Abstract
In every synapse, a large number of proteins interact with other proteins in order to carry out signaling and transmission in the central nervous system. In this study, we used interaction proteomics to identify novel synaptic protein interactions in mouse cortical membranes under native conditions. Using immunoprecipitation, immunoblotting, and mass spectrometry, we identified a number of novel synaptic protein interactions involving soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs), calcium-activated potassium channel (BKCa) alpha subunits, and dynamin-1. These novel interactions offer valuable insight into the protein-protein interaction network in intact synapses that could advance understanding of vesicle trafficking, release, and recycling.
Keywords: Exocytosis, Immunoprecipitation, Mass spectrometry, Protein - protein interactions, SNARE, Synaptic vesicles
Cellular function in higher organisms depends ultimately on interactions among proteins and other molecules. Indeed, proteins do not act in isolation but engage in complexes and dynamically interact with other proteins to fulfill their diverse cellular roles [1]. Interaction proteomics is a rapidly developing field providing crucial insight into the interactions among proteins; we have used this technique to study protein complexes in synapses, which are considered the most complicated and functionally diverse cellular organelles, with about 1,500 proteins that interact in an activity-dependent manner [2].
There is evidence that some of these diverse proteins may interact in the synapse to mediate neuronal transmission. For example, dynamin is involved in (but not sufficient for) constricting the neck of nascent vesicles and thereby promoting fission [3]. Moreover, SNARE complex protein members SNAP-25, syntaxin, and VAMP-2, are the key players in membrane fusion, and they are also involved in neurotransmitter vesicle transport and targeting [4]. Since both SNAREs and dynamin are individually involved in membrane fusion, a complex involving these proteins could theoretically assemble to help orchestrate neurotransmission in the synapse. Given that a Ca2+-dependent interaction between dynamin and synaptophysin has been hypothesized in the recycling process [5], Ca2+ may also be a factor in a dynamin/SNARE complex. Furthermore, large conductance, calcium-activated potassium channels (BKCa or maxi-K) are widely expressed in the brain and are also known to influence transmitter release, regulate intracellular Ca2+, and play important roles in neurotransmission [6]. There is evidence that the BKCa channel co-associates with syntaxin and several brain proteins [7–9]. Some of its partners can, in turn, interact among themselves and with dynamin, clathrin, and endocytosis-related proteins [10–12]. Interestingly, another type of potassium channel known as Kir2.3 has been shown to co-immunoprecipitate with alpha-adaptin (AP-2) and the internalization is clathrin- and dynamin-dependent [13], providing further evidence for protein complexes involving potassium channels and endocytic proteins such as dynamin.
The known interactions discussed above, together with some partner proteins that they share in common, suggest that there may be common players in protein-protein interactions underlying synaptic transmitter release and signaling. We used co-immunoprecipitation, immunoblotting, and mass spectrometry to investigate the potential interaction of dynamin, SNAREs, and BKCa channels as part of a functional synaptic network. Our results uncovered novel, definitive interactions among synaptic proteins, including the BKCa channel with dynamin-1 and munc-18, and the SNARE protein VAMP-2 and dynamin-1. The novel interactions reported here provide important new evidence for synaptic protein complexes that can advance the understanding of vesicle trafficking, release, and recycling.
2. Materials and Methods
2.1 Sample preparation and co-immunoprecipitation
Preparation of cortical membranes from young adult male mice was carried out as previously described [14]. Briefly, samples were homogenized in a mildly denaturing buffer (containing 1% Triton X-100) and cleared by low speed centrifugation. The soluble material was collected and used for co-immunoprecipitation assays. A detailed description is available in the Supplemental Data.
Co-immunoprecipitation was performed as described previously [7]. The soluble cortical extract was diluted and pre-cleared by using bovine serum albumin (BSA) and protein A-Sepharose (Zymed, San Francisco, CA) or A/G Agarose (Thermo Scientific, Waltham, MA) beads. Pre-cleared samples were incubated first with specific antibodies listed in Suppl. Table 1 and then with fresh beads. The co-immunoprecipitated proteins were eluted from the beads, resolved by SDS-PAGE and analyzed by Western blotting or mass spectrometry. See Supplemental Data for detailed procedures.
2.2 Mass spectrometric analysis
Synaptosomal associated protein of 25 kDa (SNAP-25), vesicle-associated membrane protein 2 (VAMP-2), and BKCa alpha subunit associated proteins were separated by 4–20% SDS–PAGE, and the bands were visualized by Coomassie staining (Thermo Scientific, Waltham, MA) (Suppl. Fig. 1). Discrete specific bands ranging from 15 to 150 kDa and their corresponding FLAG (non-specific control) bands from the adjacent lanes were excised and digested with trypsin. Tryptic peptides from proteins present in excised gel bands were subjected to liquid chromatography, and the eluted peptides were then spotted directly onto a MALDI plate (Applied Biosystems, Foster City, CA) and subjected to tandem mass spectrometry (MS/MS). The spotted peptides were analyzed using the 4700 TOF-TOF proteomics analyzer (Applied Biosystems, Foster City, CA). A job-wide interpretation method was used that collected up to 10 tandem mass spectrometry (MS/MS) spectra of the top 10 most abundant peaks in the original MS spectrum of each spot. Further details are available in the Supplemental Data.
