Abstract
Most transcripts in growing cells are ribosomal RNA precursors (pre-rRNA). Here, we show that in mammals, aberrant pre-rRNA transcripts generated by RNA polymerase I (Pol I) are polyadenylated and accumulate markedly after treatment with low concentrations of actinomycin D (ActD), which blocks the synthesis of full-length rRNA. The poly(A) polymerase-associated domain-containing protein 5 is required for polyadenylation, whereas the exosome is partly responsible for the degradation of the short aberrant transcripts. Thus, polyadenylation functions in the quality control of Pol I transcription in metazoan cells. The impact of excessive aberrant RNAs on the degradation machinery is an unrecognized mechanism that might contribute to biological properties of ActD.
Keywords: actinomycin D, exosome, non-canonical poly(A) polymerase, ribosomal RNA, RNA processing
Introduction
Diverse types of RNA generated in cells by transcription and post-transcriptional processing are subject to surveillance and quality control by proteins that track and destroy aberrant RNA molecules (Doma & Parker, 2007; Houseley & Tollervey, 2009). The exosome is a complex of nine core subunits and several associated proteins (Mitchell et al, 1997; Liu et al, 2006), and it has an important role in RNA surveillance owing to its exonucleolytic and endonucleolytic functions (reviewed in Houseley et al, 2006; Schmid & Jensen, 2008; Lykke-Andersen et al, 2009). Decay of many exosome substrates in the yeast Saccharomyces cerevisiae is stimulated by the Trf4/5-Air1/2-Mtr4 polyadenylation (TRAMP) complex, which is composed of non-canonical poly(A) polymerase (PAP), Trf4 or Trf5, RNA-binding proteins Air1/2 and the putative helicase Mtr4 (LaCava et al, 2005; Vanácová et al, 2005; Wyers et al, 2005). Trf4 and Trf5 tag exosome substrates with short poly(A) tails that are thought to act as markers for degradation, similar to the role of poly(A) tails in RNA decay in prokaryotes (Deutscher, 2006). The exosome and proteins homologous to components of the TRAMP complex exist in mammals (Raijmakers et al, 2004; Martin & Keller, 2007; Houseley & Tollervey, 2008) and were shown to participate in the degradation of certain messenger RNAs (Chen et al, 2001; Mukherjee et al, 2002; West et al, 2006), as well as cryptic promoter upstream transcripts (Preker et al, 2008), but their function in higher eukaryotic cells remains poorly characterized.
The RNA polymerase I (Pol I) generates the largest fraction of all newly synthesized RNAs in growing eukaryotic cells (Warner, 1999) by transcribing a single large ribosomal RNA (rRNA) precursor—called 47S pre-rRNA in mammalian species (Eichler & Craig, 1994)—which undergoes complex post-transcriptional processing into 18S, 5.8S and 28S rRNAs and assembly into mature ribosomal subunits (reviewed in Fromont-Racine et al, 2003; Henras et al, 2008; Staley & Woolford, 2009). Actinomycin D (ActD) is a classic DNA-binding drug and transcription inhibitor capable of blocking synthesis of rRNA by Pol I in animal cells at 50–100-fold lower concentrations than those required for inhibiting Pol II- and Pol III-mediated transcription (Perry & Kelley, 1968, 1970). ActD is an anti-cancer agent that has long been known to induce the disruption of the nucleolus and activation of the tumour suppressor p53, which is usually attributed to the lack of new rRNA synthesis (Lohrum et al, 2003; Zhang et al, 2003; Bhat et al, 2004; Gilkes et al, 2006). Here, we report a dynamic accumulation of short polyadenylated Pol I transcripts in mouse cells treated with low doses of ActD, which seems to be related to the ability of this drug to induce premature termination of pre-rRNA transcripts (Fetherston et al, 1984; Hadjiolova et al, 1995). Our data reveal that the non-canonical mammalian PAP poly(A) polymerase-associated domain-containing protein 5 (Papd5) has a key role in the polyadenylation of the abortive Pol I transcripts and also show that the exosome participates in their degradation. This is the first evidence of polyadenylation acting in the surveillance of a main class of non-coding RNA in a higher eukaryotic organism. These results also raise the possibility that some of the biological effects of ActD might be due to the interference of excessive aberrant RNAs with the normal functioning of RNA degradation systems.
