Abstract
The intracellular retinoic acid-inducible gene I-like receptors (RLRs) sense viral ribonucleic acid and signal through the mitochondrial protein mitochondrial antiviral signalling (MAVS) to trigger the production of type I interferons and proinflammatory cytokines. In this study, we report that RLR activation promotes elongation of the mitochondrial network. Mimicking this elongation enhances signalling downstream from MAVS and favours the binding of MAVS to stimulator of interferon genes, an endoplasmic reticulum (ER) protein involved in the RLR pathway. By contrast, enforced mitochondrial fragmentation dampens signalling and reduces the association between both proteins. Our finding that MAVS is associated with a pool of mitofusin 1, a protein of the mitochondrial fusion machinery, suggests that MAVS is capable of regulating mitochondrial dynamics to facilitate the mitochondria–ER association required for signal transduction. Importantly, we observed that viral mitochondria-localized inhibitor of apoptosis, a cytomegalovirus (CMV) antiapoptotic protein that promotes mitochondrial fragmentation, inhibits signalling downstream from MAVS, suggesting a possible new immune modulation strategy of the CMV.
Keywords: MAVS, innate immunity, mitochondrial dynamics, RIG-I-like receptors, virus
Introduction
After infection, viruses are recognized by the innate immune system through germline-encoded pattern-recognition receptors (Akira et al, 2006). Two classes of pattern-recognition receptors, including Toll-like receptors and retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), recognize viral components and directly activate immune cells. RLRs comprise RIG-I and melanoma differentiation-associated gene 5 (MDA5), which are helicases sensing viral RNA (Yoneyama & Fujita, 2008). Recently, it was reported that RNA polymerase III detects cytosolic DNA and activates innate immunity through RIG-I (Ablasser et al, 2009; Chiu et al, 2009). Importantly, RIG-I and MDA5 contain two caspase recruitment domains (CARDs) that are required for interaction with the CARD domain of the mitochondrial adaptor mitochondrial antiviral signalling (MAVS; also known as IPS-1, Cardif and VISA; Kawai et al, 2005; Meylan et al, 2005; Seth et al, 2005; Xu et al, 2005). MAVS then activates two cytosolic protein kinase complexes, one consisting of the ‘non-canonical' initiation κB kinase (IKK)-related TANK binding kinase 1 (TBK1) or IKK-i associated with various adaptor proteins, and the other containing IKKα, IKKβ and nuclear factor-κB (NF-κB) essential modulator (NEMO; Akira et al, 2006). The TBK1 complex leads to phosphorylation of transcription factors interferon regulatory factor (IRF) 3 and IRF7 to induce the expression of type I interferons (IFNs). The IKK complex activates NF-κB, subsequently promoting the expression of proinflammatory cytokines.
It has been reported that MAVS must be localized to mitochondria to exert its function (Seth et al, 2005), suggesting that the mitochondrial environment is required for signal transduction after activation of the RLR. Mitochondrial shape varies in living cells and can range from punctuate structures to tubular networks, and some signals such as death signals or calcium signalling have been reported to affect mitochondrial dynamics (McBride et al, 2006). Here, we report that activation of the RLR pathway promotes elongation of the mitochondrial network and that changes in mitochondrial dynamics modulate signalling downstream from MAVS. We therefore provide evidence that the functions of mitochondrial dynamics also include intracellular viral detection signalling. Interestingly, the cytomegalovirus (CMV) protein viral mitochondria-localized inhibitor of apoptosis (vMIA), which promotes fragmentation of the mitochondrial network, impedes signalling downstream from MAVS, suggesting a possible new immune modulation strategy of CMV.
