Abstract
Understanding complex genotype-by-environment interactions (GEIs) is crucial for understanding phenotypic variation. An important factor often overlooked in GEI studies is time. We measured the contribution of GEIs to expression variation in four nonlaboratory Saccharomyces cerevisiae strains responding dynamically to a 25°–37° heat shock. GEI was a major force explaining expression variation, affecting 55% of the genes analyzed. Importantly, almost half of these expression patterns showed GEI influence only during the transition between environments, but not in acclimated cells. This class reveals a genotype-by-environment-by-time interaction that affected expression of a large fraction of yeast genes. Strikingly, although transcripts subject to persistent GEI effects were enriched for nonessential genes with upstream TATA elements, those displaying transient GEIs were enriched for essential genes regardless of TATA regulation. Genes subject to persistent GEI influences showed relaxed constraint on acclimated gene expression compared to the average yeast gene, whereas genes restricted to transient GEIs did not. We propose that transient GEI during the transition between environments provides a previously unappreciated source of expression variation, particularly for essential genes.
PHENOTYPES are influenced by both genetic and environmental factors, as well as by complex interactions between the two. Predicting phenotypic variation from underlying genotypes, including those related to human disease, therefore requires an understanding of genotype-by-environment interaction (GEI) (Mackay and Anholt 2007). GEI may also be important for phenotypic evolution, as it is proposed to unmask cryptic variation for traits otherwise buffered from change through canalization (Waddington 1942; Gibson and Dworkin 2004). In this regard, GEI could support rapid evolution of phenotypes that are constrained from varying under standard conditions (Rice 1998; Gibson and Wagner 2000). Despite these important implications, elucidating the effects of GEI is particularly challenging for organisms in uncontrollable niches. Model organisms therefore provide a perfect test bed for characterizing the relative contribution of genotype, environment, and GEI to phenotypic variation.
Gene expression is an important phenotype through which this can be addressed. Recent studies comparing genomic expression within and between species reveal distinct patterns of variation (Fay and Wittkopp 2008). Genes with variable expression tend to be nonessential genes, genes with paralogs, and genes with upstream TATA elements (Denver et al. 2005; Landry et al. 2006, 2007; Tirosh et al. 2006; Kvitek et al. 2008). Many of these are responsive to mutation accumulation (Denver et al. 2005; Rifkin et al. 2005; Landry et al. 2007) and, in the case of TATA-containing genes, to environmental perturbation (Basehoar et al. 2004; Raser and O'Shea 2004; Blake et al. 2006). It has been proposed that nonessential genes and those regulated by TATA elements may be under less stringent purifying selection than the average gene, and thus their expression may be more “evolvable” (Tirosh et al. 2006; Landry et al. 2007). However, these inferences raise important questions about how expression of essential genes might evolve, given the level of constraint that maintains their expression within species and across close relatives. A major unanswered question is how essential genes can tolerate expression variation, which is a prerequisite for the evolution of gene expression.
Similar constraints have been inferred for genes whose expression is influenced by GEIs. Several studies have compared condition-specific expression variation across yeast strains responding to nutrient changes (Landry et al. 2006), copper treatment (Fay et al. 2004), or ethanol stress (Smith and Kruglyak 2008). Landry et al. (2006) reported nearly twice the genes affected by GEI vs. genetic-only effects when studying strains grown on different media; this group was enriched for dispensable genes buffered by paralogs. Likewise, Smith and Kruglyak (2008) characterized GEIs in vineyard strain RM11-1a and a lab strain grown on glucose or ethanol; although they observed fewer genes affected by GEIs compared to genetic or environmental effects, GEIs remained a significant contributor to expression variation.
An important aspect often overlooked in GEI studies is the transition between environments or states. Dynamic environmental responses are extremely important for microbes living in variable niches. In multicellular organisms, transitions between environments or through developmental stages can also have a dramatic impact on future phenotypes. A classic example is the onset of psychiatric disease, which is heavily influenced by adolescent experiences years before (Tsuang 2000). While understanding the effect of time on GEIs is hopelessly complicated in humans, budding yeast provides an opportunity to study the temporal effects of GEIs on gene expression differences. Here we measured GEIs in gene-expression variation by following Saccharomyces cerevisiae strains responding to a 25°–37° shock. Our results raise important insights into expression variation in response to fluctuating environments and point to the critical importance of timing in uncovering GEI effects on phenotypic variation.
