Summary
The albCre transgene, having Cre recombinase driven by the serum albumin (alb) gene promoter, is commonly used to generate adult mice having reliable hepatocyte-specific recombination of loxP-flanked (“floxed”) alleles. Based on previous studies, it has been unclear whether albCre transgenes are also reliable in fetal and juvenile mice. Perinatal liver undergoes a dynamic transition from being predominantly hematopoietic to predominantly hepatic. We evaluated Cre activity during this transition in albCre mice using a sensitive two-color fluorescent reporter system. From fetal through adult stages, in situ patterns of Cre-dependent recombination of the reporter closely matched expression of endogenous Alb mRNA or protein, indicating most or all hepatocytes, including those in fetal and juvenile livers, had expressed Cre and recombined the reporter. Our results indicate the albCre transgene is effective at converting simple floxed alleles in fetal and neonatal mice and is an appropriate tool for studies on hepatocyte development.
Keywords: Albumin-Cre, conditional allele, liver-specific knockout, liver development, hepatocyte, fetal albumin expression
For over a decade, albCre transgenic mice, having Cre driven by the serum albumin (alb) gene promoter, have proven useful for studies involving Cre-dependent excision of loxP-flanked (“floxed” or “conditional”) sequences in adult hepatocytes (Postic et al., 1999). The endogenous alb gene is expressed exclusively in hepatocytes, in which it is induced at differentiation. Developmentally, Alb mRNA is first detected at embryonic day 10.5 (E10.5) in hepatic primordia (Meehan et al., 1984; Murakami et al., 1987). Expression of albCre transgenes is also hepatocyte-specific; however there is some discord concerning the developmental onset of Cre expression in albCre transgenics.
Previously, two papers co-published in this journal reached different conclusions concerning the activity of albCre transgenes in fetal/juvenile liver. Whereas one study used an in situ assay to show that Cre-dependent allelic conversion initiated as early as E10.5 (Kellendonk et al., 2000), the other, using a different transgene design and a Southern-blot analysis, reported that conversion was only 40% efficient at birth (E19.5) and did not approach completion until postnatal day 42 (P42) (Postic and Magnuson, 2000). The later paper proposed that sub-threshold expression of the albCre transgene in juvenile hepatocytes resulted in incomplete conversion (Postic and Magnuson, 2000). Contradicting this interpretation, however, this paper also showed nearly complete conversion in hepatocytes of juvenile animals using a Cre-dependent lacZ reporter and an in situ assay (Postic and Magnuson, 2000). It has remained unclear what roles the differences in transgene design versus the differences in assays played in these disparate reports, and at what stage one might expect functional expression of an albCre transgene.
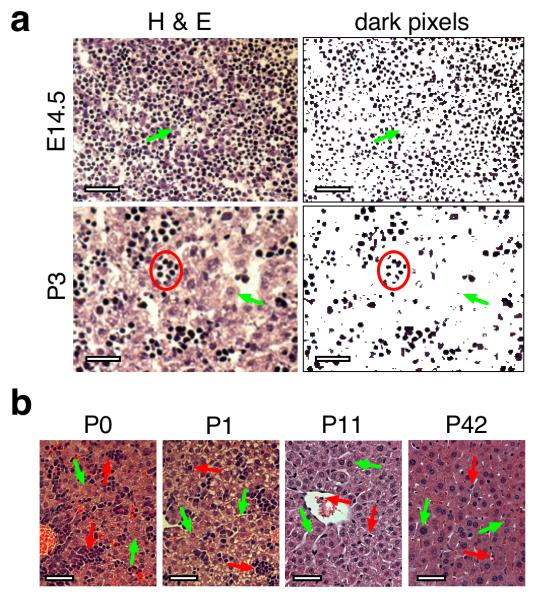
Fetal liver is a hematopoietic organ with only minority representation by hepatocytic cells (Paul et al., 1969). By hematoxylin and eosin (H&E) staining, hepatocytes are large cells with abundant cytoplasm and pale nuclei. Conversely, most hematopoietic cells are small with little cytoplasm and dark-staining nuclei. During perinatal stages, as hematopoiesis moves to the bone marrow and hepatic functions expand, the cellular composition of the liver progressively shifts to being more purely populated by hepatocytes (Zaret, 2000). Reflecting the hematopoietic nature of the fetal organ, liver sections from fetuses harvested at E14.5 exhibited a relatively uniform distribution of small cells with dark nuclei; few cells had a hepatocytic morphology (Fig. 1a). Hepatocytes became more abundant and smaller cells became clustered between E14.5 and P3. Dark pixels from the photomicrographs highlight distributions of non-hepatocytic cell nuclei (right panels). Newborn (P0 and P1) liver showed dense clusters of small cells interspersed among a modest population of hepatocytes (Fig. 1b). At P11 and P42, these clusters were reduced and hepatocytes predominated.