2.3 Strategy for protein identification
The MS/MS data were searched against the SwissProt mouse database using the GPS Explorer v.3.6 software suite (Applied Biosystems, Foster City, CA) and the MASCOT v.2.2 search engine [15], as previously described [12,16]. See Supplemental Data for detailed parameters and a brief description of the procedure.
Proteins were identified based on at least two peptide matches and were classified into high- and medium- probability hits using the following criteria: high-probability hits were proteins identified by MASCOT as having an ions probability-based Molecular Weight Search (MOWSE) score that is greater than the cutoff indicated by MASCOT software (p < 0.05; Student’s t-test) [15] for at least one matched peptide. Medium-probability hits were proteins with ions probability-based MOWSE scores lower than the cutoff, but with total MOWSE score above 40; they can be considered as possible hits since several peptides were matched and were not identified in the corresponding control samples.
In the mass spectrometric analysis, we did not detect some expected, previously known partners of synaptic proteins. Nevertheless, the absence of a protein from a large-scale analysis cannot automatically exclude that protein from the list of potential interacting partners [17]. Instead, it is possible that under the particular experimental conditions used, the protein in question is not a significant component of the complex [17].
3. Results
We used co-immunoprecipitation followed by mass spectrometry or western blotting to investigate the synaptic protein network for the candidate proteins BKCa, dynamin-1, SNAP-25, syntaxin-1A, and VAMP-2. To validate our procedure, we first characterized the antibodies and the optimal conditions by testing known interactions; then, we tested unknown interactions using the same high-quality immunoprecipitation conditions. To control for potential non-specific protein-protein interactions that may occur during co-immunoprecipitation [18], we carried out each experiment using the same cortical extract with a corresponding monoclonal or polyclonal anti-FLAG antibody (recognizing an artificial epitope tag, Suppl. Table 1) or with just the sedimentable matrix alone. No non-specific immunoreactivity was detected under either control condition for any of the interactions studied. In many cases, the specificity of the interactions found with co-immunoprecipitation was supported by both immunoblotting and LC-MS/MS.
3.1 Immunoprecipitation of syntaxin-1A