MATERIALS AND METHODS
Strains are described in supporting information, Table S1. All microarray measurements were done in biological duplicate, with replicate cell collections done on separate days. Cells were grown in rich YPD medium at 25° for more than three generations in shaker flasks, and an aliquot was removed for the 25° sample. One volume of 55° medium was added to each culture for a final temperature of 37° upon mixing. Samples were removed for microarray analysis at 5, 15, 30, 45, 60, and 120 min after shock, and RNA was collected by a standard hot-phenol lysis method (Gasch 2002). For heat-shock time courses, RNA collected at each time point was compared to RNA from the 25° sample collected for that strain, using two-color fluorescence hybridizations to 70-mer oligonucleotide (QIAGEN, Valencia, CA) arrays designed against S288c, constructed in house. Expression relative to S288c was also measured in each strain by directly comparing transcripts collected at 25° to that reference. We showed previously that <5% of measured expression differences could be affected by hybridization defects due to polymorphism (Kvitek et al. 2008). Microarray data collection and normalization were done as previously described (Kvitek et al. 2008), using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA), amino-allyl dUTP (Ambion, Austin, TX), and cyanine dyes (Flownamics, Madison, WI). Microarray data are available in the National Institutes of Health GEO database under accession no. GSE15147.
Data were normalized by regional median centering (Lyne et al. 2003) and missing data were imputed using k-nearest neighbors for genes with <50% missing values in any single strain data set, resulting in 4269 genes analyzed and 4.4% data points imputed. We constructed a two-way ANOVA model for each gene, using strains and time points post-heat shock as factors. Considering log2 Ystr as the log2 absolute expression of a single gene in strain s at time t in array replicate r, we modeled the data as
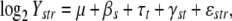 |
where μ is the mean expression across strains before heat shock, fixed effect βs is the strain-specific (genetic) difference from the average strain, fixed effect τt is the time (environment) effect in the average strain over t = 0, 5, … , 120 min, and interaction γst models the difference in expression between strain s and the average strain at time t (GEI). This absolute expression model facilitates the joint analysis of heat-shock time course data (which measures changes in expression relative to the 25° expression levels in each strain) and the relative expression of each strain at 25° compared to the S288c reference, by incorporating the S288c time course data without estimating parameters for this strain. Genes that failed Levene's test for constant variance were omitted. Genes with significant main effect (ANOVA F-tests) and no significant interactions were defined as additively affected by genotype and environment, while genes with a single significant main effect in strain or time were defined as affected by genotype or environment, respectively. Strains with a significant interaction F-test in the main terms were defined as influenced by GEI. Significance of each factor was defined at FDR < 0.05 unless otherwise noted, using the Benjamini and Hochberg correction (Benjamini and Hochberg 1995). Strain-specific conclusions for each gene (Table 1) were based on post hoc tests for each strain s.
TABLE 1.
Genes affected by G, E, and GEI
| Strains |
|||||
|---|---|---|---|---|---|
| Joint analysis (%) | K9 (%) | M22 (%) | RM11-1a (%) | YPS163 (%) | |
| Genetic | 275 (6.4) | 87 (2) | 77 (1.8) | 83 (1.9) | 77 (1.8) |
| Environmental | 3301 (77) | 2953 (69) | 3125 (73) | 3003 (70) | 3301 (77) |
| Additive | 282 (6.6) | 166 (3.9) | 131 (3.1) | 222 (5.2) | 157 (3.7) |
| GEI | 2367 (55) | 1215 (28) | 1744 (41) | 1184 (28) | 1539 (36) |
| Persistent GEI | 1129 (26) | 522 (12) | 713 (17) | 533 (12) | 720 (17) |
| Transient GEI | 1238 (29) | 693 (16) | 1031 (24) | 651 (15) | 819 (19) |
The numbers of genes (of 4269 analyzed) with genetic (G) only, environmental (E), additive genetic plus environmental, and GEI (including persistent and transient GEI) effects on expression variation are shown (FDR < 0.05). Results from the joint analysis are shown, as well as post hoc calls for each strain. Genes listed with environmental effects included those with additive and GEI effects.