Results And Discussion
RLR activation promotes mitochondrial elongation
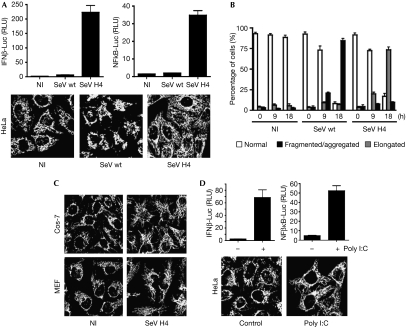
It has been reported that some intracellular signalling pathways affect mitochondrial dynamics (McBride et al, 2006). As the RLR pathway requires a mitochondrial step through the adaptor MAVS (Kawai et al, 2005; Meylan et al, 2005; Seth et al, 2005; Xu et al, 2005; Akira et al, 2006), we inferred that RLR activation might induce modifications in mitochondrial morphology. A defective Sendai virus (SeV) strain, such as H4 (Strahle et al, 2006), is a model for activating RLR (Fig 1A; supplementary Fig S1A,B online). Whereas infection of HeLa cells with the wild-type (wt) SeV strain triggered the fragmentation and aggregation of mitochondria—probably as a consequence of the cellular stress induced by high levels of viral replication—infection with SeV H4 promoted the elongation of the mitochondrial network, compared with uninfected cells (Fig 1A,B). A similar elongation of the mitochondrial network was observed in other cell lines (Fig 1C). RIG-I knockdown inhibited signalling after SeV H4 infection (supplementary Fig S2A online) and prevented mitochondrial elongation (supplementary Fig S2B online). Finally, cytosolic double-stranded RNA such as poly I:C, which activates the RLR pathway (Fig 1D; supplementary Fig S1C online; Akira et al, 2006; Yoneyama & Fujita, 2008), also resulted in elongation of the mitochondrial network (Fig 1D). Our data indicate that specific activation of the RLR promotes elongation of the mitochondrial network.
Figure 1.
RLR activation promotes elongation of the mitochondrial network. (A) HeLa cells were transfected with either an IFNβ promoter reporter or an NF-κB reporter, as well as with Renilla luciferase as an internal control. At 20 h after transfection cells were infected with SeV wt or SeV H4, or left uninfected. Luciferase assays were performed 9 h after infection and were normalized using Renilla luciferase activity. The error bars represent the standard deviation (s.d.) from the mean value obtained from three experiments. In parallel, mitochondrial morphology was assessed by immunofluorescence 18 h after infection. (B) Histogram for quantitating mitochondrial morphology in HeLa cells that were left either uninfected or infected with SeV wt or SeV H4 for 9 h or 18 h. Data are the means±s.d. of three independent experiments, with 300 cells per condition. (C) Cos-7 cells or MEFs were infected with SeV H4 for 18 h and mitochondrial morphology was observed by immunofluorescence. (D) HeLa cells were transfected as in panel (A). Twenty hours later cells were transfected with poly I:C (1 μg/ml) for 8 h and luciferase assays were performed. In parallel, mitochondrial morphology was assessed by immunofluorescence. IFN, interferon; Luc, luciferase; MEF, mouse embryonic fibroblast; NI, uninfected; NF-κB, nuclear factor-κB; RLR, RIG-I-like receptor; RLU, relative luciferase unit; SeV, Sendai virus; wt, wild type.
In various cell lines, including HeLa, human embryonic kidney 293 (HEK293; supplementary Fig S1A,B online), A549, Huh7 or Jurkat cells (data not shown), MAVS is expressed as two main isoforms (supplementary Fig S3 online). Interestingly, following SeV H4 but not SeV wt infection, the higher form was degraded (supplementary Fig S1A,B online). Degradation of the higher isoform of MAVS was concomitant with phosphorylation of both IRF3 and the NF-κB inhibitor IκBα, suggesting that this degradation might be required for downstream signalling (supplementary Fig S1A,B online).
