A. Desai and H. L. True, 2007. Prion protein repeat expansion results in increased aggregation and reveals phenotypic variability. Mol. Cell. Biol. 27 5445–5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True, H. L., and S. L. Lindquist, 2000. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407 477–483. [DOI] [PubMed] [Google Scholar]
- True, H. L., I. Berlin and S. L. Lindquist, 2004. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 431 184–187. [DOI] [PubMed] [Google Scholar]
- Tsai, I. J., D. Bensasson, A. Burt and V. Koufopanou, 2008. Population genomics of the wild yeast Saccharomyces paradoxus: quantifying the life cycle. Proc. Natl. Acad. Sci. USA 105 4957–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite, M. F., and C. S. McLaughlin, 1982. Endogenous read-through of a UGA termination codon in a Saccharomyces cerevisiae cell-free system: evidence for involvement of both a mitochondrial and a nuclear tRNA. Mol. Cell. Biol. 2 490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers, J., M. L. Madariaga and S. Lindquist, 2008. Prion switching in response to environmental stress. PLoS Biol. 6 e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov, K. V., A. Y. Aksenova, M. J. Soom, K. V. Osipov, A. V. Svitin et al., 2002. Novel non-Mendelian determinant involved in the control of translation accuracy in Saccharomyces cerevisiae. Genetics 160 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, G. P., C. H. Chiu and T. F. Hansen, 1999. Is Hsp90 a regulator of evolvability? J. Exp. Zool. 285 116–118. [DOI] [PubMed] [Google Scholar]
- Wickner, R. B., D. C. Masison and H. K. Edskes, 1995. [PSI] and [URE3] as yeast prions. Yeast 11 1671–1685. [DOI] [PubMed] [Google Scholar]
- Wolf, D. M., V. V. Vazirani and A. P. Arkin, 2005. Diversity in times of adversity: probabilistic strategies in microbial survival games. J. Theor. Biol. 234 227–253. [DOI] [PubMed] [Google Scholar]
- Zhouravleva, G., L. Frolova, X. Le Goff, R. Le Guellec, S. Inge-Vechtomov et al., 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 14 4065–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]