We first tested the well-characterized syntaxin-1A complex as a measure of our experimental technique. Native syntaxin-1A was detected as a 35 kDa immunoreactive band, which was enriched following immunoprecipitation (Fig. 1A). As shown previously [4,7,19,20], probing this immunoprecipitate for the presence of BKCa alpha subunit (Fig. 1B), munc-18 (Fig. 1C), SNAP-25 (Fig. 1D), and VAMP-2 (Fig. 1E) revealed specific immunoreactive bands. Based on our highly selective co-immunoprecipitation experiments, we conclusively showed that syntaxin 1A, under native conditions, interacts with SNAP-25 and VAMP-2 within a protein complex and co-associates with munc-18 and BKCa alpha subunit.
Figure 1–5 . Identification of synaptic protein complexes using co-immunoprecipitation.
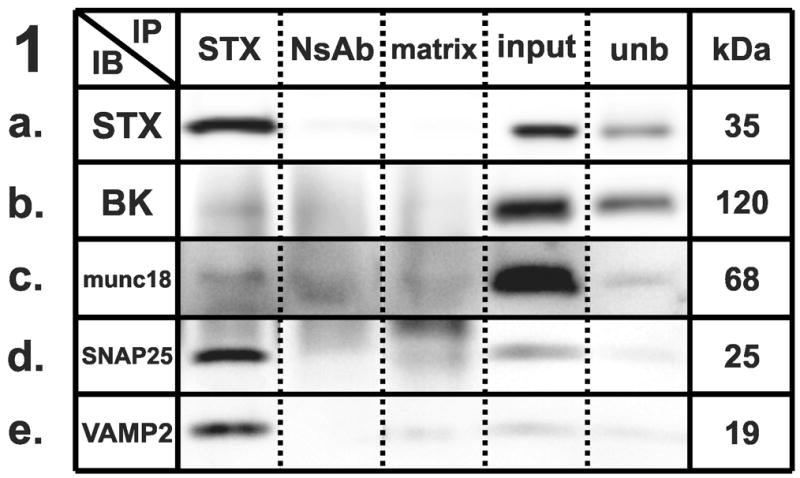
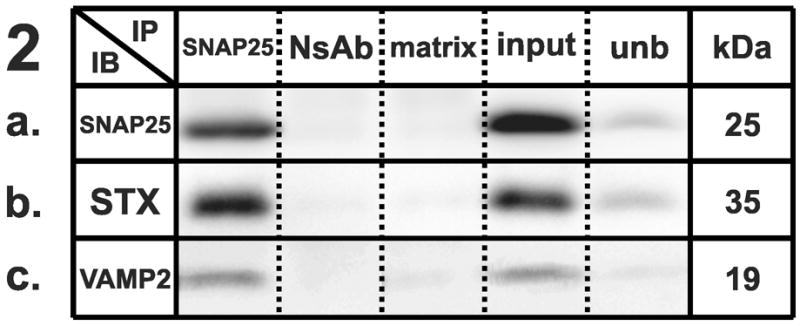
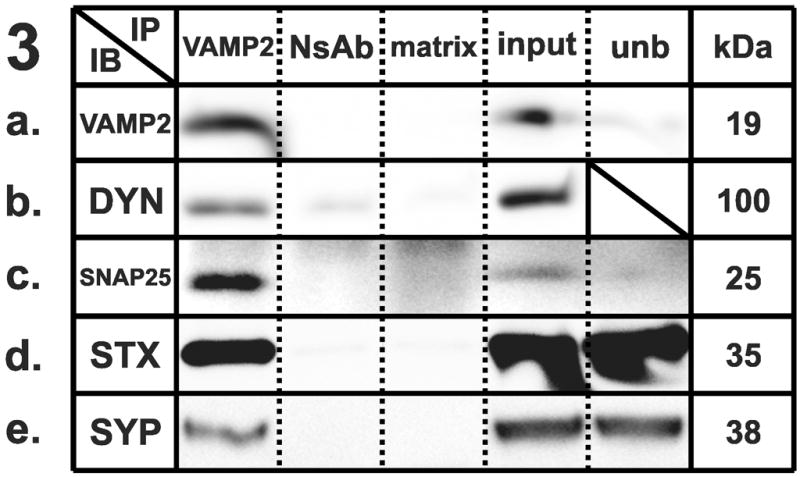
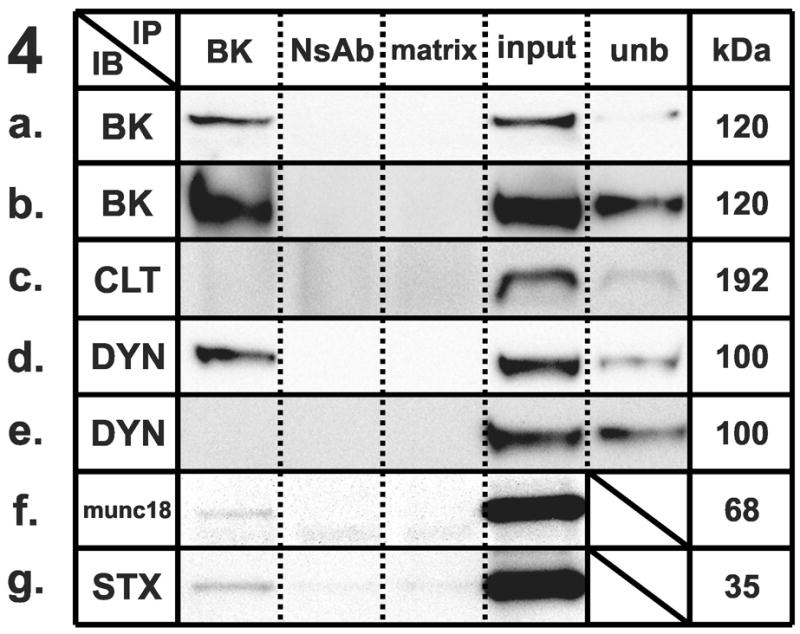
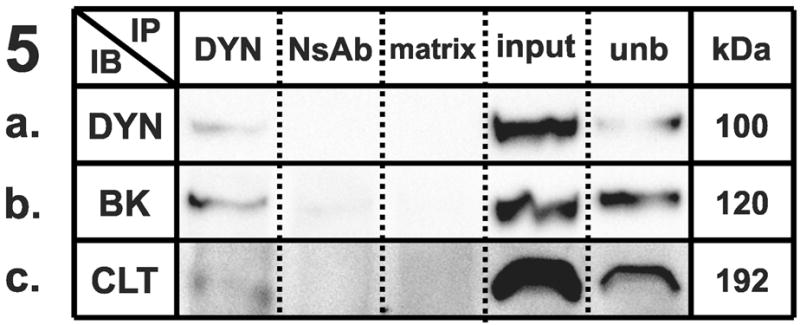
Immunoprecipitation (IP) experiments from mouse cortex membranes were performed by using (1) syntaxin-1, (2) SNAP-25, (3) VAMP-2, (4) large-conductance calcium-activated potassium channel, alpha subunit, and (5) dynamin-1 antibodies (or their respective non-specific control antibodies) followed by immunoblots (IB) with different antibodies recognizing potential protein interactions. All the experiments were performed under mildly-denaturing conditions, except 4b and 4e, where samples were immunoprecipitated under denaturing conditions. Elution from specific antibody IP was loaded in lane 1; elution from respective non-specific control antibody developed in the same species was loaded in lane 2; the matrix used to preclear the lysates subjected to IP was loaded in lane 3; total protein lysate (30 μg) was loaded in lane 4; and an aliquot of the unbound fraction of proteins after IP was loaded in lane 5. Samples were subjected to SDS-PAGE and then separated proteins were transferred to polyvinylidene fluoride membrane and blotted with different primary antibodies, followed by incubation in appropriate secondary antibodies and detection with enhanced chemiluminescence. The appropriate molecular weight for each western blot was checked with the molecular markers (not shown). Each row shows a single representative western blot. NsAb, non-specific antibody; matrix, Sepharose-A or A/G-Agarose slurry beads matrix; input, mouse cortical membrane preparation lysate (equal amounts of cortical lysates were used for each IP); unb, unbound proteins; kDa, molecular weight, expressed in kilodaltons; STX, syntaxin-1A; SNAP25, synaptosome-associated protein of 25 kilodaltons; VAMP2, vesicle-associated membrane protein-2; BK, large-conductance calcium-activated potassium channel; DYN, dynamin-1; CLT, clathrin heavy chain; munc18, syntaxin binding protein 1; SYP, synaptophysin.