Mitochondrial dynamics modulate the RLR pathway
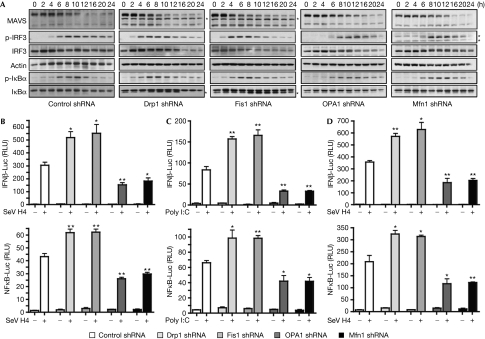
Next, we investigated the role of mitochondrial network elongation in the RLR pathway. Mitochondrial dynamics were altered by knocking down the effectors of the mitochondrial fission and fusion machinery, such as Drp1, Fis1, OPA1 or mitofusion 1 (Mfn1), with short-hairpin RNA (shRNA; supplementary Fig S4A online). Dynamin-related protein 1 (Drp1) and Fis1 are both effectors of the mitochondrial fission machinery, whereas optic atrophy type 1 (OPA1) and Mfn1 are required for mitochondrial fusion (Chan, 2006). Knockdown of Drp1 or Fis1 promoted mitochondrial fusion, whereas knockdown of OPA1 or Mfn1 induced fragmentation (supplementary Fig S4B,C online). We investigated the kinetics of signalling downstream from MAVS by studying the phosphorylation of IRF3 and IκBα at various times after SeV H4 infection in Drp1 or Fis1 shRNA-transfected cells, as compared with that in control shRNA-treated cells (Fig 2A). Interestingly, we observed that phosphorylation of IRF3 and IκBα occurred faster in cells where mitochondria are fused. Phosphorylation of IRF3 and IκBα was also assessed in OPA1 or Mfn1 shRNA-transfected cells in which mitochondria are fragmented, and in these cells phosphorylation of both factors was delayed (Fig 2A). Furthermore, MAVS degradation was accelerated in cells where mitochondria were fused, whereas it was delayed when mitochondria were fragmented, thus confirming a link between degradation of the higher form of MAVS and downstream signalling. Luciferase assays with SeV-H4-infected HeLa or HEK293 cells confirmed that elongation of the mitochondrial network by depletion of Drp1 or Fis1 either with shRNA or small interfering RNA (siRNA; supplementary Fig S5A,B online) enhances significantly the activation of the IFNβ promoter and NF-κB, whereas in cells where mitochondria were fragmented by the knockdown of OPA1 or Mfn1, both events were inhibited (Fig 2B,D; supplementary Fig S5C online). Similar results were obtained with HeLa cells transfected with poly I:C to activate the RLR (Fig 2C). Finally, activation of the IFNβ promoter was unaffected after transfection of TBK1 in cells where mitochondrial morphology was altered, whereas it was affected after transfection of MAVS (supplementary Fig S6 online). Together, our results suggest that elongation and fusion of the mitochondrial network specifically enhances MAVS-mediated signalling, whereas fragmentation of mitochondria has the opposite effect. Therefore, it seems that elongation of the mitochondrial network after RLR activation stimulates signalling downstream from MAVS.
Figure 2.
The RLR signalling pathway is modulated by alterations in mitochondrial dynamics. (A) HeLa cells were transfected with control, Drp1, Fis1, OPA1 or Mfn1 shRNA. After the selection of transfectants, cells were infected with SeV H4 and at various times after infection MAVS, p-IRF3, IRF3, p-IκBα and IκBα were analysed in cell extracts by immunoblotting. Actin was used as a protein loading control. *A probable non-specific protein band. Data shown are representative of three independent experiments. (B,C) IFNβ-Luc or NF-κB-Luc reporter plasmids were transfected into control cells or Drp1-, Fis1-, OPA1- or Mfn1-depleted cells, which were then infected with SeV H4 for 9 h (B) or transfected with poly I:C for 8 h (C), and then IFNβ induction and NF-κB activation were assessed. (D) The same conditions as in panel B, but HeLa cells were replaced by human embryonic kidney 293 cells. **0.001<P<0.01, *0.01<P<0.05. Drp1, dynamin-related protein 1; IFN, interferon; IRF, IFN regulatory factor; Luc, luciferase; MAVS, mitochondrial antiviral signalling; Mfn1, mitofusion 1; NF-κB, nuclear factor-κB; OPA1, optic atrophy type 1; RLR, RIG-I-like receptor; RLU, relative luciferase unit; SeV, Sendai virus; shRNA, short-hairpin RNA.