Results And Discussion
Treatment of mouse 3T3 cells with 20 ng/ml of ActD stops new rRNA synthesis, as can be observed by the loss of radiolabelled nucleotide incorporation into 28S and 18S rRNAs (Fig 1A). Consistent with metabolic labelling, northern hybridizations showed that the primary pre-rRNA transcript (47S) almost disappeared after 1 h of ActD treatment, and the level of its downstream processing intermediates decreased, indicating their processing and/or degradation (Fig 1B,C). In a marked contrast to normal pre-rRNAs, hybridization with probe 5′ETS-1 (5′ external transcribed spacer 1) revealed a significant accumulation of short heterogeneous RNA species derived from the 5′ region of the pre-rRNA transcript after ActD treatment (Fig 1B, left panel). This result agreed with previous studies showing that at these concentrations, ActD primarily inhibits the elongation by Pol I and prevents synthesis of complete pre-rRNA transcripts (Fetherston et al, 1984; Hadjiolova et al, 1995). To determine whether the accumulation of short RNAs resulted from abortive transcription or the impaired degradation of pre-existing transcripts, we treated cells with 2 μg/ml of ActD to shut down Pol I activity (Perry & Kelley, 1970). Short pre-rRNA species did not accumulate under these conditions, suggesting that ongoing transcription was required for their production (compare left panels in Fig 1B,D). In support of this conclusion, attenuating transcription by gradually increasing ActD concentration led to diminished accumulation of the aberrant 5′ETS transcripts (supplementary Fig S1 online).
Figure 1.
Low doses of ActD induce the accumulation of short rRNA transcripts. (A) Metabolic labelling of RNA in mouse 3T3 cells with [3H]Uridine. Cells were treated with 20 ng/ml of ActD where indicated 10 min before labelling. Incorporation of the label into mature rRNAs (28S and 18S) and their main precursors (45S, 32S) was detected by fluorography (F). Methylene blue (MB) staining of the same membrane shows equal loading. (B) Hybridization analysis of pre-rRNA in untreated cells or cells treated with 20 ng/ml of ActD for 30 or 60 min. The main precursors detected with each probe are indicated. A range of co-migrating precursors is indicated with a ‘/'; for example, 47S/45S indicates 47S, 46S and 45S. The asterisk indicates abortive transcripts that accumulate in ActD-treated cells. (C) Structure of the primary transcript (47S) synthesized by Pol I in mouse cells. The 47S pre-rRNA contains rRNA sequences flanked by ETS and separated by ITS, which are removed during processing into mature rRNAs. The main processing sites and intermediates are shown. Probes specific for spacer regions (indicated at the top) allow detection of pre-RNAs but do not hybridize to mature rRNAs. (D) Rapid decay of pre-synthesized rRNA transcripts after transcription shut-off. Cells were either untreated or treated with 20 ng/ml ActD for 30 min to induce accumulation of abortive 5′ETS transcripts. Cells were then treated with 2 μg/ml of ActD to stop further synthesis. RNA was isolated after the indicated time and analysed by northern hybridization with probe 5′ETS-1. ActD, actinomycin D; ETS, external transcribed spacer; ITS, internal transcribed spacer; pre-rRNA, rRNA precursors; rRNA, ribosomal RNA.