FIG. 1. Histological assessment of mouse liver development.
a. Perinatal development. Left panels are H&E-stained cryosections of mouse livers harvested at E14.5 (top) or P3 (bottom). Right panels are dark pixels selected from each corresponding left panel using Photoshop software to highlight the distribution of small dark nuclei at each stage. Green arrows indicate the position of typical hepatocytic cells at each stage; red circle surrounds a typical cluster of small non-hepatocytic cells. b. Postnatal development. H&E-stained paraffin sections from mouse livers harvested at P0, P1, P11, and P42. Green arrows indicate representative hepatocyte nuclei at each stage; red arrows indicate representative non-hepatocytic cell types found at each stage. Scale bars represent 25 μm in each panel.
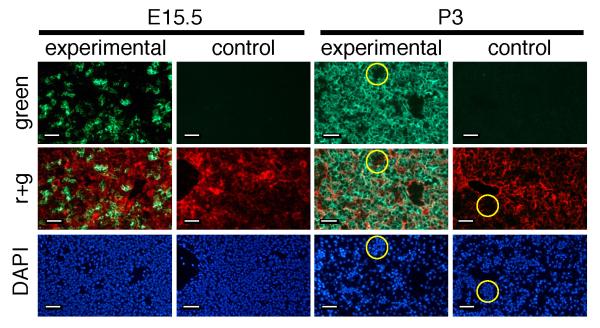
Using a marker that converts from red- to green-fluorescence following Cre-dependent recombination (Muzumdar et al., 2007), we re-evaluated perinatal Cre activity in the commonly used albCre transgenic mouse line that led to previous reports of incomplete recombination in juvenile livers (Postic and Magnuson, 2000). ROSAmT-mG/mT-mG mice (Muzumdar et al., 2007) were bred to hemizygous albCre (albCre1) mice (Postic et al., 1999) to generate pups that were either ROSAmT-mG/+;albCre1 (experimentals) or ROSAmT-mG/+;albCre0 (controls, no albCre transgene). Livers were harvested at E15.5 and P3. Green fluorescence was undetectable in controls, indicating that, in the absence of Cre expression, no GFP accumulated (Fig. 2). Conversely, in experimental animals, E15.5 livers showed a mosaic distribution of cells expressing GFP. By P3, the liver was predominated by strongly GFP-expressing cells; however close examination revealed that non-hepatocytic liver cells did not express GFP. Although the ROSAmT-mG allele is ubiquitously expressed, the intensity of fluorescent marker expression is low in small cells, likely reflecting the difference in overall gene expression between large and small cells (Schmidt and Schibler, 1995). Thus, non-hepatocytic cells in P3 livers (clustered small nuclei in DAPI-stained panel) only modestly expressed tdTomato and appeared as dark regions in the merged fluorescence panel (yellow circles). No GFP expression was observed in these clusters (Fig. 2).
FIG. 2. Perinatal recombination of ROSAmT-mG allele by albCre transgene.
ROSAmT-mG/+;albCre1 (experimental) or ROSAmT-mG/+;albCre0 (littermate control) mice were harvested at the indicated stages, stained with DAPI, and liver cryosections were analyzed by fluorescence microscopy for DAPI (bottom), green fluorescence (top) and green + red fluorescence (middle). Yellow circles in P3 samples surround a cluster of small nuclei in DAPI images, and indicate the corresponding region in micrographs above to show the relative weakness of red fluorescence and absence of green fluorescence in small hematopoietic cells. Scale bars represent 25 μm in each panel.