MAVS regulates mitochondrial morphology
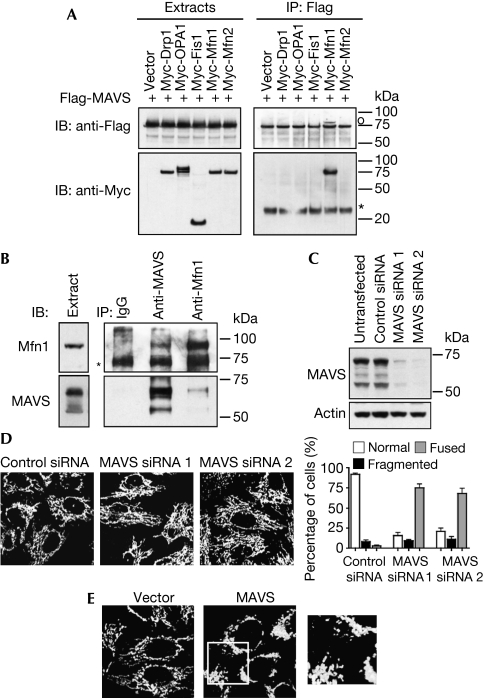
Given that RLR activation promotes mitochondrial elongation and that MAVS is a mitochondrial protein, we investigated whether MAVS might regulate mitochondrial morphology. In co-immunoprecipitation experiments, we detected an association of MAVS with Mfn1, which is a protein of the mitochondrial fusion machinery associated to the outer membrane (Chan, 2006), but not with other effectors of the mitochondrial dynamic machinery (Fig 3A). Interaction between both proteins was confirmed further by reciprocal co-immunoprecipitations of endogenous MAVS and Mfn1 (Fig 3B). As mentioned, RLR activation promotes elongation of the mitochondrial network, which is associated with degradation of the higher isoform of MAVS (supplementary Fig S1 online). To mimic MAVS degradation, MAVS expression was knocked down with siRNAs. Whereas only the higher form of MAVS was degraded after viral infection, transfection of MAVS siRNA induced a knockdown of all forms of MAVS (Fig 3C), but it similarly induced an elongation of the mitochondrial network (Fig 3D). Therefore, we propose that MAVS degradation releases Mfn1, thus allowing Mfn1 to promote the mitochondrial fusion and elongation that is observed when RLRs are activated. MAVS would then function as a negative regulator of Mfn1. In agreement with this hypothesis, MAVS expression in cells promotes mitochondrial fission (Fig 3E).
Figure 3.
MAVS binds to Mfn1 and regulates mitochondrial morphology. (A) Human embryonic kidney 293 cells were transfected with Flag-MAVS and Myc constructs, where indicated, and cell extracts and immunoprecipitates (IP) were analysed by immunoblotting (IB). *IgG light chain; °a band from a previous immunoblot. (B) Endogenous Mfn1 and MAVS were immunoprecipitated from cell extracts with specific antibodies, and the presence of both proteins in immune complexes was examined by immunoblotting. *IgG heavy chain. (C,D) Control siRNA or siRNAs targeting MAVS were transfected into HeLa cells. Knockdown of MAVS was confirmed by immunoblotting (C) and mitochondrial morphology was analysed by immunofluorescence (D). (E) Control vector or MAVS-expressing vector was transfected into HeLa cells. Twenty-four hours later, mitochondrial morphology was analysed by immunofluorescence. An enlarged view of the boxed area is shown to the right. Drp1, dynamin-related protein 1; IgG, immunoglobulin G; OPA1, optic atrophy type 1; MAVS, mitochondrial antiviral signalling; Myc, myelocytose; siRNA, small interfering RNA.