Phosphorimager quantification showed that 5′ETS transcripts generated in the presence of low doses of ActD are degraded rapidly, with only about 5% remaining in cells 15 min after transcription shut-off (Fig 1D, right panel). In yeast, many types of aberrant RNAs are degraded by the exosome after their transient polyadenylation by the TRAMP complex (Kadaba et al, 2004; LaCava et al, 2005; Vanácová et al, 2005; Wyers et al, 2005; Schneider et al, 2007), prompting us to investigate whether a similar pathway operates in mammalian cells. We isolated poly(A)+ RNA from untreated and ActD-treated mouse cells through two consecutive rounds of selection with oligo(dT)-conjugated magnetic particles. Control experiments showed that this stringent purification eliminated more than 99.9% non-polyadenylated rRNA from the final poly(A)+ preparation (supplementary Fig S2 online). As shown in Fig 2A, normal pre-rRNAs detectable in total RNA samples were absent from the highly purified poly(A)+ RNA (except a 47S-like band, see below). Instead, hybridizations revealed a smear of heterogeneous RNAs with a distinct cut-off that for each probe corresponded to the distance between the transcription start site and the site of probe hybridization (supplementary Table S1 online). This pattern is inconsistent with random degradation, but rather indicates a population of molecules that possess a fixed 5′ end and heterogeneous 3′ ends (Fig 2B). Control hybridizations showed the absence of significant degradation of mRNA in poly(A)+ fractions, also arguing against degradation during or after lysis (supplementary Fig S2 online). Furthermore, treatment of cells with a low dose of ActD markedly increased the amount of 3′-truncated pre-rRNA species recovered in the poly(A)+ fraction (Fig 2A), indicating that abortive Pol I transcripts undergo polyadenylation. Sequencing analysis of randomly cloned 5′ETS sequences showed wide distribution of 3′ ends, as would be expected from abortive synthesis (supplementary Fig S3 online).
Figure 2.
Polyadenylated pre-rRNAs in normal conditions and after ActD treatment. (A) Poly(A)+ RNA was prepared as described in the Methods section and analysed by northern hybridizations with probes, as indicated in Fig 1C. (B) The site of probe hybridization determines the minimal length of molecules detected in a population of RNAs with a fixed 5′ end and heterogeneous 3′ ends. The asterisks denote RNAs that would be detectable with the probe. ActD, actinomycin D; pre-rRNA, precursor rRNA; rRNA, ribosomal RNA.
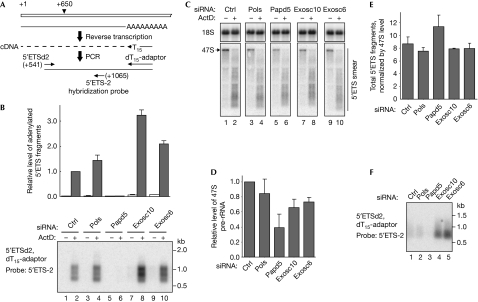
The factors that polyadenylate Pol I transcripts in mammalian cells are unknown. Two mouse proteins, DNA polymerase sigma (Pols) and Papd5, display 30–40% amino-acid sequence similarity to the S. cerevisiae proteins Trf4 and Trf5 (Martin & Keller, 2007). We used small interfering RNA (siRNA) pools to downregulate Pols and Papd5 in mouse 3T3 fibroblasts (supplementary Fig S4A online). To assess polyadenylation of Pol I transcripts, we synthesized complementary DNA from total RNA by using an oligo(dT)-anchor primer and amplified 5′ETS regions with the anchor and another primer annealing at nucleotide position +541 using a low number of cycles (Fig 3A). The resulting PCR products were visualized by Southern hybridization with the internal probe 5′ETS-2. As a complete primary rRNA transcript undergoes cleavage rapidly at nucleotide position +650 in mouse cells and all subsequent pre-rRNA intermediates lack the 5′ proximal sequence (Kass et al, 1987), the hybridization signal in this assay reflects the level of unprocessed nascent transcripts. Treatment of cells with ActD markedly increased the amount of polyadenylated RNAs derived from 5′ETS (Fig 3B, lanes 1–2), consistent with data from the northern analysis (Fig 2A). The knockdown of Pols did not significantly change the level of these RNAs relative to the control (Fig 3B, lane 4). By contrast, knockdown of Papd5 almost completely abolished their hybridization signal (Fig 3B, lane 6), indicating that Papd5 provides the main polyadenylase activity for the short Pol I transcripts generated in the presence of ActD. Analysis of total RNA by northern hybridizations showed that knockdown of Papd5 also had an inhibitory effect on Pol I transcription, as indicated by lower levels of 47S pre-rRNA (Fig 3C, lane 5; Fig 3D), but 5′ETS fragments still accumulated after ActD treatment in these cells (Fig 3C, lane 6). When normalized by the 47S signal in untreated cells, 5′ETS RNAs showed a higher accumulation after Papd5 depletion compared with control cells (Fig 3E), which is consistent with a role of Papd5 in their turnover.