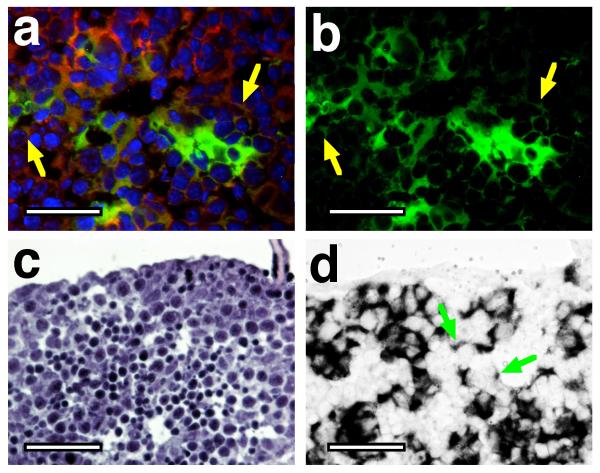
Although the P3 livers exhibited a fairly uniform distribution of albCre-converted (green) cells, the pattern of GFP fluorescence seen in livers of E15.5 fetal ROSAmT-mG/+;albCre1 animals was mosaic (Fig. 2). This could have had two underlying causes: (1) only a mosaic subpopulation of differentiated hepatocytes at E15.5 had functionally expressed Cre and converted the ROSAmT-mG allele or (2) fetal liver contained only a mosaic distribution of differentiated hepatocytes, all of which had converted the ROSAmT-mG allele. To distinguish between these, we assessed the distribution of cells expressing endogenous Alb mRNA, a marker of differentiated hepatocytes (Liao et al., 1980; Meehan et al., 1984; Murakami et al., 1987). Sections from E14.5 ROSAmT-mG/+;albCre1 livers were stained with DAPI, with H&E, or by in situ hybridization for Alb mRNA (Fig. 3). Results showed similar patterns of GFP fluorescence and Alb mRNA expression. In a few cells, GFP fluorescence or Alb mRNA expression was weak (Fig. 3, arrows); however, only following Cre-dependent recombination can any GFP be expressed from ROSAmT-mG (Fig. 2)(Muzumdar et al., 2007), and only in hepatocytes can any endogenous Alb mRNA be expressed (Meehan et al., 1984; Murakami et al., 1987). Thus, these weak expressing cells were likely newly differentiated hepatocytes that had recently activated expression of alb, albCre, and the GFP cistron of ROSAmT/mG, but had not yet accumulated high levels of GFP protein or Alb mRNA.
FIG. 3. Correlation between distribution of fetal cells having converted ROSAmT-mG allele by albCre transgene and cells expressing endogenous Alb mRNA.
Cryosections from an E14.5 fetal liver were either stained with DAPI and photographed by fluorescent microscopy (a and b), stained with H&E (c), or stained by in situ hybridization for endogenous Alb mRNA (d). Panel a shows a merged image of red (tdTomato fluorescence, non-recombined ROSAmT-mG allele), green (GFP fluorescence, recombined ROSAmT-mG allele), and blue (DAPI, nuclei) channels. Panel b shows only the green channel from the same image. Yellow arrows indicate weakly GFP-expressing cells, which have converted the ROSAmT-mG allele but not accumulated much GFP; more intensely green cells have likely been accumulating GFP protein for a longer duration (see text). Panel c is an H&E-stained cryosection taken from near the cryosection used in panel d. Panel d shows in situ hybridization for endogenous Alb mRNA. Green arrows indicate hepatocytes that have accumulated only low levels of Alb mRNA; more intensely black cells have likely been accumulating Alb mRNA for a longer duration. Scale bars represent 25 μm in each panel.
To compare expression of the endogenous alb gene with the recombination state of ROSAmT-mG in individual cells, we used immunofluorescence for nascent intracellular Alb protein in neonatal livers. Alb protein is secreted via the trans-Golgi through the constitutive pathway (Webb et al., 2005), and therefore exhibits a punctate intracellular distribution. To reduce background, P4 ROSAmT-mG/+;albCre1 pups were perfused to flush the capillaries. Cryosections were stained using anti-mouse albumin antibody and a blue-fluorescent secondary antibody. Results showed albumin in green cells but not within clusters of red cells (Fig. 4, yellow arrows). The incidence of red cells expressing Alb protein was very low, verifying that most hepatocytes had functionally expressed albCre.
FIG. 4. Intracellular expression of nascent Alb protein in neonatal ROSAmT-mG;albCre1 mouse liver.