3.2 Immunoprecipitation of SNAP-25
We successfully performed immunoprecipitations using SNAP-25 (Fig. 2A) as a bait protein. Consistent with previous reports [21], syntaxin-1A and VAMP-2 immunoreactivities were clearly detected in SNAP-25 immunoprecipitates (Figs. 2B and 2C), but not in the precipitates generated by the anti-FLAG antibody or the matrix beads.
3.3 Immunoprecipitation of VAMP-2
We also successfully performed immunoprecipitations using VAMP-2 (Fig. 3A) as bait protein. Consistent with previous reports [22], SNAP-25 and syntaxin-1A immunoreactivities were clearly detected in VAMP-2 immunoprecipitates (Figs. 3C and 3D), but not in the precipitates generated by the anti-FLAG antibody or the matrix beads.
Furthermore, we showed a novel co-association between VAMP-2 and dynamin-1 (Fig. 3B); functional evidence for this interaction is also supported by a web database dedicated to protein-protein interaction showing that putative homologs of dynamin-1 and VAMP-2 were found to interact in other species (STRING, http://string.embl.de). Our results shown in Fig. 3E also corroborated the previously reported interaction between VAMP-2 and synaptophysin [2,22].
3.4 Co-immunoprecipitation of BKCa/dynamin complex
We performed immunoprecipitation experiments using the BKCa channel alpha subunit as a bait protein to co-precipitate its interacting partners. The native BKCa was detected as a 120 kDa immunoreactive band, which was enriched following immunoprecipitation under mildly-denaturing (Fig. 4A) and denaturing (Fig. 4B) conditions with a polyclonal antibody. Probing the BKCa channel alpha subunit immunoprecipitate for the presence of syntaxin revealed a specific immunoreactive band (Fig. 4G), as shown previously [7,20].
We tested the hypothesis that BKCa could co-associate with dynamin in mouse cortex. Probing the BKCa alpha subunit immunoprecipitate for the presence of dynamin-1 revealed a specific immunoreactive band of 100 kDa (Fig. 4D), showing a robust interaction that was disrupted under denaturing conditions (Fig. 4E). The reciprocal co-immunoprecipitation performed by using dynamin-1 as a bait protein (Fig. 5A) to co-precipitate BKCa yielded a 120 kDa band (Fig. 5B), confirming the interaction between the two proteins. We tested the possibility that the BKCa-dynamin interaction could be due to some nonspecific binding which could occur among proteins in the same subcellular compartment. However, we found that BKCa does not interact with the endocytic protein clathrin (Fig. 4C) under the same conditions where the well-established dynamin/clathrin interaction was observed (Fig. 5C).
Furthermore, we evaluated the hypothesis that BKCa and munc-18 could co-associate. Probing the BKCa alpha subunit immunoprecipitate for the presence of munc-18 revealed a weak but specific immunoreactive band of 68 kDa (Fig. 4F), showing that BKCa can interact not only with syntaxin, but also with its associated protein. We failed to show a VAMP-2 interaction with dynamin using dynamin as a bait protein (data not shown).
3.5 Mass spectrometric analysis of SNAP-25, VAMP-2, and BKCa co-immunoprecipitated proteins
Following SDS-PAGE and Coomassie staining, specific bands were excised from the gels (Suppl. Fig. 1), and proteins therein were identified by LC-MS/MS. The specificity of the immunoprecipitation procedure was evident since the bait proteins were readily detected by mass spectrometry, except in the case of SNAP-25, due to the overlap of the bait specific band with the antibody immunoglobulin light chain band (25 kDa).
Following immunoprecipitation using SNAP-25 as bait protein, we specifically identified synaptophysin (Suppl. Table 2A), a SNARE complex interacting protein [4]. With the same technique, we identified a number of other partner proteins, including cytoskeletal proteins such as tubulin, microtubule-associated proteins, kinesin which is involved in vesicular transport, and AP-2 complex subunit proteins which play a key role in clathrin-mediated endocytosis (Suppl. Table 2A).
Dynamin-1 was specifically detected after precipitations using VAMP-2 as bait protein, as well as different syntaxin isoforms and synaptophysin (Suppl. Table 2B), which was consistent with our findings from co-immunoprecipitation experiments (Fig. 3). A number of other interacting proteins were also identified (Suppl. Table 2B).
Using BKCa as bait protein, dynamin-1 was specifically detected as the most abundant protein by excising a discrete band in the 100 kDa range (Suppl. Table 2C), consistent with co-immunoprecipitation experiments (Fig. 4). We identified many other proteins (Suppl. Table 2C), including kinesin (heavy and light chains isoforms), tubulin, and actin-like proteins, which are well-established cytoskeletal/motor proteins, as well as different subunits of phosphoinositide-3-kinase and sodium/potassium-transporting ATPase.
4. Discussion
We examined synaptic protein complexes isolated from a mildly-denaturing extract of freshly dissected mouse brain cortex, using a gentle extraction procedure that maintains the integrity of the protein complexes. Using co-immunoprecipitation, immunoblotting, and mass spectrometry, we successfully detected novel protein-protein interactions in cortical synapses. Although some of the interactions shown here have been reported previously, they provide not only a validation of our procedure, but in some cases are the first evidence for SNARE protein interactions in the absence of GST-tagged proteins as baits or outside of in vitro systems. Though the use of artificial systems is useful to explore protein interactions, it is imperative to study synaptic protein interactions under native conditions as shown here. Our protocol also provided appropriate matrix and antibody controls to avoid potential non-specific protein-protein interactions that may occur during co-immunoprecipitation [18], and we further provided mass spectrometric evidence for the interaction of SNARE proteins. Our experiments do not distinguish between diverse cell compartments since we used membranes from both pre- and postsynaptic fractions. However, using a simplified synaptosomal preparation as described by Ling et al. [7], we were still able to show the same interactions in both mouse cortex and midbrain (data not shown). Thus, the preparation that we used provides reproducible synaptic interactions that are not dependent on a particular micro-environment and ensures a physiological system that precludes non-specific interactions.
Many proteins interact in the synapses to mediate and maintain neuronal transmission, and they control vesicle-related processes by forming complexes. Synapses have different endocytic mechanisms at their disposal, including clathrin-dependent and -independent pathways, in addition to other mechanisms [23]. Also, components of SNARE complexes mediate membrane fusion in all of the trafficking steps of the secretory pathway. As we showed here, syntaxin-1A, SNAP-25, and VAMP-2 can establish reciprocal interactions in agreement with previous reports [22], and these interactions have been shown to mediate neurotransmitter vesicle transport and targeting [4]. SNAREs and dynamin, both involved in membrane fusion, could be linked in a common protein complex to finely regulate synaptic neurotransmission by controlling the vesicle recycling process. Indeed, other molecules and proteins could connect dynamin and SNAREs, forming a macromolecular complex. For example, synaptophysin can be co-immunoprecipitated using VAMP-2 as a bait protein, as we verified with multiple techniques, in agreement with previous reports [2,22] and can also bind dynamin in a Ca2+-dependent fashion [5], suggesting that a dynamin/synaptophysin/Ca2+ complex may participate in recycling of synaptic vesicles. A definitive link between endocytic machinery and SNAREs is represented by our finding that dynamin-1 can be co-immunoprecipitated using VAMP-2 as a bait protein. This interaction was assessed with both mass spectrometry and immunoblotting. Thus, VAMP-2 and dynamin may exert their functions as components of the same complex. However, this interaction was not seen using dynamin as a bait, and no interactions were detected between dynamin and syntaxin or SNAP25 (data not shown). These observations are consistent with the current model of synaptic neurotransmission, where VAMP-2 is anchored in the vesicular membrane and can thus be re-internalized by endocytotic processes while syntaxin and SNAP-25 reside in the cell membrane and do not take part in the recycling process. This model of synaptic neurotransmission implicates the disassembly of the SNARE complex or a time/location shift before dynamin/VAMP-2 interaction takes place. VAMP-2 and dynamin could interact during the vesicle recycling process that elapses between exo- and endo-cytosis, but their interaction may be transient and not linked to the start of endocytosis. In addition, other mechanisms have been hypothesized [4,23] and we cannot exclude that interactions between dynamin and SNAP-25 or syntaxin could be identified by using different experimental conditions.
We also evaluated the possibility that the BKCa channel could operate as a link between the endocytic machinery and SNAREs. In fact, it has been recently shown that a different potassium channel, the Kir2.3, can be internalized from the plasma membrane via vesicle-mediated endocytosis in a dynamin-dependent manner [13]. We also confirmed that the BKCa channel can co-associate with syntaxin-1A, as previously reported [7,20]. Above all, our reciprocal co-immunoprecipitations conclusively showed that the BKCa channel and the GTP-ase, dynamin-1, established a strong interaction, which was disrupted under denaturing conditions and was not driven by nonspecific compartment binding, given that the BKCa channel did not interact with clathrin under identical conditions. Mass spectrometric analysis of the BKCa interactome confirmed the presence of dynamin among co-immunoprecipitated proteins; the sensitivity of the mass spectrometer could ensure the absence of dynamin in the corresponding FLAG control band, thus showing specificity of the BKCa-dynamin interaction in mouse cortex. BKCa α-subunit may act as a link between calcium-activated potassium channel activity and the cell’s endocytic machinery, given that it can interact with both syntaxin and dynamin. We speculate that the assembly and density of BKCa channels could be regulated by endocytosis via the interaction of its alpha subunit with dynamin. Additional experiments are required to verify the functional significance of this interaction.
BKCa channel association with the SNARE complex is not limited to its interaction with syntaxin-1A. Immunoblots and mass spectrometry also provided novel evidence for BKCa channel interaction with syntaxin binding protein, munc-18, a protein known to co-associate with syntaxin [19], as we confirmed, and to play a role in its conformational switch during exocytosis. Recent evidence suggests that both syntaxin-dependent and -independent functions may exist for this protein; indeed, munc-18 may be able to exert its role in two additional ways, by binding to the N-terminus of syntaxin or to the assembled SNARE complex [24]. The mechanisms of transition between the different modes of binding are poorly understood. In this context, the novel interaction between BKCa channel and munc-18 that we report here may be transient and take place during one of the diverse modes cited above. Given that we found that the BKCa channel does not interact with SNAP-25 and VAMP-2 (data not shown), more experimental information is needed to elucidate the molecular mechanism and significance underlying this novel and selective interaction with munc-18.
The sensitivity of mass spectrometry allowed us to identify a number of other protein-protein interactions. In Suppl. Table 2C, we report additional partners of BKCa channels: phosphoinositide-3-kinase subunits, sodium/potassium-transporting ATPase subunits, cytoskeletal proteins like actin and tubulin, and kinesin heavy and light chains which are involved in vesicular transport. The above BKCa partners, with the exception of kinesin, have also been recently identified by Kathiresan et al. [9] in the mouse cochlea. Previous studies in the brain reported many partners of BKCa channels, including actin, tubulin, calcium channels, microtubule proteins [6], and syntaxin [7,20]. SNARE proteins (syntaxin and VAMP), dynamin-1, clathrin, actin, and microtubule proteins have also been identified as interacting with N-type calcium channels [11,22]. Taken together, these interactions may help to depict a general synaptic protein-protein network with calcium channels, SNAREs, and cytoskeletal proteins as possible links between BKCa and dynamin (Fig. 6). The hypothesized network would be consistent with our mass spectrometric analysis of synaptic protein partners, which detected kinesin heavy chains, tubulin, microtubule associated proteins, and AP-2 complex subunit proteins as SNAP-25 partners and sodium/potassium-transporting ATPase subunits as VAMP-2 partners.
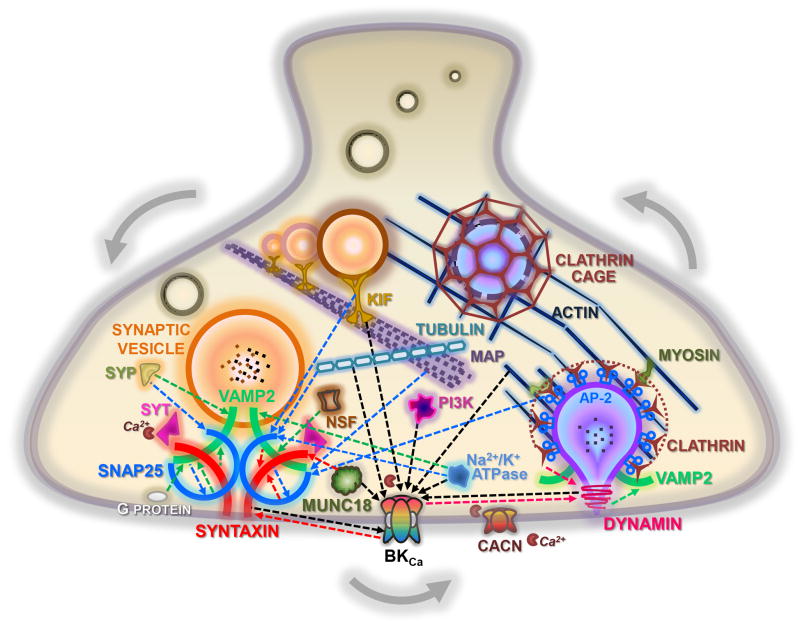
Figure 6. Provisional representation of a putative synaptic protein network.
A summary of the interactions shown in this study, derived from the BKCa channel alpha subunit, dynamin-1, SNAP-25, syntaxin-1A, and VAMP-2 immunoprecipitations from mouse cortex. Proteins are shown, as players in exocytic (left) and endocytic (right) pathways. Interactions are indicated by dashed arrows, whose colors and heads refer to the bait protein used to co-IP the partner. The calcium channels, synaptotagmin, and myosin proteins were not found in our studies, but are known to exist in a complex with one or more proteins that were identified in our screen. Additional experiments are required to verify the topology, molecular mechanisms, and functional significance of these interactions. CACN, voltage-gated calcium channels; KIF, kinesin family proteins; MAP, microtubule associated proteins; NSF, N-ethylmaleimide sensitive fusion attached proteins; PI3K, phosphoinositide 3-kinase protein family; SyP, synaptophysin; Syt, synaptotagmin.
In summary, we identified novel synaptic protein complexes in mouse cortical membranes using an interaction proteomics approach, which included co-immunoprecipitation followed by immunoblotting or LC-MS/MS; the experimental conditions combined high-specificity with a native system that resembled the synaptic environment. This is the first report to uncover protein interactions between 1) dynamin and the BKCa channel, 2) dynamin and VAMP-2, and 3) the BKCa channel and munc-18 in synapses. These newly identified interactions could elucidate some of the missing links in synaptic protein complexes and contribute to better understanding of synaptic vesicle trafficking, release, and recycling.
Structured summary
MINT-7543319:
Snap-25 (uniprotkb:P60879) physically interacts (MI:0914) with Tubulin beta-5 chain (uniprotkb:P99024), V-type proton ATPase subunit d 1 (uniprotkb:P51863), Zinc finger homeobox protein 3 (uniprotkb:Q61329), Tubulin beta-2A chain (uniprotkb:Q7TMM9), Synaptophysin (uniprotkb:Q62277), Gapdh (uniprotkb:P16858), Basement membrane-specific heparan sulfate proteoglycan core protein (uniprotkb:Q05793), Tubulin alpha-4A chain (uniprotkb:P68368), Tubulin alpha-1A chain (uniprotkb:P68369), Microtubule-associated protein 6 (uniprotkb:Q7TSJ2), AP-2 complex subunit beta (uniprotkb:Q9DBG3), Phosphofurin acidic cluster sorting protein 1 (uniprotkb:Q8K212), AP-2 complex subunit alpha-1 (uniprotkb:P17426), Kinesin-1 heavy chain (uniprotkb:Q61768), Kinesin heavy chain isoform 5C (uniprotkb:P28738), Sodium/potassium-transporting ATPase subunit alpha-1 (uniprotkb:Q8VDN2) and Nck-associated protein 1 (uniprotkb:P28660) by anti bait coimmunoprecipitation (MI:0006)
MINT-7543636:
Calcium-activated potassium channel subunit alpha-1 (uniprotkb:Q08460) physically interacts (MI:0914) with AMP deaminase 2 (uniprotkb:Q9DBT5), Gamma-tubulin complex component 4 (uniprotkb:Q9D4F8), Gamma-tubulin complex component 2 (uniprotkb:Q921G8), Sodium/potassium-transporting ATPase subunit alpha-1 (uniprotkb:Q8VDN2), Phosphoinositide 3-kinase regulatory subunit 4 (uniprotkb:Q8VD65), Beta-centractin (uniprotkb:Q8R5C5), KIAA1107 (uniprotkb:Q80TK0), Sodium/potassium-transporting ATPase subunit alpha-2 (uniprotkb:Q6PIE5), Sodium/potassium-transporting ATPase subunit alpha-3 (uniprotkb:Q6PIC6), Phosphatidylinositol 3-kinase catalytic subunit type 3 (uniprotkb:Q6PF93), KH domain-containing, RNA-binding, signal transduction-associated protein 1 (uniprotkb:Q60749), Tubulin gamma-1 chain (uniprotkb:P83887), Heat shock cognate 71 kDa protein (uniprotkb:P63017), Alpha-centractin (uniprotkb:P61164), Gamma-tubulin complex component 3 (uniprotkb:P58854), Dynamin-1 (uniprotkb:P39053), Kinesin heavy chain isoform 5C (uniprotkb:P28738), Elongation factor 1-alpha 1 (uniprotkb:P10126), Kinesin light chain 2 (uniprotkb:O88448), Activated CDC42 kinase 1 (uniprotkb:O54967) and Syntaxin-binding protein 1 (uniprotkb:O08599) by anti bait coimmunoprecipitation (MI:0006)
MINT-7544031:
Calcium-activated potassium channel subunit alpha-1 (uniprotkb:Q08460) physically interacts (MI:0914) with Syntaxin-binding protein 1 (uniprotkb:O08599), Syntaxin-1A (uniprotkb:O35526) and Dynamin-1 (uniprotkb:P39053) by anti bait coimmunoprecipitation (MI:0006)
MINT-7543287:
Syntaxin-1A (uniprotkb:O35526) physically interacts (MI:0914) with Vamp2 (uniprotkb:P63044), Snap-25 (uniprotkb:P60879), munc-18 (uniprotkb:O08599) and BKCa alpha subunit (uniprotkb:Q08460) by anti bait coimmunoprecipitation (MI:0006)
MINT-7543972:
Vamp-2 (uniprotkb:P63044) physically interacts (MI:0914) with Dynamin-1 (uniprotkb:P39053), Snap-25 (uniprotkb:P60879), Syntaxin-1A (uniprotkb:O35526) and Synaptophysin (uniprotkb:Q62277) by anti bait coimmunoprecipitation (MI:0006)
MINT-7543728:
Dynamin-1 (uniprotkb:P39053) physically interacts (MI:0914) with Clathrin heavy chain 1 (uniprotkb:Q68FD5) and Calcium-activated potassium channel subunit alpha-1 (uniprotkb:Q08460) by anti bait coimmunoprecipitation (MI:0006)
MINT-7543905:
Snap-25 (uniprotkb:P60879) physically interacts (MI:0914) with Syntaxin-1A (uniprotkb:O35526) and Vamp-2 (uniprotkb:P63044) by anti bait coimmunoprecipitation (MI:0006)
MINT-7543476:
Vamp-2 (uniprotkb:P63044) physically interacts (MI:0914) with Syntaxin-7 (uniprotkb:O70439), Neuronal membrane glycoprotein M6-a (uniprotkb:P35802), Syntaxin-1B (uniprotkb:P61264), Beta-soluble NSF attachment protein (uniprotkb:P28663), Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-3 (uniprotkb:Q61011), Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 (uniprotkb:P62874), Guanine nucleotide-binding protein G(o) subunit alpha (uniprotkb:P18872), V-type proton ATPase subunit d 1 (uniprotkb:P51863), Zinc transporter 3 (uniprotkb:P97441), Sodium/potassium-transporting ATPase subunit alpha-2 (uniprotkb:Q6PIE5), Sodium/potassium-transporting ATPase subunit alpha-3 (uniprotkb:Q6PIC6), Sodium/potassium-transporting ATPase subunit alpha-1 (uniprotkb:Q8VDN2), Potassium-transporting ATPase alpha chain 1 (uniprotkb:Q64436), Synaptophysin (uniprotkb:Q62277), Syntaxin-1A (uniprotkb:O35526) and Dynamin-1 (uniprotkb:P39053) by anti bait coimmunoprecipitation (MI:0006)itation (MI:0006)
Supplementary Material
Acknowledgments
This work was supported by NIAAA grant AA016648.
List of abbreviations
- SNAREs
soluble N-ethylmaleimide-sensitive factor attachment protein receptors
- BKCa
calcium-activated potassium channel
- VAMP-2
vesicle-associated membrane protein-2
- munc-18
syntaxin binding protein 1
- AP-2
alpha-adaptin
- SNAP-25
synaptosomal associated protein of 25 kDa
- LC-MS/MS
liquid chromatography followed by tandem mass spectrometry
- MOWSE
Molecular Weight Search
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pandey A, Mann M. Proteomics to study genes and genomes. Nature. 2000;405:837–46. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 2.Klemmer P, Smit AB, Li KW. Proteomics analysis of immuno-precipitated synaptic protein complexes. J Proteomics. 2009;72:82–90. doi: 10.1016/j.jprot.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Ungewickell EJ, Hinrichsen L. Endocytosis: clathrin-mediated membrane budding. Curr Opin Cell Biol. 2007;19:417–25. doi: 10.1016/j.ceb.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–43. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 5.Daly C, Sugimori M, Moreira JE, Ziff EB, Llinas R. Synaptophysin regulates clathrin-independent endocytosis of synaptic vesicles. Proc Natl Acad Sci U S A. 2000;97:6120–5. doi: 10.1073/pnas.97.11.6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu R, Alioua A, Kumar Y, Eghbali M, Stefani E, Toro L. MaxiK channel partners: physiological impact. J Physiol. 2006;570:65–72. doi: 10.1113/jphysiol.2005.098913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling S, Sheng JZ, Braun JE, Braun AP. Syntaxin 1A co-associates with native rat brain and cloned large conductance, calcium-activated potassium channels in situ. J Physiol. 2003;553:65–81. doi: 10.1113/jphysiol.2003.051631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loane DJ, Lima PA, Marrion NV. Co-assembly of N-type Ca2+ and BK channels underlies functional coupling in rat brain. J Cell Sci. 2007;120:985–95. doi: 10.1242/jcs.03399. [DOI] [PubMed] [Google Scholar]
- 9.Kathiresan T, Harvey M, Orchard S, Sakai Y, Sokolowski B. A protein interaction network for the large conductance Ca2+-activated K+ channel in the mouse cochlea. Mol Cell Proteomics. 2009;8:1972–87. doi: 10.1074/mcp.M800495-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins MO, Husi H, Yu L, Brandon JM, Anderson CN, Blackstock WP, Choudhary JS, Grant SG. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J Neurochem. 2006;97(Suppl 1):16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- 11.Khanna R, Zougman A, Stanley EF. A proteomic screen for presynaptic terminal N-type calcium channel (CaV2.2) binding partners. J Biochem Mol Biol. 2007;40:302–14. doi: 10.5483/bmbrep.2007.40.3.302. [DOI] [PubMed] [Google Scholar]
- 12.Maiya R, Ponomarev I, Linse KD, Harris RA, Mayfield RD. Defining the dopamine transporter proteome by convergent biochemical and in silico analyses. Genes Brain Behav. 2007;6:97–106. doi: 10.1111/j.1601-183X.2006.00236.x. [DOI] [PubMed] [Google Scholar]
- 13.Mason AK, Jacobs BE, Welling PA. AP-2-dependent internalization of potassium channel Kir2.3 is driven by a novel di-hydrophobic signal. J Biol Chem. 2008;283:5973–84. doi: 10.1074/jbc.M709756200. [DOI] [PubMed] [Google Scholar]
- 14.Lu J, Helton TD, Blanpied TA, Racz B, Newpher TM, Weinberg RJ, Ehlers MD. Postsynaptic positioning of endocytic zones and AMPA receptor cycling by physical coupling of dynamin-3 to Homer. Neuron. 2007;55:874–89. doi: 10.1016/j.neuron.2007.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 16.Dennis MD, Person MD, Browning KS. Phosphorylation of plant translation initiation factors by CK2 enhances the in vitro interaction of multifactor complex components. J Biol Chem. 2009;284:20615–28. doi: 10.1074/jbc.M109.007658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steen H, Mann M. The ABC’s (and XYZ’s) of peptide sequencing. Nature Reviews Molecular Cell Biology. 2004;5:699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- 18.Fletcher S, Bowden SE, Marrion NV. False interaction of syntaxin 1A with a Ca(2+)-activated K(+) channel revealed by co-immunoprecipitation and pull-down assays: implications for identification of protein-protein interactions. Neuropharmacology. 2003;44:817–27. doi: 10.1016/s0028-3908(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 19.Hata Y, Slaughter CA, Sudhof TC. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 1993;366:347–51. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- 20.Cibulsky SM, Fei H, Levitan IB. Syntaxin-1A binds to and modulates the Slo calcium-activated potassium channel via an interaction that excludes syntaxin binding to calcium channels. J Neurophysiol. 2005;93:1393–405. doi: 10.1152/jn.00789.2004. [DOI] [PubMed] [Google Scholar]
- 21.Chapman ER, An S, Barton N, Jahn R. SNAP-25, a t-SNARE which binds to both syntaxin and synaptobrevin via domains that may form coiled coils. J Biol Chem. 1994;269:27427–32. [PubMed] [Google Scholar]
- 22.el Far O, Charvin N, Leveque C, Martin-Moutot N, Takahashi M, Seagar MJ. Interaction of a synaptobrevin (VAMP)-syntaxin complex with presynaptic calcium channels. FEBS Lett. 1995;361:101–5. doi: 10.1016/0014-5793(95)00156-4. [DOI] [PubMed] [Google Scholar]
- 23.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 24.Burgoyne RD, Barclay JW, Ciufo LF, Graham ME, Handley MT, Morgan A. The functions of Munc18-1 in regulated exocytosis. Ann N Y Acad Sci. 2009;1152:76–86. doi: 10.1111/j.1749-6632.2008.03987.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.