Figure 3.
Polyadenylation of aberrant Pol I transcripts involves Papd5 and the exosome. (A) RT–PCR assay for semiquantitative analysis of polyadenylated pre-rRNA transcripts. (B) Hybridization of RT–PCR products to detect polyadenylated 5′ETS transcripts before and after 30 min treatment with 20 ng/ml of ActD in cells transfected with the indicated siRNAs. Average values of the signal from phosphorimager quantification of two separate PCR assays with primers 5′ETSd2 and 5′ETSd4 is shown relative to the signal in ActD-treated cells transfected with a non-targeting siRNA pool. Amplification using primer 5′ETSd2 (17 PCR cycles) is shown as an example. (C) RNA samples used for the assay shown in (B) were analysed by northern hybridization with probe 5′ETS-1 to detect nascent transcripts; 18S rRNA was used as a loading control. (D) Levels of 47S pre-rRNA in untreated cells were quantified by phosphorimager analysis of hybridizations with probe 5′ETS-1. The hybridization signal in each sample was normalized to 18S rRNA to compensate for loading variations. The data are average values from three independent transfections; one set used for the quantification is shown in (C). (E) Ratio of the 5′ETS fragments accumulating after ActD treatment to the 47S primary transcript level in the same cells before treatment. Analysis was performed as in (D). (F) Polyadenylated pre-rRNA fragments detected under normal conditions. The same RNA samples from untreated cells as in (B) were assayed using 22 PCR cycles. Error bars in all graphs indicate s.e. values. ActD, actinomycin D; Ctrl, control; ETS, external transcribed spacer; Exosc, exosome component; Papd5, poly(A) polymerase-associated domain-containing protein 5; Pols, DNA polymerase sigma; pre-rRNA, precursor rRNA; rRNA, ribosomal RNA; RT–PCR, reverse transcriptase PCR; siRNA, small interfering RNA.
To investigate nucleases involved in the decay of polyadenylated Pol I transcripts, we used siRNAs to downregulate exosome component 6 (Exosc6), the mouse homologue of the S. cerevisiae core exosome subunit Mtr3, and Exosc10 (PM/Scl100), the homologue of the yeast nuclear exonuclease Rrp6 that associates with the core exosome (Allmang et al, 1999; Burkard & Butler, 2000). The steady-state levels of polyadenylated 5′ETS RNAs were increased by knockdown of Exosc10 and, to a lesser extent, Exosc6 (Fig 3B, compare lanes 2, 8 and 10), indicating that the exosome is involved in their degradation. Surprisingly, the knockdowns showed no discernible effect on the smear intensity in total RNA after ActD treatment (Fig 3E). The level of depletion achieved for each exonuclease was functionally significant, as judged by increased accumulation of the A′–A0 fragment (supplementary Fig S4B online) excised during early pre-rRNA processing and degraded by the exosome (Kent et al, 2009). Therefore, additional mechanisms must exist in mammalian cells for the decay of aberrant pre-rRNAs, and the exosome provides only one of the redundant activities. This result also raises the possibility that polyadenylation of aberrant RNAs might have additional roles in these cells besides facilitating the degradation by the exosome.