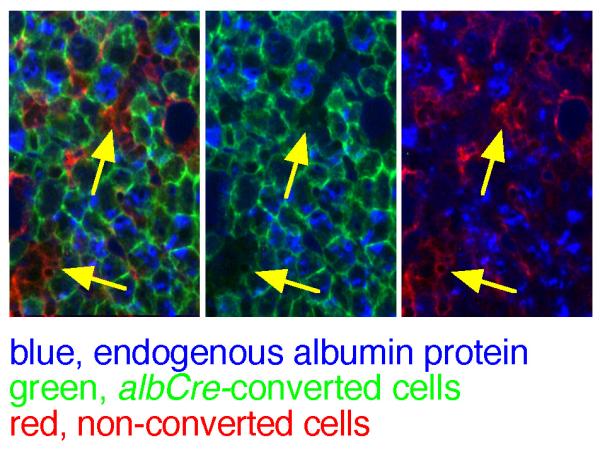
Perfused mouse livers from P4 pups were crysectioned and stained for albumin protein using a blue fluorescent secondary antibody. Panels from left to right show the same region of a single section under the blue + red + green, blue + green, or blue + red fluorescent channels. Yellow arrows draw attention to clusters of red cells that lack substantial Alb protein.
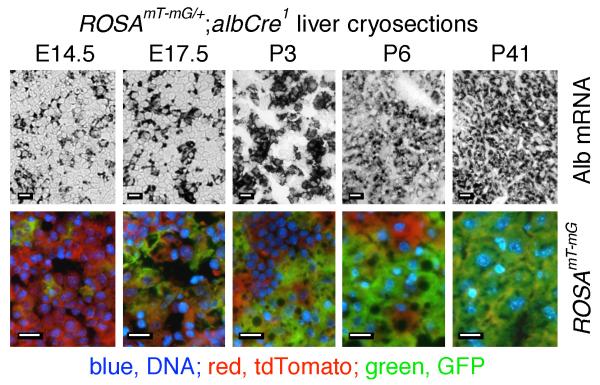
Between E14.5 and P41, the proportion of GFP-expressing cells progressively increased, which was matched by an increase in cells expressing Alb mRNA (Fig. 5). The correlation between GFP and Alb mRNA expression favored the second possibility above, that most or all differentiated hepatocytes functionally express albCre and, in fetal liver, these cells are relatively rare and distributed in a mosaic pattern.
FIG. 5. Developmental expansion of Alb mRNA-expressing hepatocytes correlates with developmental increase in liver cells that have functionally expressed Cre in albCre mice.
ROSAmT-mG/+;albCre1 mice were harvested at the indicated ages and livers were prepared for cryosectioning. Top panels, in situ hybridization for Alb mRNA (black). Lower panels, merged red (tdTomato), green (GFP, Cre-recombined cells), and blue (DAPI, nuclei) fluorescence. At each developmental time point, in situ hybridization and fluorescence microscopy were performed on separate sections from the same liver. Scale bars represent 10 μm in each panel.
Interestingly, although only a minority of cells in fetal livers were Alb mRNA-expressing hepatocytes, most of these exhibited strong Alb mRNA expression (Figs 3 & 5). We infer that the weak-expressers (Fig. 3, arrows) rapidly become high-expressing cells, even in fetal stages, suggesting the weak expressers are newly differentiated and still in the process of accumulating high levels of Alb mRNA.
In summary, in the livers of fetal and neonatal mice, most cells are hematopoietic (Paul et al., 1969); differentiated hepatocytes are a minority sub-population. During perinatal development, the liver matures into a more strictly hepatic organ (Zaret, 2000). In adult liver, ~80% of all genomes reside in hepatocytes, with the remainder being in endothelial, Kupffer, and other cell types (Duncan, 2000; Schmidt and Schibler, 1995; Zaret, 2000). Of these, only hepatocytes express Alb mRNA or protein (Meehan et al., 1984). We show that, within this milieu of developmental cell population transitions, the albCre transgene is functionally expressed in close correlation with activation of the endogenous alb gene during hepatocyte differentiation. We attribute previous reports of progressively efficient allelic conversion by albCre during this period (Postic and Magnuson, 2000) to the progressive decrease in the proportion of non-hepatocytic cell genomes in liver and not to a protracted lag between hepatocyte differentiation and functional Cre expression. We conclude that the common albCre transgene (Postic et al., 1999) is a valid and useful tool for studying most stages, including late fetal and neonatal, of hepatocyte development, especially when used in combination with a Cre activity marker like ROSAmT-mG for in situ analyses.