Mitochondrial dynamics regulate MAVS–STING association
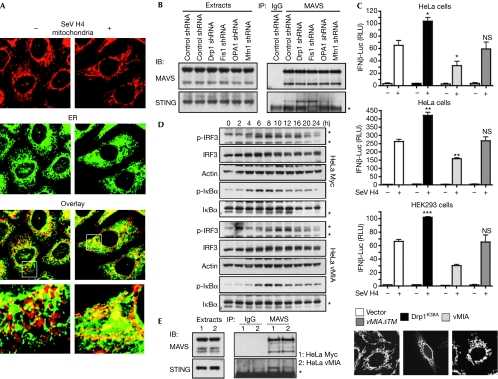
We examined the morphology of mitochondria and endoplasmic reticulum (ER) in uninfected or SeV-H4-infected cells. In infected cells, a ‘dereticulation' of the ER was observed, which is associated with mitochondrial elongation (Fig 4A). Importantly, ER was found to be associated more to elongated mitochondria compared with control cells (Fig 4A; supplementary Fig S7 online). We then hypothesized that elongation of mitochondria after RLR activation favours the binding of MAVS to an ER factor required for signalling. Recently, such a factor was uncovered through identification and characterization of stimulator of interferon genes (STING; also called MITA, MPYS or ERIS), an ER protein (supplementary Fig S8 online) that was reported to be associated with MAVS and is crucial for type I IFN production and NF-κB activation after viral infection (Ishikawa & Barber, 2008; Zhong et al, 2008; Sun et al, 2009).
Figure 4.
Mitochondrial dynamics modulate the MAVS–STING association. (A) HeLa cells were infected or not with SeV H4. After 18 h, mitochondrial and ER morphology were examined by immunofluorescence. (B) HeLa cells were transfected with control, Drp1, Fis1, OPA1 or Mfn1 shRNA. After the selection of transfectants, MAVS was immunoprecipitated (IP) from cell extracts and the association with STING in each condition was examined by immunoblotting (IB). *IgG light chain. Data shown are representative of three independent experiments. (C) IFNβ-Luc or NF-κB-Luc reporter plasmids, as well as vMIA, vMIAΔTM, Drp1K38A or the vector alone, were transfected into the indicated cells. Sixteen hours later cells were infected with SeV H4 for 9 h or transfected with poly I:C for 8 h, and then IFNβ induction and NF-κB activation were assessed. ***P<0.001, **0.001<P<0.01, *0.01<P<0.05. NS, not significant (P>0.05). Mitochondrial morphology was also analysed by immunofluorescence. (D) Control HeLa cells (HeLa Myc) or HeLa cells stably expressing Myc-tagged vMIA (HeLa vMIA) were infected with SeV H4. At various time points after infection, p-IRF3, IRF3, p-IκBα and IκBα were analysed in cell extracts by immunoblotting. Actin was used as a protein loading control. *A probable non-specific protein band. (E) MAVS was immunoprecipitated from cell extracts of control HeLa cells or HeLa cells stably expressing vMIA and the association with STING was examined by immunoblotting. *IgG light chain. Data shown are representative of three independent experiments. Drp1, dynamin-related protein 1; ER, endoplasmic reticulum; HEK293, human embryonic kidney 293; IFN, interferon; IgG, immunoglobulin G; IRF, IFN regulatory factor; Luc, luciferase; MAVS, mitochondrial antiviral signalling; Myc, myelocytose; NF-κB, nuclear factor-κB; OPA1, optic atrophy type 1; RLU, relative luciferase unit; SeV, Sendai virus; shRNA, short-hairpin RNA; STING, stimulator of interferon genes; vMIA, viral mitochondria-localized inhibitor of apoptosis.
Given that STING has been reported to be a MAVS-interacting protein (Ishikawa & Barber, 2008; Zhong et al, 2008), we explored whether changes in mitochondrial dynamics might affect the association between both proteins. Interestingly, the interaction was more prominent in cells with elongated mitochondria, whereas it was inhibited with fragmented mitochondria (Fig 4B; supplementary Fig S5D online), suggesting that modifications in mitochondrial dynamics regulate the MAVS–STING association. As MAVS is found in a complex with STING and Mfn1 (supplementary Fig S9 online), we propose that after viral infection, at the sites of ER–mitochondrial tethering, activated MAVS transduces the signals leading to production of type I IFNs and cytokines. Simultaneously, the higher isoform of MAVS is then degraded selectively, releasing Mfn1, which in turn promotes mitochondrial fusion to enhance ER–mitochondrial tethering for further associations between MAVS and STING, and ensuing new initiations of signalling downstream from MAVS. Consequently, initiation of MAVS-mediated signalling propagates all along the ER–mitochondrial junctions through changes in mitochondrial dynamics. Interestingly, while our paper was in revision, it was described that Mfn2, a close homologue of Mfn1, inhibits mitochondrial antiviral immunity (Yasukawa et al, 2009). So, although homologous proteins Mfn1 and Mfn2 have a similar function in mitochondrial fusion (Chan, 2006), it seems that they have opposite roles in viral innate immunity. As Mfn1 and Mfn2 also engage homotypic as well as heterotypic interactions on the mitochondrial outer membrane, associations with their partners might ensure fine-tuning of MAVS-mediated signalling, in addition to regulation of mitochondrial fusion.