Genes with significant GEI in the joint analysis were subsequently classified as persistently affected by GEI if the final, acclimated time point (120 min post-heat shock) was significant using a liberal cutoff of P < 0.05 in the single degree of freedom test: 90% of these genes showed significant GEI (FDR < 0.05) over the intervening time points. The subset of genes that failed this test, but that had significant GEI influence over all time points, was identified as transiently affected by GEI. This classification was not driven by genes with poorly measured data, since genes in the two classes had replicates at the final time point that were of similar correlation (data not shown). To further ensure that our results were not specific to this threshold, we also conducted a sensitivity analysis (Figure S2) showing that enrichment results are robust to different cutoffs for defining genes subject to persistent vs. transient GEI effects.
Tests of functional enrichment were performed using Fisher's exact test for essential genes (Winzeler et al. 1999), genes with paralogs (defined as genes with a BLAST hit of E < 10−10), genes with upstream TATA elements (Basehoar et al. 2004), and other functional groups defined by the Saccharomyces Genome Database (Hong et al. 2008). To ensure that enrichment of essential genes and those with upstream TATA elements was not driven by enrichment of some other functional group, we repeated the test by systematically removing all genes in each of the nonredundant functional categories from the analysis. In all cases, reported enrichments remained significant at Bonferroni-corrected P < 0.05. Vg/Vm ratios were calculated for the 1174 genes in our data set that varied in Landry et al. (2007) and for which Vg could be estimated. Vg was estimated as the between-strain variance in a random-effects model (Bates et al. 2008) among the 25° array data and compared to Vm reported by Landry et al. (2007). Relative differences in Vg/Vm ratios between groups were assessed using a Wilcoxon rank-sum test. Although we caution against interpreting precise values for individual genes, that we recapitulate results of other studies for key gene groups (Denver et al. 2005; Tirosh et al. 2006; Landry et al. 2007) (Figure 3) validates comparing relative ratios across different gene groups.
Figure 3.—
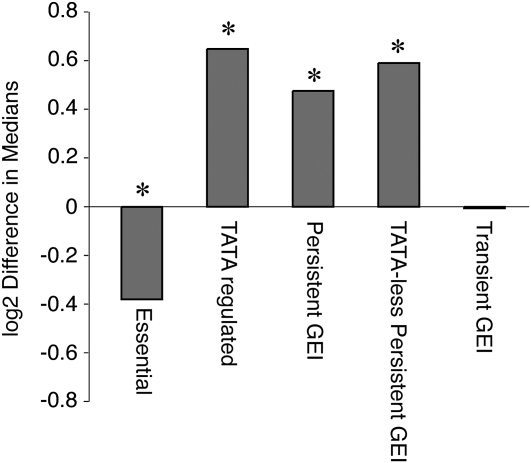
Genes affected by GEI show different levels of purifying selection. The log2 difference in the median Vg/Vm was measured for each indicated group vs. all remaining genes analyzed. Negative values indicate smaller ratios and positive values represent larger ratios than the comparison group. TATA-less persistent-GEI genes were scored against all remaining TATA-less genes. Statistically significant differences (P < 0.05, Wilcoxon's rank-sum test) are indicated with an asterisk. See text for P-values.
RESULTS
To study the contribution of GEIs to expression variation, we characterized the dynamic gene expression response of four nonlaboratory strains to a 25°–37° heat shock. We examined the sake-producing strain K9, vineyard isolates M22 and RM11-1a, and a Pennsylvania oak-soil strain (YPS163), in addition to an S288c-derived lab strain (Table S1). Expression was compared across strains acclimated to 25° and to 37° for 2 hr (approximately two generations). We also followed the dynamic response to sudden heat shock, measuring expression at 5, 15, 30, 45, and 60 min after the temperature shift.