MATERIALS AND METHODS
Mice bearing the (ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo (Muzumdar et al., 2007) (“ROSAmT-mG”) allele and Tg(Alb-cre)21Mgn (Postic et al., 1999) (“albCre”) mice were purchased from Jackson Laboratories (stocks 007576 and 003574, respectively). ROSAmT-mG (Muzumdar et al., 2007) is a “knock-in” of the ubiquitously expressed ROSA26 locus (Soriano, 1999). ROSAmT-mG contains a floxed cistron encoding a membrane-targeted red fluorescent protein (tdTomato) followed by a cistron encoding a membrane-targeted GFP (Muzumdar et al., 2007). In its non-recombined state, ROSAmT-mG causes red fluorescence in all cells. Following Cre-dependent recombination, cells convert from red- to green-fluorescence (Muzumdar et al., 2007).
In situ hybridizations used a digoxygenin-labeled cRNA probe recognizing +1553 to +1865 of Alb mRNA (NM_009654). The procedure obliterates GFP and tdTomato so within figures, in situ hybridization and fluorescence images are of different sections from the same liver. For detection of intracellular Alb protein, anesthetized pups were sacrificed and perfused via cardiac puncture with 2 ml saline followed by 2 ml 4% paraformaldehyde/PBS. Immunofluorescence used goat-anti-mouse albumin antibody (Bethyl #A90-134A) and Alexa Fluor 350-labeled donkey-anti-goat secondary antibody (Molecular Probes #A21081). Animal protocols were approved by the Montana State University Institutional Animal Care and Use Committee.
ACKNOWLEDGEMENTS
We thank M. Rollins and Y. Piao for their contributions to this study. Supported by an NSF CAREER award, an NIH grant, and an appointment by the Montana Agricultural Experiment Station to EES, and an NIH COBRE grant to VMB.
LITERATURE CITED
- Duncan SA. Transcriptional regulation of liver development. Devel Dynam. 2000;219:131–142. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1051>3.3.co;2-e. [DOI] [PubMed] [Google Scholar]
- Kellendonk C, Opherk C, Anlag K, Schutz G, Tronche F. Hepatocyte-specific expression of Cre recombinase. Genesis. 2000;26:151–153. doi: 10.1002/(sici)1526-968x(200002)26:2<151::aid-gene17>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Liao WS, Conn AR, Taylor JM. Changes in rat alpha 1-fetoprotein and albumin mRNA levels during fetal and neonatal development. J Biol Chem. 1980;255:10036–10039. [PubMed] [Google Scholar]
- Meehan RR, Barlow DP, Hill RE, Hogan BL, Hastie ND. Pattern of serum protein gene expression in mouse visceral yolk sac and foetal liver. Embo J. 1984;3:1881–1885. doi: 10.1002/j.1460-2075.1984.tb02062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Yasuda Y, Mita S, Maeda S, Shimada K, Fujimoto T, Araki S. Prealbumin gene expression during mouse development studied by in situ hybridization. Cell Differ. 1987;22:1–9. doi: 10.1016/0045-6039(87)90408-8. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Paul J, Conkle D, Freshney RI. Erythropooietic cell population changes in the hepatic phase of erthropoiesis in the foetal mouse. Cell Prolif. 1969;2:283–294. [Google Scholar]
- Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, Shelton KD, Lindner J, Cherrington AD, Magnuson MA. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- Schmidt EE, Schibler U. Cell size regulation, a mechanism that controls cellular RNA accumulation: consequences on regulation of the ubiquitous transcription factors Oct1 and NF-Y and the liver-enriched transcription factor DBP. J Cell Biol. 1995;128:467–483. doi: 10.1083/jcb.128.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Webb RJ, Judah JD, Lo LC, Thomas GM. Constitutive secretion of serum albumin requires reversible protein tyrosine phosphorylation events in trans-Golgi. Am J Physiol Cell Physiol. 2005;289:C748–756. doi: 10.1152/ajpcell.00019.2005. [DOI] [PubMed] [Google Scholar]
- Zaret KS. Liver specification and early morphogenesis. Mech Dev. 2000;92:83–88. doi: 10.1016/s0925-4773(99)00326-3. [DOI] [PubMed] [Google Scholar]