Several observations indicate that low levels of polyadenylated pre-rRNAs are present in growing cells that are not treated with inhibitors. Northern hybridization of poly(A)+ fractions reveals a low-intensity smear of 3′ heterogeneous pre-rRNAs in untreated cells (Fig 2A, ‘-' lanes). In addition, heterogeneous polyadenylated pre-rRNAs are readily detectable in these cells by the oligo(dT)-primed reverse transcriptase PCR (RT–PCR), and their levels are substantially reduced by the depletion of Papd5 and increased by knockdown of either Exosc10 or Exosc6 (Fig 3F). These results indicate that polyadenylation-dependent pre-rRNA surveillance occurs under normal conditions, which might explain some of the polyadenylated rRNA fragments previously cloned from human cells by PCR-based approaches (Slomovic et al, 2006). Interestingly, a 47S-like band, which we refer to as 47S(A)+, was also detected in poly(A)+ fractions from untreated cells (Fig 2A, ‘-' lanes). As 47S(A)+ RNA is detectable using all pre-rRNA-specific probes including a 3′ETS probe (supplementary Fig S2 online), this RNA extends beyond the 28S rRNA sequence (Fig 1C). Unlike the normal primary transcript, 47S(A)+ RNA does not give rise to smaller processing intermediates, implying that it does not enter the normal processing pathway. These observations suggest that in addition to incomplete pre-rRNAs, a polyadenylation-mediated pathway might also target full-length, but presumably non-functional, transcripts. Further study will be required to understand the molecular details of this type of pre-rRNA surveillance.
In summary, these data, to the best of our knowledge, demonstrate for the first time that polyadenylation acts as a surveillance mechanism for Pol I transcripts in metazoan cells. Low levels of polyadenylated pre-rRNAs observed under normal conditions might reflect surveillance of occasional premature termination events by Pol I or degradation of mis-assembled pre-rRNA-processing complexes, similar to the known functions of the TRAMP–exosome pathway in yeast rRNA transcription (Schneider et al, 2007; Wery et al, 2009). Our results also reveal intensive polyadenylation of aberrant RNA species generated in mammalian cells by treatment with low doses of ActD, which is commonly used to block rRNA synthesis. Given the high activity of Pol I, many cycles of abortive elongation induced by ActD seem to overload the RNA degradation machinery that normally deals with a much lower level of defective transcripts, leading to accumulation of short polyadenylated RNAs. The effects of ActD on RNA degradation need to be considered as a possible factor in the cytotoxicity of this drug and its ability to induce stress responses.
Methods
Cell culture and transfections. The NIH3T3 cells were maintained in sub-confluent state in Dulbecco's modified Eagle medium (DMEM) containing 10% calf serum. Stock solution of ActD (Calbiochem, San Diego, CA, USA) was prepared in ethanol and verified by spectrophotometer (A420=162 for 10 mg/ml solution in 95% ethanol). For knockdowns, cells were seeded one day before transfection in a 6-well plate in 2 ml of DMEM at a density of 4 × 104 cells per well. A 100 μl solution of an optimized amount (∼0.5 μg) of siGENOME siRNA SMARTpool (Dharmacon, Lafayette, CO, USA) in 0.25 M CaCl2 was mixed with 100 μl 2 × BBS (Chen & Okayama 1988), incubated for 1–2 min and added to cells. Cells were washed with phosphate buffered saline and fed with fresh medium after 24 h. Knockdowns were analysed at 48 h after transfection and their efficiency was monitored by RT–PCR with gene-specific and control RPL23A primers, followed by gel quantification using a Kodak Gel Logic 100 system (Rochester, NY, USA). Transfections in which mRNA levels were estimated to decrease by at least 75% relative to cells transfected with Dharmacon's non-targeting siRNA pool #2 were used for further analysis.
RNA techniques. Total RNA was isolated with TRIzol (Invitrogen, Carlsbad, CA, USA). Metabolic labelling and hybridizations of rRNA were performed as described previously (Pestov et al, 2008). For poly(A)+ RNA isolation, cells grown to 80–90% confluency on three 150 mm tissue culture dishes were collected by trypsinization and pelleted at 1,000g for 5 min at 4°C. Cell lysis and the first round of poly(A) purification were carried out using a small-scale μMACS mRNA isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's protocol. The RNA was eluted from the oligo(dT) microbeads in 75 μl elution buffer, ethylenediaminetetraacetic acid was added to 5 mM, the mixture was heated to 70°C for 1 min and passed through a Microspin P30 column (Bio-Rad, Hercules, CA, USA). The RNA was ethanol precipitated after adding sodium acetate (NaOAc) to 0.3 M and 3 μg glycogen carrier (Fermentas, Burlington, ON, Canada); the pellet was washed with 80% ethanol, dried and dissolved in 12 μl of formamide by incubation on ice for 2–3 h with occasional mixing followed by 5 min at 70°C. The RNA was mixed with 250 μl MACS lysis buffer before a second round of poly(A) purification with 25 μl oligo(dT) microbeads. The resulting RNA was precipitated using NaOAc/glycogen/ethanol as above and re-suspended in 7.5 μl of formamide. The RNA concentration was measured using the fluorimetric Quant-iT RNA assay (Invitrogen).
RT–PCR analysis of polyadenylated pre-rRNA. To reliably examine changes in pre-RNAs after siRNA-mediated knockdowns, we developed an assay in which RT–PCR products were quantified by hybridization with an internal probe. Using hybridization ensured specificity of detection and allowed us to use PCR with a low number of cycles, thereby avoiding the problem of non-linear amplification rates at later cycles. First, total RNA (2.5 μg) was treated with 0.5 U of RNase-free DNAse (Worthington, Lakewood, NJ, USA) to remove contaminating genomic DNA in 100 μl of 10 mM Tris–HCl (pH 7.5), 2.5 mM MgCl2 and 0.1 mM CaCl2 for 7 min at 37°C, purified by phenol–chloroform extraction and precipitated with isopropanol. The RNA pellet was washed with 80% ethanol, air-dried and dissolved in 10 μl of formamide. About one-tenth of the RNA was mixed with 10 pmoles of primer BR3T-T15 in 10 μl of water, heated to 70°C for 5 min and chilled on ice. The cDNA was synthesized in a 20 μl reaction using Moloney murine leukaemia virus reverse transcriptase (Promega, Madison, WI, USA) in the manufacturer-supplied buffer supplemented with 20 U RNase inhibitor (Fermentas) and 0.5 mM dNTPs at 42°C for 1 h. The 5′ETS regions of pre-rRNA were amplified with primers 5′ETSd2 or 5′ETSd4 and BR3T-T15 using 17–22 cycles of PCR; both sets of primers yielded similar results. The PCR products were separated on an agarose gel and analysed by Southern hybridization with probe 5′ETS-2. PCR with primers designed to amplify a fragment of the mouse ribosomal protein Rpl23a mRNA was used to monitor the efficiency of cDNA synthesis in all samples. See supplementary Tables S1 and S2 online for oligonucleotide sequences.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
This study was supported by the National Institutes of Health grant GM074091 to D.P.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P (1999) The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes Dev 13: 2148–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KP, Itahana K, Jin A, Zhang Y (2004) Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J 23: 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard KT, Butler JS (2000) A nuclear 3′–5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol Cell Biol 20: 604–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CA, Okayama H (1988) Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. Biotechniques 6: 632–638 [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M (2001) AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464 [DOI] [PubMed] [Google Scholar]
- Deutscher MP (2006) Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res 34: 659–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R (2007) RNA quality control in eukaryotes. Cell 131: 660–668 [DOI] [PubMed] [Google Scholar]
- Eichler DC, Craig N (1994) Processing of eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol 49: 197–239 [DOI] [PubMed] [Google Scholar]
- Fetherston J, Werner E, Patterson R (1984) Processing of the external transcribed spacer of murine rRNA and site of action of actinomycin D. Nucleic Acids Res 12: 7187–7198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Senger B, Saveanu C, Fasiolo F (2003) Ribosome assembly in eukaryotes. Gene 313: 17–42 [DOI] [PubMed] [Google Scholar]
- Gilkes DM, Chen L, Chen J (2006) MDMX regulation of p53 response to ribosomal stress. EMBO J 25: 5614–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolova KV, Hadjiolov AA, Bachellerie JP (1995) Actinomycin D stimulates the transcription of rRNA minigenes transfected into mouse cells. Implications for the in vivo hypersensitivity of rRNA gene transcription. Eur J Biochem 228: 605–615 [DOI] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y (2008) The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65: 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D (2006) RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539 [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D (2008) The nuclear RNA surveillance machinery: The link between ncRNAs and genome structure in budding yeast? Biochim Biophys Acta 1779: 239–246 [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D (2009) The many pathways of RNA degradation. Cell 136: 763–776 [DOI] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J (2004) Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S, Craig N, Sollner-Webb B (1987) Primary processing of mammalian rRNA involves two adjacent cleavages and is not species specific. Mol Cell Biol 7: 2891–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent T, Lapik YR, Pestov DG (2009) The 5′ external transcribed spacer in mouse ribosomal RNA contains two cleavage sites. RNA 15: 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D (2005) RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD (2006) Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127: 1223–1237 [DOI] [PubMed] [Google Scholar]
- Lohrum MAE, Ludwig RL, Kubbutat MHG, Hanlon M, Vousden KH (2003) Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3: 577–587 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S, Brodersen DE, Jensen TH (2009) Origins and activities of the eukaryotic exosome. J Cell Sci 122: 1487–1494 [DOI] [PubMed] [Google Scholar]
- Martin G, Keller W (2007) RNA-specific ribonucleotidyl transferases. RNA 13: 1834–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 91: 457–466 [DOI] [PubMed] [Google Scholar]
- Mukherjee D, Gao M, O'Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J (2002) The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J 21: 165–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RP, Kelley DE (1968) Persistent synthesis of 5S RNA when production of 28S and 18S ribosomal RNA is inhibited by low doses of actinomycin D. J Cell Physiol 72: 235–246 [DOI] [PubMed] [Google Scholar]
- Perry RP, Kelley DE (1970) Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J Cell Physiol 76: 127–139 [DOI] [PubMed] [Google Scholar]
- Pestov DG, Lapik YR, Lau LF (2008) Assays for ribosomal RNA processing and ribosome assembly. Curr Protoc Cell Biol Chapter 22: Unit 22.11 [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH (2008) RNA exosome depletion reveals transcription upstream of active human promoters. Science 322: 1851–1854 [DOI] [PubMed] [Google Scholar]
- Raijmakers R, Schilders G, Pruijn GJM (2004) The exosome, a molecular machine for controlled RNA degradation in both nucleus and cytoplasm. Eur J Cell Biol 83: 175–183 [DOI] [PubMed] [Google Scholar]
- Schmid M, Jensen TH (2008) The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci 33: 501–510 [DOI] [PubMed] [Google Scholar]
- Schneider DA, Michel A, Sikes ML, Vu L, Dodd JA, Salgia S, Osheim YN, Beyer AL, Nomura M (2007) Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol Cell 26: 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic S, Laufer D, Geiger D, Schuster G (2006) Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Res 34: 2966–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Woolford JLJ (2009) Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr Opin Cell Biol 21: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanácová S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W (2005) A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440 [DOI] [PubMed] [Google Scholar]
- Wery M, Ruidant S, Schillewaert S, Leporé N, Lafontaine DLJ (2009) The nuclear poly(A) polymerase and Exosome cofactor Trf5 is recruited cotranscriptionally to nucleolar surveillance. RNA 15: 406–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Gromak N, Norbury CJ, Proudfoot NJ (2006) Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol Cell 21: 437–443 [DOI] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle J, Dufour M, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B et al. (2005) Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y (2003) Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 23: 8902–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information