We devised a linear model to quantify the genetic, environmental, and GEI effects on expression differences, relative to the mean across all strains, to estimate the contribution of each factor to expression variation (see materials and methods). Of 4269 genes analyzed, 3301 genes (77%) showed a significant environmental response [false discovery rate (FDR) < 0.05]. This response was overall similar to that in our previous studies of the lab strain (Gasch et al. 2000) and includes some genes whose expression change is unique to conditions inflicted by heat shock (e.g., protein-folding chaperones and mitochondrial genes) and many genes whose expression is altered by diverse types of stress (i.e., genes in the environmental stress response, ESR). The response was initiated within 5 min after heat shock, with most genes showing maximal expression differences at 15 min before relaxing to new steady-state expression levels thereafter. Consistent with our previous results, expression differences for most genes were acclimated by 45–60 min after the shift: there were only 82 genes (2.5%) with significant expression differences at 120 min vs. 60 min (P < 0.05) postshock. Furthermore, most expression differences seen at 60 min postshift are observed in cells growing long-term at 37° (Gasch et al. 2000).
Focusing on expression patterns that differed across strains revealed that GEI provides a major source of variation (Table 1). A total of 275 genes (6.4%) showed expression variation due to genetic differences only, independent of the environment, whereas an additional 282 genes showed an additive environmental response. In contrast, >2000 genes (2367, or 55%) showed a complex interaction between genotype and environment. The number of genes subject to GEI effects was more than fourfold the number of genes affected by genetic effects alone, revealing a much greater contribution of GEIs than measured in other yeast studies (Fay et al. 2004; Landry et al. 2006; Smith and Kruglyak 2008). Thus, heat shock uncovers a substantial amount of variation in gene expression across strains.
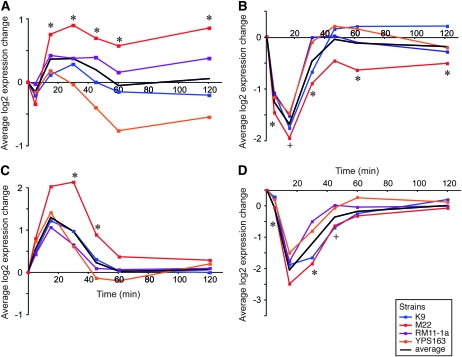
However, considering the timing of GEI effects revealed that a striking proportion of GEI was manifest only during the transition between environments. To further investigate this, we partitioned genes with GEI influences across all time points into two groups. Those for which GEI was also measured at the final, acclimated time point (using a liberal cutoff of P < 0.05, see materials and methods) were defined as showing “persistent” GEI even after cells had acclimated to the new temperature (Figure 1, A and B). In contrast, the subset of GEI-affected genes with no significant GEI at the final, acclimated time point was defined as having “transient” GEI effects only during the transition between environments (Figure 1, C and D). These transient GEI effects essentially represent a genotype-by-environment-by-time interaction, for which expression differences are revealed only during the appropriate temporal window. Of the 2367 genes with GEIs, over half (1238 genes) did not show observable variation across strains once cells had acclimated to 37°. Thus, fully half of the genes subject to GEI effects would have been missed by examining acclimated cells only.
Figure 1.—
Expression patterns of representative genes subject to persistent GEI or transient GEI effects. Gene groups were manually selected from hierarchically clustered expression data, clustered over all genes with persistent GEI or with transient GEI effects. The average log2 expression change across genes in each group is shown for each strain and for the mean of all strains, according to the key in the bottom right. Expression was normalized to the 25° expression levels in each strain for clarity. Average expressions are shown of (A) 30 genes enriched for mitochondrial ribosomal protein genes and displaying persistent GEI, (B) 49 genes enriched for rRNA processing factors and displaying persistent GEI, (C) 34 genes enriched for proteasome subunits and showing transient GEI effects, and (D) 74 genes enriched for ribosomal protein genes and displaying transient GEI influence. In all cases, functional enrichment was significant at a Bonferroni-corrected P < 0.05 (see Table 2). Time points with statistically significant variation (P < 0.01, ANOVA) are indicated with an asterisk; two time points of marginal significance (P = 0.055) are indicated with a +.
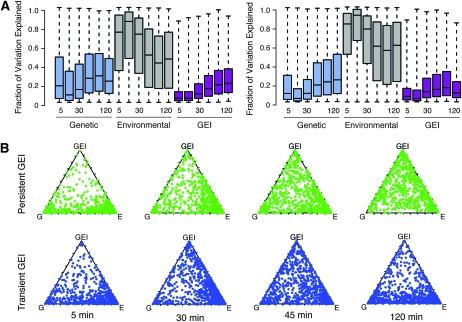
To explore the temporal contribution of GEI to expression variation, relative to genetic- and environment-only effects, we quantified the proportion of each gene's variation explained by the three factors at each time point. Not surprisingly, the relative contributions were different depending on timing. Variation explained by environment was maximal (median of ∼85%) at 15 min before relaxing to ∼60% upon acclimation. GEI explained ∼14% of the variation 45 min postshock for both persistent and transient GEI gene groups. However, the interaction of GEI with time was significantly different for the two classes. The GEI contribution increased continuously for genes in the persistent-GEI group, peaking at a median of 20% variation explained by 120 min after shock. In contrast, the maximum contribution of GEI to genes in the transient-GEI group was similar from 30 to 60 min and decreased to a median level of 9% variation explained at the final time point. This trend was also evident on a gene-by-gene basis, most clearly seen in individual strains such as M22 (Figure 2B and Figure S1). Although by definition genes in the transient-GEI class have minimal GEI effects at the last time point, the different temporal patterns across other time points confirm the distinction between groups. Thus, there is a fundamental difference in the interaction of genotype, environment, and time for genes in the two classes.
Figure 2.—
Contribution of genetic, environmental, and GEI effects to expression variation. The proportion of variance explained by genetic (G), environmental (E), and GEI factors was computed from the ANOVA sums of squares. (A) Box plots show the contribution of G, E, and GEI to expression variation at each time point, for all genes with persistent (left) and transient (right) GEI effects. (B) Triangle plots show the contribution of G, E, and GEI to expression variation of 1744 genes from M22 (scored against the mean of all strains), for genes in the persistent GEI class (top) and the transient GEI class (bottom) at 5, 30, 45, and 120 min after heat shock. Each gene is represented as a single point whose coordinates indicate the proportion of variance explained by each factor.
We next investigated if certain functional groups were particularly affected by GEI. In fact, different gene classes were subject to persistent vs. transient GEI effects. Consistent with numerous studies, we found that the 1129 genes displaying a persistent GEI effect were enriched for genes with upstream TATA elements (P = 3 × 10−16) and nonessential genes (P = 3 × 10−7), as well as genes encoding mitochondrial proteins (P = 3 × 10−14) and genes induced in the ESR (P = 2 × 10−6) (Table 2). In contrast, the 1238 genes with transient GEI effects were enriched for distinct functional groups, including ribosomal protein genes (P = 2 × 10−9), protein-synthesis genes repressed as part of the yeast ESR (P = 2 × 10−4), and genes encoding proteasome subunits (P = 4 × 10−3) and purine biosynthetic enzymes (P = 3 × 10−3). In stark contrast with the previous group, genes subject to transient GEI were enriched for essential genes (P = 4 × 10−4) and showed no preference for TATA-regulated genes. This could not be fully explained by any other enriched functional group, since enrichment of essential genes remained significant (P < 0.002) when each of the other functional groups was removed from the analysis (see materials and methods). These enrichments were also robust to different cutoffs used to define genes in the persistent-GEI vs. transient-GEI groups, as shown by sensitivity analysis (Figure S2). Thus, essential genes are prone to transient-GEI effects while nonessential and TATA-containing genes tend to show persistent GEI on expression variation.
TABLE 2.
Functional enrichments
| Functional category | Enrichment P-value |
|---|---|
| Persistent GEI | |
| TATA Containing | 3.32E-16 |
| Mitochondrial ribosomal proteins | 2.84E-14 |
| Nonessential | 2.68E-07 |
| Induced with the ESR | 1.56E-06 |
| Transient GEI | |
| Ribosomal proteins | 1.58E-09 |
| Protein synthesis genes repressed in the ESR | 1.88E-04 |
| Essential | 3.50E-04 |
| Genes with paralogs | 6.12E-04 |
| Purine biosynthesis | 3.05E-03 |
| Proteasome subunits | 3.38E-03 |
Nonredundant functional groups enriched with statistical significance in the 1129 genes with persistent GEI or the 1238 genes with transient GEI are shown. ESR, environmental stress response. All P-values were significant (P < 0.05) after stringent Bonferroni correction.
We wondered if “steady-state,” acclimated expression of genes with persistent vs. transient GEI was subject to different levels of constraint. One measure of this is the level of variation seen in nature compared to the level of variation induced by mutation in the absence of selection. We compared expression variation in our strains acclimated to 25° (Vg) to expression variation resulting from mutation accumulation in the absence of selection, measured by Landry et al. (2007) (Vm). Genes whose expression is under the most stringent purifying selection are expected to have the smallest Vg/Vm ratios, since deleterious expression variation induced by mutation would be removed by natural selection. Consistent with previous studies (Denver et al. 2005), we found essential genes had smaller ratios compared to nonessential genes (P = 0.036), whereas TATA-containing genes had significantly larger ratios than TATA-less genes (P = 2 × 10−5) (Figure 3). This recapitulates previous results suggesting steady-state expression of essential genes is under strong purifying selection while TATA-regulated genes display relaxed constraint (Denver et al. 2005; Tirosh et al. 2006; Landry et al. 2007).
Genes whose heat-induced expression variation was subject to persistent GEI also displayed higher-than-expected Vg/Vm (P = 2 × 10−6, Figure 3). This was true also for genes in the group lacking TATA elements (1 × 10−5), revealing that other factors besides TATA regulation contribute to high expression variation. Thus, genes with persistent GEI are under relaxed constraint in acclimated cells. In contrast, genes in the transient GEI group did not show higher variation in acclimated expression compared to the average gene, even though they showed significant variation during the transition between environments. Since the expression of most genes is thought to be under strong purifying selection (Denver et al. 2005; Rifkin et al. 2005), this suggests that expression of genes affected by transient GEI is also constrained in acclimated cells.
DISCUSSION
Understanding how and when expression varies across individuals is of major importance, in terms of understanding both phenotypic variation and evolution. In particular, the role of GEIs in human disease susceptibility and prognosis has attracted renewed attention, yet proves to be extremely challenging to elucidate. Our results demonstrate that a large fraction of genes are influenced by genotype-by-environment-by-time interactions, such that over half the GEI effects we see are not observed in cells growing continuously in distinct environments. For these genes, and even many for which GEI effects persist in acclimated cells, most of the variation occurs during the transition between environments. The phenomenon may have masked a significant amount of GEI in prior studies. For example, Sambandan et al. (2008) observed surprisingly few expression differences in adult flies reared in distinct environments, despite wide variation in adult olfactory behavior; the authors proposed that their results may represent a general trend, in which few genes contribute to GEIs. However, we suggest that significant expression variation may have been missed by characterizing only acclimated adults. Our results demonstrate the challenges in identifying GEIs that could significantly affect subsequent variation and underscore the importance of considering the timing of analysis.
Despite their transience, expression differences during the acclimation to new conditions could have profound impacts on fitness. Subtle differences in transcript levels could significantly affect corresponding protein abundance and function, leading to dramatic phenotypic differences. Furthermore, differences in the dynamics of gene expression change are known to have important phenotypic consequences. We showed previously that a primary role of stress-activated gene expression changes in yeast is in preparation for impending environmental stress (Berry and Gasch 2008). The response provoked by a mild stress pretreatment renders cells tolerant to subsequent severe stress; this acquisition of stress tolerance is heavily dependent on the initial expression response to the mild stressor. A faster gene expression response during the initial acclimation results in faster acquisition of subsequent stress tolerance (Berry and Gasch 2008). Given that environmental changes occur frequently in nature and often in tandem, the differences in expression dynamics seen here could have profound impacts on stress survival in the wild (Berry and Gasch 2008; Mitchell et al. 2009). Thus, although much of the variation we see is likely neutral, a fraction of these phenotypic differences may be physiologically relevant.
It is particularly striking that essential genes are more likely to display transient GEI effects while nonessential and TATA-containing genes are prone to persistent GEIs. The latter genes display relaxed constraint on steady-state expression, while expression of essential genes is subject to strong constraint in acclimated cells (Figure 3). We propose that the period of transition to a new environment may provide a temporary window of reduced constraint, allowing expression of essential genes and others under strong constraint to vary. Environmental stress and GEIs in particular are proposed to uncover hidden variation that could influence phenotypes under canalizing selection (Waddington 1942; Gibson and Wagner 2000; Gibson and Dworkin 2004; Gibson 2009). However, we find that neither persistent GEI nor heat stress alone is sufficient to uncover this variation; rather, it is transient GEI during the shift between environments that uncovers this variation.
Even if largely neutral, the variation revealed by transient GEI effects may provide a route through which expression of essential genes can evolve. We and others previously showed that regulation of proteasomal and ribosomal protein genes has evolved significantly across fungi, despite the apparent constraint to maintain their coregulation (Gasch et al. 2004; Tanay et al. 2005; Hogues et al. 2008; Tuch et al. 2008). A major unresolved question is the process that allowed this regulatory change. Indeed, these gene groups show little expression variation in acclimated cells; however, they are significantly enriched in the set of genes displaying transient GEI effects (Figure 2 and Table 2). It is intriguing to speculate that the higher rate of expression variation for these genes—specifically during environmental transitions—contributed to their regulatory evolution. Given the frequency of environmental fluctuations experienced by free-living microbes in nature, transient expression variation may indeed be a common occurrence. Identifying the underlying genetic determinants of such variation is an exciting next step in understanding how expression of essential genes evolves.
Acknowledgments
The authors thank Jeff Lewis for experimental assistance, Bret Payseur for stimulating discussions, and Christian Landry for providing Vm data. This work was supported by National Science Foundation grants CAREER:0447887 (to A.P.G.) and DMS004597 (to S.K.), by National Institutes of Health grant HG003747 (to S.K. and K.H.E.), and by a University of Wisconsin-Madison Hilldale Award (to D.J.K.).
Microarray data from this article have been deposited with the National Institutes of Health GEO Database under accession no. GSE15147.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.107268/DC1.
References
- Basehoar, A. D., S. J. Zanton and B. F. Pugh, 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116 699–709. [DOI] [PubMed] [Google Scholar]
- Bates, D., M. Maechler and B. Dai, 2008. Ime4: linear mixed-effects models using S4 classifiers. R package version 0.999375–15. http://Ime4.r-forge.r-project.org.
- Benjamini, Y., and Y. Hochberg, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57 289–300. [Google Scholar]
- Berry, D. B., and A. P. Gasch, 2008. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol. Biol. Cell 19 4580–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake, W. J., G. Balazsi, M. A. Kohanski, F. J. Isaacs, K. F. Murphy et al., 2006. Phenotypic consequences of promoter-mediated transcriptional noise. Mol. Cell 24 853–865. [DOI] [PubMed] [Google Scholar]
- Denver, D. R., K. Morris, J. T. Streelman, S. K. Kim, M. Lynch et al., 2005. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat. Genet. 37 544–548. [DOI] [PubMed] [Google Scholar]
- Fay, J. C., and P. J. Wittkopp, 2008. Evaluating the role of natural selection in the evolution of gene regulation. Heredity 100 191–199. [DOI] [PubMed] [Google Scholar]
- Fay, J. C., H. L. McCullough, P. D. Sniegowski and M. B. Eisen, 2004. Population genetic variation in gene expression is associated with phenotypic variation in Saccharomyces cerevisiae. Genome Biol. 5 R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch, A. P., 2002. Yeast genomic expression studies using DNA microarrays. Methods Enzymol. 350 393–414. [DOI] [PubMed] [Google Scholar]
- Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen et al., 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch, A. P., A. M. Moses, D. Y. Chiang, H. B. Fraser, M. Berardini et al., 2004. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2 e398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, G., 2009. Decanalization and the origin of complex disease. Nat. Rev. Genet. 10 134–140. [DOI] [PubMed] [Google Scholar]
- Gibson, G., and I. Dworkin, 2004. Uncovering cryptic genetic variation. Nat. Rev. Genet. 5 681–690. [DOI] [PubMed] [Google Scholar]
- Gibson, G., and G. Wagner, 2000. Canalization in evolutionary genetics: A stabilizing theory? BioEssays 22 372–380. [DOI] [PubMed] [Google Scholar]
- Hogues, H., H. Lavoie, A. Sellam, M. Mangos, T. Roemer et al., 2008. Transcription factor substitution during the evolution of fungal ribosome regulation. Mol. Cell 29 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, E. L., R. Balakrishnan, Q. Dong, K. R. Christie, J. Park et al., 2008. Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 36 D577–D581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitek, D. J., J. L. Will and A. P. Gasch, 2008. Variations in stress sensitivity and genomic expression in diverse S. cerevisiae isolates. PLoS Genet. 4 e1000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry, C. R., J. Oh, D. L. Hartl and D. Cavalieri, 2006. Genome-wide scan reveals that genetic variation for transcriptional plasticity in yeast is biased towards multi-copy and dispensable genes. Gene 366 343–351. [DOI] [PubMed] [Google Scholar]
- Landry, C. R., B. Lemos, S. A. Rifkin, W. J. Dickinson and D. L. Hartl, 2007. Genetic properties influencing the evolvability of gene expression. Science 317 118–121. [DOI] [PubMed] [Google Scholar]
- Lyne, R., G. Burns, J. Mata, C. J. Penkett, G. Rustici et al., 2003. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 4 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, T. F., and R. R. Anholt, 2007. Ain't misbehavin'? Genotype-environment interactions and the genetics of behavior. Trends Genet. 23 311–314. [DOI] [PubMed] [Google Scholar]
- Mitchell, A., G. H. Romano, B. Groisman, A. Yona, E. Dekel et al., 2009. Adaptive prediction of environmental changes by microorganisms. Nature 460 220–224. [DOI] [PubMed] [Google Scholar]
- Raser, J. M., and E. K. O'Shea, 2004. Control of stochasticity in eukaryotic gene expression. Science 304 1811–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, S. H., 1998. The evolution of canalization and the breaking of von Baer's laws: modeling the evolution of development with epistasis. Evolution 52 647–656. [DOI] [PubMed] [Google Scholar]
- Rifkin, S. A., D. Houle, J. Kim and K. P. White, 2005. A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature 438 220–223. [DOI] [PubMed] [Google Scholar]
- Sambandan, D., M. A. Carbone, R. R. Anholt and T. F. Mackay, 2008. Phenotypic plasticity and genotype by environment interaction for olfactory behavior in Drosophila melanogaster. Genetics 179 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, E. N., and L. Kruglyak, 2008. Gene-environment interaction in yeast gene expression. PLoS Biol. 6 e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay, A., A. Regev and R. Shamir, 2005. Conservation and evolvability in regulatory networks: the evolution of ribosomal regulation in yeast. Proc. Natl. Acad. Sci. USA 102 7203–7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh, I., A. Weinberger, M. Carmi and N. Barkai, 2006. A genetic signature of interspecies variations in gene expression. Nat. Genet. 38 830–834. [DOI] [PubMed] [Google Scholar]
- Tsuang, M., 2000. Schizophrenia: genes and environment. Biol. Psychiatry 47 210–220. [DOI] [PubMed] [Google Scholar]
- Tuch, B. B., D. J. Galgoczy, A. D. Hernday, H. Li and A. D. Johnson, 2008. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 6 e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington, C. H., 1942. Canalization and the inheritance of an acquired character. Nature 150 563–565. [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285 901–906. [DOI] [PubMed] [Google Scholar]