Viruses have evolved strategies to interfere with antiviral signalling pathways (Arnoult et al, 2009). vMIA is an antiapoptotic protein encoded by CMV that triggers fragmentation of the mitochondrial network (Fig 4C; McCormick et al, 2003). In agreement with our findings, we observed that vMIA expression dampens signalling downstream from MAVS (Fig 4C,D) by inducing a reduction in the MAVS–STING association (Fig 4E). vMIA expression also delayed the degradation of the higher isoform of MAVS (data not shown). As expected, expression of a dominant-negative mutant of Drp1 (Drp1K38A) generated not only mitochondrial fusion but also increased the activation of the IFNβ promoter (Fig 4C). Finally, expression of a vMIA mutant (vMIAΔTM) that does not alter mitochondrial morphology (Fig 4C) did not modulate signalling downstream from MAVS (Fig 4C). The observation that vMIA is an antiapoptotic protein that also inhibits MAVS-mediated signalling by promoting mitochondrial fission, probably contributes to the requirement of vMIA for CMV replication and pathogenesis in vivo.
In conclusion, our findings shed new light on mitochondrial function during viral infection. We provide evidence that the functions of mitochondrial dynamics are not restricted to regulation of the transmission of mitochondrial DNA, calcium signalling, cellular energetics or apoptosis (Chan, 2006; McBride et al, 2006), but also include intracellular viral detection signalling, thereby opening a new direction for the functional roles of mitochondria within cells.
Methods
Luciferase assay. HeLa or HEK293 cells were seeded in 24-well plates at a cell density of 0.5 × 105 or 1 × 105 cells per well, respectively. On the second day, cells were co-transfected with 50 ng of firefly luciferase constructs under the control of the IFNβ promoter or driven by three copies of an NF-κB enhancer, and 10 ng of the Renilla luciferase pRL-TK plasmid (Promega, Madison, WI, USA). The next day, cells were either infected by SeV or transfected by poly I:C (1 μg/ml; Invivogen, San Diego, CA, USA) with oligofectamine (Invitrogen) for a few hours. The lysates were analysed using the dual-luciferase reporter assay (Promega) on a Fluorostar Optima (BMG Labtech, Champigny-sur-Marne, France). Each experiment was performed in triplicate. For each sample, to obtain relative fluorescence units firefly luciferase fluorescence units were normalized to Renilla luciferase fluorescence units.
Protein extraction and antibodies. Cells were lysed in buffer-A (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 20 mM β-glycerophosphate, 1 mM sodium orthovanadate, 10% glycerol, 0.5 mM EGTA, 0.2% NP-40, 1 mM dithiothreitol) supplemented with the protease inhibitor mixture, Complete (Roche Molecular Biochemicals, Meylan, France). After incubation on ice for 10 min, a soluble extract was collected after centrifugation at 20,000g for 15 min at 4°C. The lysate (30 μg) was boiled in sodium dodecyl sulphate sample buffer and resolved by sodium dodecyl sulphate–polyacrylamide gel electrophoresis.
The antibodies used in immunoblotting were: mouse monoclonal anti-RIG-I (Alexis Biochemicals, Lausen, Switzerland; clone Alme-1; 1:2,000); mouse monoclonal anti-Cardif/MAVS (Alexis Biochemicals; clone Adri-1; 1:2,000); mouse monoclonal anti-actin (Sigma-Aldrich, Saint Louis, MI, USA; clone AC-40; 1:5,000); rabbit monoclonal anti-phospho-IRF3 (Cell Signaling, Beverly, MA, USA; clone 4D4G; 1:1,000); rabbit polyclonal anti-IRF3 (Santa Cruz Biotechnology, Santa Cruz, CA, USA; FL-425; 1:2,000); mouse monoclonal anti-phospho-IκBα (Cell Signaling; clone 5A5; 1:2,000); rabbit polyclonal anti-IκBα (Santa Cruz Biotechnology; C-21; 1:2,000); mouse monoclonal anti-Drp1/DLP1 (BD Biosciences Pharmingen, Franklin Lakes, NJ, USA; clone 8; 1:1,000); mouse monoclonal anti-OPA1 (BD Biosciences Pharmingen, clone 18; 1:1,000); rabbit polyclonal anti-Fis1 (Alexis Biochemicals; 1:500); rabbit polyclonal anti-STING (Ishikawa & Barber, 2008; 1:2,000); rabbit polyclonal anti-Mfn1, mouse monoclonal anti-Hsp60 (BD Biosciences Pharmingen, clone 24; 1:4,000); mouse monoclonal anti-calreticulin (BD Biosciences Pharmingen, clone 16; 1:2,000); mouse monoclonal anti-FLAG (Sigma-Aldrich, clone M2; 1:4,000) and mouse monoclonal anti-Myc (Sigma-Aldrich, clone 9E10; 1:2,000).
Details on all other procedures are in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary information
Acknowledgments
We are grateful to M. Rojo (Institut de Biochimie et Génétique Moléculaire, Bordeaux, France), C. Blackstone (National Institute of Neurological Disorders and Stroke/National Institutes of Health, Bethesda, MD, USA), S.J. Martin (Smurfit Institute, Dublin, Ireland), R.J. Youle (NIH/NINDS, Bethesda, MD, USA), V. Goldmacher (Immunogen Inc., Cambridge, MA, USA) and G.N. Barber (University of Miami School of Medicine, Miami, FL, USA) for providing reagents. This work was supported by grants from Agence Nationale de La Recherche sur le SIDA, Fondation pour La Recherche Médicale and La Ligue contre le Cancer (équipe labellisée) and from Université Paris Sud.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V (2009) RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol 10: 1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O (2006) Pathogen recognition and innate immunity. Cell 124: 783–801 [DOI] [PubMed] [Google Scholar]
- Arnoult D, Carneiro L, Tattoli I, Girardin SE (2009) The role of mitochondria in cellular defense against microbial infection. Semin Immunol 21: 223–232 [DOI] [PubMed] [Google Scholar]
- Chan DC (2006) Mitochondria: dynamic organelles in disease, aging, and development. Cell 125: 1241–1252 [DOI] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ (2009) RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138: 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN (2008) STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455: 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S (2005) IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6: 981–988 [DOI] [PubMed] [Google Scholar]
- McBride HM, Neuspiel M, Wasiak S (2006) Mitochondria: more than just a powerhouse. Curr Biol 16: R551–R560 [DOI] [PubMed] [Google Scholar]
- McCormick AL, Smith VL, Chow D, Mocarski ES (2003) Disruption of mitochondrial networks by the human cytomegalovirus UL37 gene product viral mitochondrion-localized inhibitor of apoptosis. J Virol 77: 631–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J (2005) Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437: 1167–1172 [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ (2005) Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122: 669–682 [DOI] [PubMed] [Google Scholar]
- Strahle L, Garcin D, Kolakofsky D (2006) Sendai virus defective-interfering genomes and the activation of interferon-β. Virology 351: 101–111 [DOI] [PubMed] [Google Scholar]
- Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z (2009) ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci USA 106: 8653–8658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB (2005) VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 19: 727–740 [DOI] [PubMed] [Google Scholar]
- Yasukawa K, Oshiumi H, Takeda M, Ishihara N, Yanagi Y, Seya T, Kawabata S, Koshiba T (2009) Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal 2: ra47. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T (2008) Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity 29: 178–181 [DOI] [PubMed] [Google Scholar]
- Zhong B et al. (2008) The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29: 538–550 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary information