Abstract
The transcription factor Pit-1/Pou1f1 regulates GH and prolactin (PRL) secretion in the pituitary gland. Pit-1 expression and GH regulation by Pit-1 have also been demonstrated in mammary gland. However, no data are available on the role of Pit-1 on breast PRL. To evaluate this role, several human breast cancer cell lines were transfected with either the Pit-1 expression vector or a Pit-1 small interference RNA construct, followed by PRL mRNA and protein evaluation. In addition, transient transfection of MCF-7 cells by a reporter construct containing the proximal PRL promoter, and ChIP assays were performed. Our data indicate that Pit-1 regulates mammary PRL at transcriptional level by binding to the proximal PRL promoter. We also found that Pit-1 raises cyclin D1 expression before increasing PRL levels, suggesting a PRL-independent effect of Pit-1 on cell proliferation. By using immunohistochemistry, we found a significant correlation between Pit-1 and PRL expression in 94 human breast invasive ductal carcinomas. Considering the possible role of PRL in breast cancer disorders, the function of Pit-1 in breast should be the focus of further research.
Introduction
The transcription factor Pit-1/Pou1f1 was first described in the pituitary gland, where it acts in cell differentiation during organogenesis of the anterior pituitary in mammals and as a transcriptional activator for pituitary gene transcription (Lefevre et al. 1987, Nelson et al. 1988). Mice with inactivating mutations or deletions of the Pit-1 gene fail to generate somatotrophs, lactotrophs, and thyrotrophs, and consequently exhibit anterior pituitary hypoplasia and dwarfism (Li et al. 1990), thereby demonstrating the importance of Pit-1 in the ontogeny of the pituitary gland. While its pituitary function has been extensively evaluated, Pit-1 is also expressed in several extrapituitary tissues and cell lines, including the mammary gland, where its role is still unclear (Delhase et al. 1993, Bamberger et al. 1995, Gil-Puig et al. 2002, 2005). Moreover, as in the pituitary gland, Pit-1 regulates GH expression in the MCF-7 human breast adenocarcinoma cell line (Gil-Puig et al. 2005).
Prolactin (PRL) and its corresponding receptor have also been found in extrapituitary tissues, including normal human breast, mammary tumors, and breast cell lines (Clevenger et al. 1995, Ginsburg & Vonderhaar 1995, Mertani et al. 1998), but the regulation of extrapituitary PRL is not yet understood. Human PRL gene is regulated at a transcriptional level by two different promoters (Berwaer et al. 1994, Gellersen et al. 1994). First, the proximal pituitary promoter is located in the exon 1b and contains Pit-1 response elements; the human pituitary PRL gene contains only this promoter. Secondly, the distal or extrapituitary promoter is located at 5.8 kb upstream of the above mentioned proximal PRL promoter and does not contain any Pit-1-binding sites. The distal promoter is present in PRL gene at extrapituitary sites, such as placenta, lymphoid cells, normal and neoplastic human breast, and human prostate (Berwaer et al. 1994, Gellersen et al. 1994, Shaw-Bruha et al. 1997, Dagvadorj et al. 2007). However, both proximal pituitary and distal extrapituitary PRL promoters are expressed in a subset of normal and tumoral breast tissues, breast cell lines, and in prostate tissue (Shaw-Bruha et al. 1997, Dagvadorj et al. 2007). PRL transcripts arising from either the distal or proximal promoter encode identical proteins, which gives the PRL gene a tissue-specific regulation. The effects of PRL on milk production and on mammary growth and differentiation are well known (Freeman et al. 2000). In addition, PRL provides a proliferative stimulus to rodent mammary tumorogenesis (Welsch & Nagasawa 1977). Several researchers have related endocrine and/or autocrine/paracrine PRL production with mammary disorders, including tumorogenesis (Vonderhaar 1999, Wennbo & Törnell 2000, Ben-Jonathan et al. 2002, Clevenger et al. 2003). For instance, recent prospective studies have linked PRL levels with breast cancer etiology in women, particularly in estrogen-positive tumors (Tworoger et al. 2007). The administration of anti-PRL antibodies, PRL antisense oligonucleotides, or PRL antagonists suppresses the mitogenic activity of locally produced PRL and inhibits growth in breast tumor cell lines (Fuh & Wells 1995, Llovera et al. 2000). In addition, transgenic mice that overexpress the PRL gene develop mammary carcinoma, and this tumor development is caused by the activation of PRL receptor (Wennbo et al. 1997).
To explore the regulation of autocrine breast PRL, in the present study, several human breast cell lines were used to evaluate the effect of Pit-1 overexpression and/or knockdown on PRL expression. In addition, we evaluate the effect of Pit-1 and PRL on cell proliferation. Finally, we evaluate the relationships between Pit-1 and PRL expression in human breast tumors.
Materials and methods
Cell culture
Human breast carcinoma cell lines (MCF-7, SK-BR-3, MDA-MB-231, BT474, and T47D), NIH-3T3 (mouse embryonic fibroblast), and HeLa (human cervix carcinoma) cells lines were grown in 90-mm petri dishes in DMEM supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine in an air–CO2 (95:5) atmosphere at 37 °C. Confluent cells were washed twice with PBS and harvested by a brief incubation with trypsin–EDTA solution (Sigma–Aldrich) in PBS.
Plasmids, transfections, site-directed mutagenesis, and treatments
Cells were cultured as described above. Twelve to 24 h before transfection, 106 or 2×105 cells/well were seeded in 90-mm petri dishes or 6-well plates respectively and allowed to attach overnight. To evaluate PRL and Pit-1 mRNA and protein expression after Pit-1 overexpression or after Pit-1 knockdown, cells were a) transfected with 4 μg of pRSV-hPit-1 expression construct (obtained from Dr J L Castrillo) or with 4 μg of pRc/RSV empty vector (employed as a control), using FuGene (Roche Molecular Biochemicals); or b) transfected with 20 nM Pit-1 small interference RNA (siRNA) or with 20 nM mis-sense siRNA (employed as a control), using jetSI (jetSI siRNA transfection reagent, Polyplus-Transfection, Illkirch, France) as previously described (Seoane et al. 2007). Cells were incubated for varying periods of time as indicated, and total RNA isolation (for RT-PCR or real-time PCR assays) and protein extraction (for western blot assays) were then performed.
To evaluate PRL, Pit-1 and cyclin D1 protein expression 48 h after both PRL knockdown and Pit-1 overexpression, MCF-7 cells were cotransfected using jetSI-ENDO (Polyplus-Transfection) with two PRL siRNAs, which hybridise to the PRL gene at 521 and 933 bp (20 nM, Sigma–Aldrich), and with 2.5 μg pRSV-hPit-1 overexpression vector.
The proximal promoter region of the human PRL gene (−216/+1; pGL2B-hPRL−216/+1) was synthetized by PCR using the oligonucleotides: forward, 5′-CCGCTCGAGCCTAATTAATCAAAATCCTTC-3′; and reverse, 5′-AAGCTTTTCTCTTTCCCAGATATTG-3′. The PCR product was subcloned into the XhoI and HindIII site of the pGL2-basic plasmid. In addition, the pGL2B-hPRL−216/+1 vector was mutated at two Pit-1 consensus binding sites, positioned between −151 and −149 bp (pGL2B-hPRLmut2), and −108 and −106 bp (mut1) respectively from the transcription start site on the hPRL gene. Site-directed mutagenesis was performed with the QuikChange kit (Stratagene, La Jolla, CA, USA) under the conditions recommended by the manufacturer. The mutagenised oligonucleotide primers were as follows (mutagenised bases on the sense strand indicated by lowercase letters): 5′-ATGTCTTCCTGAATATGcccAAAGAAATAAA-3′ and 5′-TTGAAATTATGGGGGTcccCTCAATGACGG-3′ (designated mut2 and mut1 respectively). Subsequently, a double mutant was constructed using the same method (designated pGL2B-hPRLmut1+2). All the nucleotide sequences of the cloned inserts were confirmed by sequencing.
Transfections were carried out in wells containing 1 μl Fugene (Roche Molecular Biochemicals) and 1 μg total DNA (750 ng pGL2-basic vector, pGL2B-hPRL−216/+1, pGL2B-hPRLmut2, pGL2B-hPRLmut1, or pGL2B-hPRLmut1+2 constructs, and 250 ng Rous sarcoma virus β-galactosidase (pRSV-gal)). The cells were cotransfected with 1 μg Pit-1 (pRSV-hPit-1) or pRc–RSV empty vector (control cells). Each experiment was performed in triplicate cultures. The cells were harvested in buffer (5× lysis buffer; Promega), and luciferase activity was then measured. β-Galactosidase activity was measured at 420 nm using a using o-nitrophenyl-β-d-galactopyranoside as substrate.
For PRL treatment, 24 h after transfection, MCF-7 cells were treated with 10 nM human PRL (hPRL, from Dr Parlow, NIDDK, USA) for 24 h. MCF-7 cells were also treated with AG-490 (a potent inhibitor of the Jak2 tyrosine kinase; Calbiochem, San Diego, CA, USA). AG-490 was dissolved in dimethyl sulfoxide (DMSO) and used at 30 μM for 24 h. As positive control for Jak2 activation, we used H2O2 (500 μM) for 1 h.
RNA isolation, RT-PCR, and quantitative PCR
Isolation of total RNA from the cell lines was performed with the TRIzol reagent (Invitrogen), according to the manufacturer's instructions. cDNA synthesis was performed as described elsewhere (Gil-Puig et al. 2005). Five microliters of the cDNA were amplified by PCR using 2 units of Taq polymerase (Gene Craft, Koln, Germany) in a buffer containing 50 mM KCl, 10 mM Tris–HCl (pH 8.4), and 1.5 mM MgCl2, with each dNTP at 0.2 mM and each of two oligonucleotide primers at 25 pM, to a total volume of 50 μl. The sample was denatured at 94 °C for 1 min, annealed at 54, 58, or 60 °C for 1 min (PRL, Pit-1, and 18S respectively) and extended at 72 °C for 1 min, for a total of 30, 32, or 36 cycles (for 18S, Pit-1, and PRL respectively) with an extension step of 10 min at 72 °C in the final cycle.
PRL and 18S mRNA levels from MCF-7, BT474, SK-BR-3, T47D, and MDA-MB-231 cell lines were quantified using real-time PCR. Reactions were done using iQ SYBR Green Supermix (Bio-Rad) on iCycler equipment (Bio-Rad). The samples (PRL and 18S) were denatured at 94 °C for 10 s, annealed at 58 °C for 10 s, and extended at 72 °C for 10 s, for a total of 40 cycles. The samples were quantified using the Optical System Software 2.0 (Bio-Rad), with 18S as normalization control.
Primer sequences
Primer sequences for PRL PCR amplification were as follows: primer Ex1a (5′-GAGACACCAAGAAGAATCGGAACATAC-3′) was a 27-mer corresponding to exon 1a of PRL cDNA; primer Ex1b (5′-AGACTTCCAGATCTTCTCTGGTGAAG-3′) was a 26-mer corresponding to exon 1b of PRL cDNA; primer Ex2 (5′-TCAAACAGGTCTCGAAGGGTCACCT-3′) was an antisense 25-mer corresponding to exon 2 of PRL cDNA; primer Ex4 (5′-AGCAAACCAAACGGCT-3′) was a 16-mer corresponding to exon 4 of PRL cDNA; and primer Ex5 (5′-AGGCGAGACTCTTCAT-3′) was an antisense 16-mer corresponding to exon 5 of PRL cDNA. The lengths of the PCR products obtained were 275 bp (Ex1a–Ex2), 194 bp (Ex1b–Ex2), and 133 bp (Ex4–Ex5). For PRL, quantitative PCR evaluations were used the Ex4–Ex5 primers. Pit-1 primer sequences were as follows: forward primer, 5′-GTGTCTACCAGTCTCCAACC-3′; and reverse primer, 5′-ACTTTTCCGCCTGAGTTCCT-3′. The length of the product obtained was 247 bp. Human 18S rRNA was used as internal reference. Primer sequences were as follows: forward primer, 5′-GTAACCCGTTGAACCCCATT-3′; and reverse primer, 5′-CCATCCAATCGCTA GTAGCG-3′. The length of the PCR product obtained was 131 bp.
Western blot analysis
Cells were lysed at 4 °C in 300 μl lysis buffer (50 mM HEPES, pH 7.5; 150 mM NaCl; 5 mM EGTA; 1.5 mM MgCl2; 1% SDS; 10% glycerol; 1% Triton X-100; 10 mM sodium orthovanadate; 4 mM phenylmethylsulphonyl fluoride (PMSF), and 50 μg/ml aprotinin). The cell lysate was then centrifuged at 14 000 g for 5 min at 4 °C, the resulting supernatant was collected, and protein concentration was determined by the Bradford method. Western blotting of Pit-1 from MCF-7 cells was carried out as described elsewhere (Seoane & Perez-Fernandez 2006). Briefly, 70 μg total protein were subjected to 12% (for Pit-1, cyclin D1, and β-actin) or 15% (for PRL) SDS–PAGE electrophoresis. Proteins were transferred to a nitrocellulose membrane that was blocked and washed. The blot was immunolabeled overnight at 4 °C with a polyclonal anti-Pit-1 antiserum (1:500, Santa Cruz Biotechnology, Santa Cruz, CA, USA), or with a polyclonal anti-PRL antiserum (1:5000, from Dr Parlow, NIDDK), then incubated with goat anti-rabbit IgG (1:5000, for Pit-1, and PRL, see below) or with anti-mouse IgG (1:5000, for cyclin D1 and β-actin) peroxidase-conjugated second antibody, using the ECL western blotting analysis system (GE Healthcare, Piscataway, NJ, USA), and visualized by placing the blot in contact with standard X-ray film, as per the manufacturer's instructions. Membranes were stripped by incubation in 0.2 M glycine, pH 2.2, containing 0.1% SDS and 1% Tween 20 at room temperature for 1 h, and then reprobed with a monoclonal anti-cyclin D1 antibody (1:400, Santa Cruz Biotechnology) and monoclonal anti-β-actin antiserum (1:2000, Sigma–Aldrich).
The optical density of immunolabeling on autoradiographic film was quantified using the UN-SCAN-IT program, version 6.1. To determine the relative amounts of Pit-1, cyclin D1, PRL, and β-actin in each sample, absolute amounts of Pit-1, cyclin D1, and PRL were expressed relative to β-actin amounts.
ChIP assays
Chromatin immunoprecipitation (ChIP) assays were performed using the protocol of Upstate (Charlottesville, VA, USA) as previously described (Seoane & Perez-Fernandez 2006). Diluted soluble chromatin fractions were immunoprecipitated with 1 μg polyclonal anti-Pit-1 antibody (Santa Cruz Biotechnology) or control human IgG (Sigma–Aldrich). The histone-DNA crosslinks were reversed by 4-h incubation at 65 °C. The DNA from these samples was extracted through phenol/chloroform and ethanol precipitated with 20 μg glycogen. The DNA extracted was then dissolved in 30 μl H2O. PCR was used to analyze the DNA fragments from ChIP assays. Five microliters of assayed DNA sample and 5 μl of input/start material were used in each 50-μl reaction. The PCR was run for 60 s at 95, 60, and 72 °C within each cycle, for a total of 35 cycles. The pairs of PRL primers were as follows: (A) forward, 5′-CCGCTCGAGCCTAATTAATCAAAATCCTTC-3′ and reverse, 5′-CCCAAGCTTTTCTCTTTCCCAGATATTG-3′; PCR product is 217 bp in length (from −216 to +1 bp, with respect to the start transcription site in the proximal PRL promoter).
Bromodeoxyuridine incorporation
MCF-7 cells (50×103 cells/well) were seeded in 24-well dishes with coverslides, and allowed to attach overnight. To evaluate bromodeoxyuridine (BrdU) incorporation after Pit-1 overexpression or after Pit-1 knockdown, cells were cotransfected using the pEPuro construct (that confers puromycin resistance), and the pRSV-hPit-1 construct (500 ng) or Pit-1 siRNA (20 nM) respectively and selected (1 μg/ml of puromycin). Forty-eight hours later, resistant cells were labeled with 10 μM BrdU for 1 h. Cells were then fixed 15 min in formaldehyde 4%, 5 min in PBS, and overnight in methanol, permeabilized in 0.07 M NaOH; and incubated overnight at 4 °C with 1:100 α-BrdU (BD Biosciences, San Diego, CA, USA), followed by 1:150 F (ab) IgG FICT (Jackson Immunoresearch, West Grove, PA, USA) plus 4,6-diamino-2-phenylindone (DAPI) for 45 min at 37 °C in darkness in a humidified chamber.
Breast cancer samples and immunohistochemistry
Formalin-fixed, paraffin-embedded breast tissue sections were obtained from 94 patients with histological diagnosis of invasive ductal carcinoma of the breast, which underwent surgery at Fundación Hospital de Jove of Gijón (Spain), between 1995 and 2006. The tissue samples were obtained at the time of surgery. Prior informed consent was obtained from patients. The study adhered to National regulations and was approved by our Institution's Ethics and Investigation Committee.
Serial 5-μm sections were consecutively cut with a microtome (Leica Microsystems GmbH, Wetzlar, Germany) and transferred to adhesive-coated slides. Immunohistochemistry was done on these sections using a TechMate TM50 autostainer (Dako, Glostrup, Denmark). Antibodies for Pit-1 and PRL were obtained from Santa Cruz Biotechnology and from Dr Parlow respectively. We used human pituitary gland (obtained from autopsy) as positive control for both PRL and Pit-1.
Tissue sections were deparaffinized in xylene, and then rehydrated in decreasing concentrations of ethanol (100, 96, 80, 70%, then water). To enhance antigen retrieval, sections were microwave treated (H2800 Microwave Processor, EBSciences, East Granby, CT, USA) in citrate buffer (Target Retrieval Solution, Dako) at 99 °C for 1 min. Endogenous peroxidase activity was blocked by incubating the slides in peroxidase-blocking solution (Dako) for 5 min. The EnVision Detection Kit (Dako) was used as the staining detection system. Sections were counterstained with hematoxylin, dehydrated with ethanol, and permanently coverslipped.
A case was considered as positive if at least 10% of tumoral cells showed positive immunostaining. In addition, as the intensity of staining varied among tumors, positive cases were classified as weak (1+), moderate (2+), and intense (3+) staining.
Statistical analysis
Each experiment was performed at least thrice. Values are expressed as means±s.d. Means were compared by unpaired t-tests or one-way ANOVA with the Tukey–Kramer multiple comparison test for post-hoc comparisons. Differences in percentages in the immunohistochemistry analyses were calculated with the χ2 test. Statistical significance is taken to be indicated by P<0.05.
Results
Effect of Pit-1 overexpression and knockdown on PRL levels
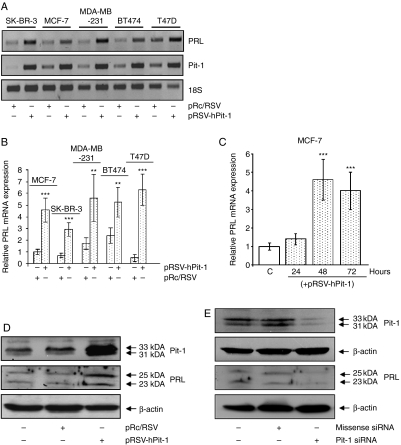
PCR amplification of cDNA prepared from SK-BR-3, MCF-7, MDA-MB-231, BT474, and T47D human breast adenocarcinoma cells using primers Ex4 and Ex5 yielded a 133-bp PCR product corresponding to PRL (Fig. 1A). In order to evaluate PRL regulation by Pit-1, pRSV-hPit-1 vector was transfected in each cell line, and PRL mRNA was evaluated by both RT-PCR and quantitative PCR after 48 h. As shown in Fig. 1A, a clear increase of PRL mRNA expression in all mammary cell lines was found after Pit-1 overexpression. Quantitative PCR revealed a significant PRL increase in all cell lines evaluated after Pit-1 transfection as compared with cells transfected with the control vector (Fig. 1B). In addition, MCF-7 cells transfected with pRSV-hPit-1 vector were analyzed by quantitative PCR at 24, 48, and 72 h (Fig. 1C). The results of these analyses indicated a significant (P<0.001) increase in the PRL mRNA expression at 48 and 72 h compared with untransfected MCF-7 cells; however, this was not the case at 24 h.
Figure 1.
Pit-1 regulates PRL expression in human mammary cell lines. (A) PRL, Pit-1, and 18S mRNA before and after 48 h of Pit-1 overexpression in the SK-BR-3, MCF-7, MDA-MB-231, BT47, and T47D cell lines. (B) Real-time PCR of PRL mRNA expression, with respect to 18S mRNA levels, evaluated in control and pRSV-hPit-1-transfected SK-BR-3, MCF-7, MDA-MB-231, BT47, and T47D cell lines. (C) MCF-7 cells were transfected with either empty pRc/RSV vector (control, C) or pRSV-hPit-1 construct at 24, 48, and 72 h respectively. Values represent means±s.d. from four independent determinations. (D) Western blots of Pit-1, PRL and β-actin in controls, and pRc/RSV (empty vector) and pRSV-hPit-1-transfected MCF-7 cells for 48 h. The major 31 and 33 kDa immunoreactive bands corresponding to Pit-1, as well as glycosylated (25 kDa) and nonglycosylated PRL (23 kDa), are indicated by arrows. (E) Western blots of Pit-1, PRL, and β-actin in control MCF-7 cells, and in cells transfected with 20 nM siRNA negative control and 20 nM Pit-1 siRNA for 48 h. (***P<0.001 and **P<0.01 versus control cells).
Figure 1D shows the western blotting of Pit-1, PRL, and β-actin (used as loading control) in MCF-7 cells transfected with pRSV-hPit-1 or pRc/RSV (used as a control). Pit-1 has two major immunoreactive bands that are readily visible in samples from human cell lines and human mammary glands. These bands that arise from two alternative translation–initiation codons in Pit-1 mRNA have previously been referred to as the 31 and 33 kDa bands (Voss et al. 1991, Gil-Puig et al. 2005). PRL also shows two immunoreactive bands corresponding to the glycosylated (25 kDa of molecular weight) and nonglycosylated (23 kDa) forms (Markoff & Lee 1987, Shinha 1995). As expected, transfection of MCF-7 cells with pRSV-hPit-1 construct induced a significant increase in Pit-1 protein expression (upper panel). PRL was detectable in control (untransfected) MCF-7 cells, as well as in pRc/RSV-transfected cells. Higher PRL expression was observed at 48 h in pRSV-hPit-1 construct compared with pRc/RSV-transfected cells. To evaluate whether Pit-1 knockdown induces changes in PRL, siRNA was used. MCF-7 cells were transfected with a missense siRNA or Pit-1 siRNA. As shown in Fig. 1E, transfection of MCF-7 cells with 20 nM missense siRNA failed to modify Pit-1 or PRL expression levels compared with the untransfected control cells. However, knockdown of Pit-1 by transfection with 20 nM Pit-1 siRNA resulted in markedly reduced expression of both Pit-1 and PRL.
MCF-7 cells contain only the PRL proximal pituitary promoter
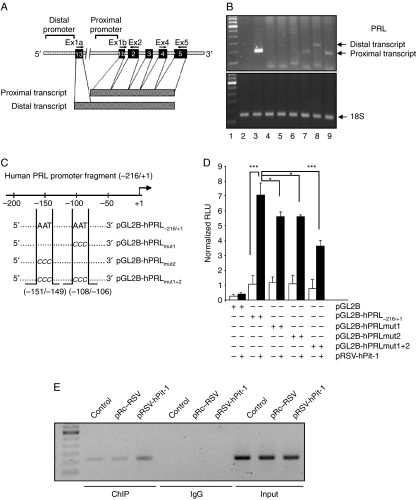
PRL mRNA transcripts in extrapituitary tissues have been shown to be different from those of pituitary origin, thus suggesting several mechanisms of PRL gene regulation (Fig. 2A). Hence, using RT-PCR, we evaluated the type of PRL mRNA that was transcribed in our experimental cell line model. As shown in Fig. 2B, human pituitary tissue (used as positive control, lanes 2 and 3), NIH 3T3 cells (used as negative control, lanes 4 and 5), MCF-7 cells (lanes 6 and 7), and HeLa cells (lanes 8 and 9) were analyzed by RT-PCR, using specific primers for the PRL extrapituitary (primers Ex1a–Ex2) and pituitary (primers Ex1b–Ex2) transcripts (see Fig. 2A). While HeLa cells express both pituitary and extrapituitary PRL mRNA transcripts, the MCF-7 cells showed only one band of 194 bp (lane 7), as is observed in the PCR of human pituitary extract (lane 3; Fig. 2B).
Figure 2.
PRL from MCF-7 cell line is identical to PRL of pituitary origin, and it is transcriptionally regulated by Pit-1. (A) Diagram of human PRL gene and promoter showing the location of the primers used in the RT-PCR assay as well as the proximal pituitary transcript and the distal extrapituitary transcript. (B) PCR products obtained from human pituitary gland, NIH-3T3, MCF-7, and HeLa cell lines using primers that amplified proximal pituitary transcript (194 bp) or distal extrapituitary transcript (275 bp). Lane 1, molecular weight marker; lanes 2 and 3, PCR product from human pituitary using PRL extrapituitary (negative control) or pituitary (positive control) primers respectively; lanes 4 and 5, PCR products from NIH-3T3 cells (used as negative control) using PRL extrapituitary or pituitary primers respectively; lanes 6 and 7, PCR products from MCF-7 cells using PRL extrapituitary or pituitary primers respectively; lanes 8 and 9, PCR products from HeLa cells using PRL extrapituitary or pituitary primers respectively (positive control). (C) Diagram of the proximal PRL gene promoter construct (pGL2B-hPRL−216/+1) and the mutated constructs (pGL2B-hPRLmut1, pGL2B-hPRLmut2, and pGL2B-hPRLmut1+2) showing the location of the Pit-1-binding sites and the mutated bases. (D) Either the wild proximal pituitary PRL promoter or a mutant PRL promoter fused to the pGL2 basic vector was cotransfected with the pRSV-hPit-1 expression vector into MCF-7 cells using FuGene reagent, and cells were then cultured for 48 h. Normalized relative luciferase units (RLU) were calculated as the ratio of luciferase activity in cells transfected with the wild or mutated constructs to the activity in the corresponding control cells. (*P<0.05; ***P<0.001 versus control cells). (E) Soluble chromatin prepared from control MCF-7 cells, cells transfected with the pRSV-hPit-1 vector, and cells transfected with the control vector (pRc/RSV) were immunoprecipitated with an anti-Pit-1 antibody or control IgG. The immunoprecipitated DNA was amplified by PCR using specific primers corresponding to the proximal PRL promoter (from −216 to +1, with respect to the start transcription site).
Pit-1 regulates PRL expression in MCF-7 cells by binding to the PRL proximal promoter
To evaluate the possible effect of Pit-1 on PRL transcriptional activity, MCF-7 cells were transfected with either a construct linking 217 bp of the proximal PRL gene promoter to a luciferase reporter vector (pGL2B-hPRL−216/+1, Fig. 2C) or the pGL2-basic vector, and then cotransfected with the pRSV-hPit-1 vector. At 48 h after transfection, cells were harvested for measurement of luciferase activity. As shown in Fig. 2D, cotransfection with the pGL2B-hPRL−216/+1 and the pRSV-hPit-1 led to a significant increase in luciferase activity (P<0.001). In order to specify the location of Pit-1 binding to the PRL promoter, we carried out a site-directed mutagenesis on two known Pit-1-binding sites (Peers et al. 1991; Fig. 2C), and then cotransfected these constructs with the pRSV-hPit-1 expression vector. As shown in Fig. 2D, the transcriptional increase obtained using the wild-type pGL2B-hPRL−216/+1 construct was significantly reduced when the mutated PRL promoter constructs were cotransfected with the pRSV-hPit-1 vector, demonstrating that these elements are functional.
To confirm in vivo interaction of Pit-1 with the hPRL proximal promoter, a ChIP assay was carried out. As shown in Fig. 2E, the ChIP PCR product was detected with the Pit-1 antibody and specific primers to the proximal PRL promoter in control, untransfected, and pRSV-hPit-1-transfected MCF-7 cells, suggesting that endogenous Pit-1 specifically binds to the regulatory region (−216/+1) of the PRL promoter.
Effect of overexpression and silencing of Pit-1 on the proliferation of MCF-7 cells
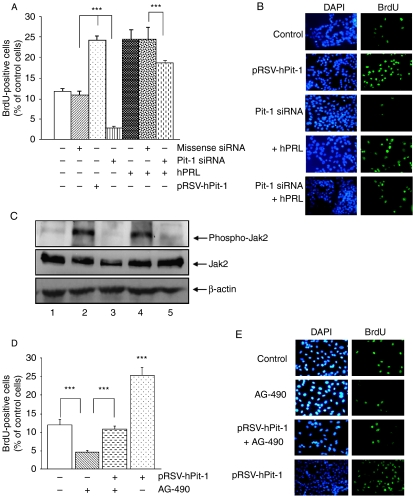
To evaluate the effect of Pit-1 overexpression on MCF-7 cell proliferation, these cells were transfected with pRSV-hPit-1 expression vector for 48 h along with a puromycin-resistance-encoding vector (pEPuro) and selected with 1 μg/ml puromycin. As reported by Gil-Puig et al. (2005), cells transfected with pRSV-hPit-1 showed an increase in BrdU uptake with respect to control cells (23.98±1.20 vs 11.64±0.72%, P<0.001; Fig. 3A and B). To explore the role of endogenous Pit-1 on MCF-7 cell proliferation, we used the siRNA approach. As shown in Fig. 3A and B, MCF-7 cells were transfected with 20 nM Pit-1 siRNA (or 20 nM of a missense siRNA, negative control) along with a puromycin-resistance-encoding vector (pEPuro), and selected with 1 μg/ml puromycin. We found that siRNA-mediated Pit-1 knockdown significantly (P<0.001) reduced BrdU uptake as compared with control siRNA-treated cells (1.93±0.30 vs 10.49±1.23%).
Figure 3.
Both PRL and Pit-1 increase cell proliferation. (A) Incorporation of bromodeoxyuridine (BrdU) in each cell group. MCF-7 cells were transfected with pRSV-hPit-1 vector, or 20 nM missense siRNA, or 20 nM Pit-1 siRNA, along with a puromycin-resistance-encoding vector (pEPuro) and selected with 1 μg/ml puromycin. hPRL treatment (10 nM) was administered at 24 h, and 24 h later were subjected to BrdU pulse labeling (10 μmol/l) for 1 h. Incorporation of BrdU was determined at 48 h by fluorescence microscopy of cells stained with a FITC-conjugated anti-BrdU antibody and counterstained with 4,6-diamine-2-phenylindole (DAPI). Proliferation rate was calculated as the percentage of BrdU-positive nuclei with respect to DAPI-positive nuclei. Values represent means±s.d. from four independent determinations (***P<0.001). (B). Representative set of fluorescence microphotographs from the experiment described in (A). Original magnifications ×20. (C) Western blot of Jak2, Jak2 phosphorylated, and β-actin in control MCF-7 cells (lane 1), and after administration of H2O2 (500 μM during 1 h, lane 2), H2O2 plus AG-490 (30 μM, Jak2 tyrosine-kinase inhibitor, lane 3), after transfection of MCF-7 cells with pRSV-hPit-1 (lane 4), and pRSV-hPit-1-transfected cells plus AG-490 (lane 5). (D) MCF-7 cells were transfected, selected, and BrdU labeled as in (A). Marked cells (+) also received AG-490 (30 μM) at 24 h. Proliferation rate and values calculated and carried out as in (A). (E) Representative set of fluorescence microphotographs from the experiment described in (D). Original magnifications ×20.
To explore the effect of PRL administration on cell proliferation after Pit-1 siRNA knockdown, MCF-7 cells were transfected either with Pit-1 siRNA or control siRNA and 24 h later treated with 10 nM hPRL. After 24 h, cells were labeled with BrdU. Results are shown in Fig. 3A. As previously reported by Brockman et al. (2002), administration of hPRL to cells induced a significant increase (P<0.001) in cell proliferation with respect to untreated control cells. However, exogenous hPRL administration to cells after Pit-1 siRNA transfection significantly (P<0.001) reduced cell proliferation with respect to the cells treated with hPRL alone. Figure 3B shows a representative panel of DAPI and BrdU in control, PRL-treated and/or transfected MCF-7 cells.
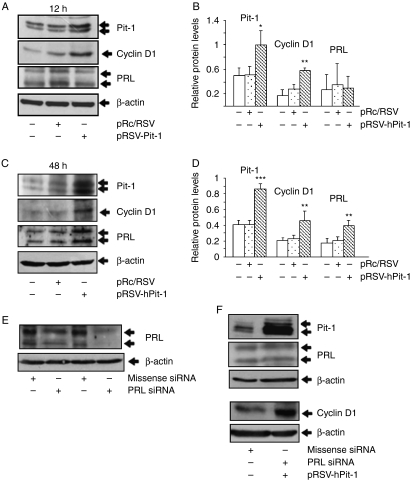
In order to evaluate the mechanisms of action of Pit-1-increased cell proliferation (i.e. independently or PRL-mediated), we administrated AG-490, a potent inhibitor of the Jak2 tyrosine kinase, which is critical for transmission of PRL-induced proliferative signals (Brockman et al. 2002). As shown in Fig. 3C (lane 4), Pit-1 overexpression induced an increase in Jak2 phosphorylation, which was blocked (lane 5) after AG-490 administration. Thus, AG-490 administration significantly reduced MCF-7 cell proliferation (4.6±0.4%; P<0.001) compared with control cells (11.9±1.5%; Fig. 3D). However, cell proliferation returned to normal values when the cells were treated with the AG-490 drug together with overexpressed Pit-1 transcription factor (10.8+0.8%; Fig. 3D). A panel with representative images of BrdU incorporation in control and treated and/or transfected MCF-7 cells is shown in Fig. 3E. To analyze more closely whether the increase in cell proliferation after Pit-1 was PRL dependent, total cell extracts were taken from MCF-7 cell controls and from those transfected with pRc/RSV or pRSV-hPit-1 vector at 12 and 48 h and, then, subjected to western blotting using specific antibodies. Western blots of Pit-1, PRL, cyclin D1 (as indicator of cell proliferation), and β-actin (used as loading control) are shown in Fig. 4A and C. Transfection of cells with the pRSV-hPit-1 vector for 12 h induced a significant increase in Pit-1 (1.0±0.24), and cyclin D1 (0.59±0.03) immunoreactive levels with respect to untransfected cells (0.50±0.1 and 0.18±0.09 respectively, P<0.05). At this stage, PRL protein levels remained unmodified (0.30±0.19) compared with untransfected (0.27±0.25) and pRc/RSV (0.36±0.33) transfected cells (Fig. 4B). However, 48 h after Pit-1 overexpression, a significant increase in PRL immunoreactive expression levels was observed (see Fig. 1B), in addition to the increase in Pit-1 and cyclin D1 protein levels (Fig. 4C and D).
Figure 4.
Pit-1 leads to a PRL-independent increase in cyclin D1. (A and C) Western blots of Pit-1, PRL, cyclin D1, and β-actin (used as loading control) were performed on control, pRc/RSV, and pRSV-hPit-1-transfected MCF-7 cells at 12 and 48 h respectively. (B and D) Relative protein levels in control MCF-7 cells and cells transfected with the pRc/RSV or pRSV-hPit-1 vector after 12 and 48 h respectively were calculated as the ratio of Pit-1, cyclin D1, and PRL with respect to the β-actin protein levels. Values represent mean±s.d. from densitometric evaluation of three western blots from independent experiments. (E) MCF-7 cells were transfected with 20 nM missense siRNA or 20 nM PRL siRNA. Forty-eight hours later, PRL protein immunoreactivity was evaluated by western blot. (F) MCF-7 cells were cotransfected with the pRSV-hPit-1 overexpression vector (2.5 μg) together with PRL siRNA (20 nM), and 48 h later, Pit-1, PRL, cyclin D1, and b-actin (as loading control) were evaluated by western blot.
To further explore if Pit-1 could increase cyclin D1 levels independently of PRL, endogenous PRL levels of MCF-7 cells were knocked down by transfecting a PRL siRNA (20 nM). Simultaneously, the pRSV-hPit-1 overexpression vector was also transfected. Figure 4E shows the effect at 48 h of PRL siRNA transfection on PRL immunoreactive protein levels in MCF-7 cells. Also at 48 h, Fig. 4F shows a visible increase in cyclin D1 and Pit-1, but not PRL, after Pit-1 overexpression and PRL siRNA cotransfection. These data suggest a PRL-independent effect of Pit-1 on cyclin D1.
Pit-1 and PRL protein expression in human breast tumors
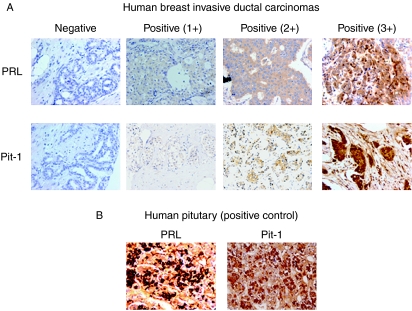
To further evaluate a possible relationship between Pit-1 and PRL expression in human breast tissue, both Pit-1 and PRL proteins were analyzed by immunohistochemistry in 94 invasive ductal carcinomas of the breast. Figure 5 shows representative examples of tumors with immunostaining for each protein evaluated. Pit-1 protein expression was detected mainly in nuclei of tumor or control epithelial cells, whereas PRL immunoreactivity showed cytoplasmic staining in both cases (Fig. 5A and B). A total of 81 tumors (86.1%) showed positive immunostaining for Pit-1, whereas 73 tumors (77.6%) showed positive immunostaining for PRL. Nevertheless, as can be seen in Table 1, there was variability in staining intensity for these factors. Our results also show a high concordance in the expression of Pit-1 and PRL. Thus, a total of 84 tumors (89.3%) showed concordance in the crude expressions of Pit-1 and PRL (P<0.0001). Significant concordance was also found between these two proteins with respect to grades of immunostaining intensity (P<0.0001; Table 1).
Figure 5.
Immunohistochemical detection of Pit-1 and PRL expression in human breast tissue. (A) Representative set of negative and positive (1+, 2+, and 3+) PRL and Pit-1 immunodetection in human breast invasive ductal carcinomas. (B) Human pituitary gland was used as positive control to detect PRL and Pit-1 immunopositivity. Original magnification ×20.
Table 1.
Relationships between Pit-1 and prolactin expression in 94 human breast invasive ductal carcinomas
| Prolactin | |||||
|---|---|---|---|---|---|
| Pit-1 | 0 | 1+ | 2+ | 3+ | Total cases |
| 0 | 12 | 0 | 1 | 0 | 13 |
| 1+ | 4 | 27 | 2 | 0 | 33 |
| 2+ | 5 | 14 | 20 | 1 | 40 |
| 3+ | 0 | 5 | 2 | 1 | 8 |
| Total cases | 21 | 46 | 25 | 2 | 94 |
Intensity of staining: 0, not staining; 1, weak; 2, moderate; 3, intense. χ2=69.36; P<0.0001.
Discussion
To explore the possible regulation of PRL by Pit-1 in human breast, several tumoral mammary cell lines were transfected with the Pit-1 expression vector. Our results demonstrate a significant increase in PRL mRNA 48 and 72 h after transfection with the pRSV-hPit-1 expression vector. In addition, a significant increase in PRL protein expression was observed in the MCF-7 cell line after Pit-1 overexpression. Conversely, a significant decrease in PRL protein expression was observed after Pit-1 knockdown. Taken together, these data seem to suggest that either endogenous or exogenous Pit-1 regulates PRL expression, at least in some mammary cell lines. Moreover, the luciferase reporter and ChIP assays reveal that this regulation is produced by Pit-1 binding to a proximal PRL promoter region.
At the pituitary level, it is well known that Pit-1 binds to the proximal PRL promoter and induces PRL expression. However, some extrapituitary tissues and cell lines, such as prostate cancer cells and the human breast adenocarcinoma cells, SK-BR-3, do not express Pit-1, yet PRL transcripts corresponding to the proximal promoter are present (Shaw-Bruha et al. 1997, Manfroid et al. 2005, Dagvadorj et al. 2007). This seems to support that Pit-1-independent PRL regulation in nonpituitary tissues can occur. For example, in a subclone of the SKUT-1B uterine sarcoma cell line, Pit-1 transfection does not modify endogenous PRL levels (Gellersen et al. 1995). However, in SK-BR-3 cells that contain PRL but not Pit-1, overexpression of Pit-1 induces the activation of transfected constructs containing the proximal pituitary PRL promoter (Manfroid et al. 2005) and the induction of PRL mRNA, as observed in our study. Pit-1 induction has also been demonstrated to lead to PRL or GH expression in other extrapituitary tissues. For instance, adenoviral delivery of Pit-1 in mouse liver induces the transient differentiation of adult hepatic stem cells into PRL-producing cells (Lee et al. 2005), and transfection of the corticotropic AtT-20 and the gonadotropic αT3-1 cell lines with the Pit-1 overexpression vector induces GH gene activation (Kurotani et al. 2002).
It has been demonstrated that PRL transcriptionally increases the levels of cyclin D1 that is a key factor in cell cycle. This further indicates that PRL directly induces cell proliferation (Brockman et al. 2002). In the present study, we also found a significant increase in BrdU uptake after PRL administration. However, when AG-490 was administered to block the Jak2 pathway and, thus, the possible effect of exogenous or Pit-1-induced endogenous PRL, overexpression of Pit-1 to the MCF-7 cells restored cell proliferation to the basal levels, suggesting a PRL-independent effect of Pit-1 on cell cycle. In fact, Pit-1 led to an increase in cyclin D1 expression levels before the increase in PRL levels was observed, and knockdown of endogenous PRL together with overexpression of Pit-1 clearly raised cyclin D1 protein levels. These data seem to suggest that Pit-1 increases cell proliferation independently of PRL probably by inducing cyclin D1 levels. With respect to this, it has been previously demonstrated that Pit-1 leads to a significant increase in cell proliferation in both pituitary (Castrillo et al. 1991) and extrapituitary cell lines (Gaiddon et al. 1999, Gil-Puig et al. 2005).
To study the relationships between Pit-1 and PRL in human breast cancer, we also evaluated the Pit-1 and PRL expression in 94 invasive ductal carcinomas. Our results indicate a significant correlation in the expression of both proteins. It has previously been demonstrated that Pit-1 is expressed in both normal and tumoral human breast, with Pit-1 mRNA levels being higher in tumors. This suggests that depending on its level, the expression of Pit-1 may be involved in normal mammary development, breast disorders, or both (Gil-Puig et al. 2005). The role of PRL in inducing mammary tumorogenesis has been thoroughly discussed (Vonderhaar 1999, Wennbo & Törnell 2000, Ben-Jonathan et al. 2002, Clevenger et al. 2003). However, few studies have evaluated PRL expression by immunohistochemistry in series of human mammary tumors (Bhatavdekar et al. 2000a,b). These authors have shown that PRL immunoreactivity was present in 78% (87 out of 111) and 82% (76 out of 93) of tumors, similar to our own finding (77%, 73/94), and that increased PRL positivity was significantly associated with increased tumor size, higher stage, nodal involvement, and worse overall survival in univariate analyses, although the association was attenuated after adjustment for stage (Bhatavdekar et al. 2000a). No data about determination of Pit-1 expression by immunohistochemistry in human breast tumor series is currently available. At least with respect to invasive ductal carcinomas of the breast, the statistically significant correlation between PRL and Pit-1 expression observed in the present study seems to suggest that Pit-1 could be involved in the pathogenesis of breast tumors either as a regulator of PRL or independently.
In summary, the present study demonstrates that Pit-1 regulates PRL mRNA and protein expression in human mammary cell lines, that Pit-1 increases cyclin D1 independently of PRL, and that a significant correlation exists between Pit-1 and PRL expression in human breast invasive ductal carcinomas. Taken together, our data seem to suggest that Pit-1 through PRL and/or independently of PRL could be related to mammary cell transformation.
Declaration of interest
The authors declare that there is no conflict of interest that would prejudice the impartiality of this work.
Funding
This study was supported by a grant from the Xunta de Galicia (PGIDIT06PXIB208028PR), Spain.
Acknowledgements
The pRSV-hPit-1 expression vector was a kind gift from Dr J L Castrillo. PRL and PRL antibody was obtained from Dr Parlow.
References
- Bamberger AM, Bamberger CM, Pu LP, Puy LA, Loh YP, Asa SL. Expression of Pit-1 messenger ribonucleic acid and protein in the human placenta. Journal of Clinical Endocrinology and Metabolism. 1995;80:2021–2026. doi: 10.1210/jcem.80.7.7608249. [DOI] [PubMed] [Google Scholar]
- Ben-Jonathan N, Liby K, McFarland M, Zinger M. Prolactin as an autocrine/paracrine growth factor in human cancer. Trends in Endocrinology and Metabolism. 2002;13:245–250. doi: 10.1016/s1043-2760(02)00603-3. [DOI] [PubMed] [Google Scholar]
- Berwaer M, Martial JA, Davis JRE. Characterization of an up-stream promoter directing extrapituitary expression of the human prolactin gene. Molecular Endocrinology. 1994;8:635–642. doi: 10.1210/mend.8.5.8058071. [DOI] [PubMed] [Google Scholar]
- Bhatavdekar JM, Patel DD, Shah NG, Vora HH, Suthar TP, Chikhlikar PR, Gosh N, Trivedi TI. Prognostic significance of immunohistochemically localized biomarkers in stage II and stage III breast cancer: a multivariate analysis. Annals of Surgical Oncology. 2000a;7:305–311. doi: 10.1007/s10434-000-0305-5. [DOI] [PubMed] [Google Scholar]
- Bhatavdekar JM, Patel DD, Shah NG, Vora HH, Suthar TP, Ghosh N, Chikhlikar PR, Trivedi TI. Prolactin as a local growth promoter in patients with breast cancer: GCRI experience. European Journal of Surgical Oncology. 2000b;26:540–547. doi: 10.1053/ejso.2000.0943. [DOI] [PubMed] [Google Scholar]
- Brockman JL, Schroeder MD, Schuler LA. PRL activates the cyclin D1 promoter via the Jak2/Stat pathway. Molecular Endocrinology. 2002;16:774–784. doi: 10.1210/mend.16.4.0817. [DOI] [PubMed] [Google Scholar]
- Castrillo JL, Theill L, Karin M. Function of the homeodomain protein GHF1 in pituitary cell proliferation. Science. 1991;253:97–199. doi: 10.1126/science.1677216. [DOI] [PubMed] [Google Scholar]
- Clevenger CV, Chang WP, Ngo W, Pasha TL, Montone KT, Tomaszewski JE. Expression of prolactin and prolactin receptor in human breast carcinoma. Evidence for an autocrine/paracrine loop. American Journal of Pathology. 1995;146:695–705. [PMC free article] [PubMed] [Google Scholar]
- Clevenger CV, Furth PA, Hankinson SE, Schuler LA. The role of prolactin in mammary carcinoma. Endocrine Reviews. 2003;24:1–27. doi: 10.1210/er.2001-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagvadorj A, Collins S, Jomain J-B, Abdulghani J, Karras J, Zellweger T, Li H, Nurmi M, Alanen K, Mirtti T, et al. Autocrine prolactin promotes prostate cancer cell growth via janus kinase-2-signal transducer and activator of transcription-5a/b signaling pathway. Endocrinology. 2007;148:3089–3101. doi: 10.1210/en.2006-1761. [DOI] [PubMed] [Google Scholar]
- Delhase M, Vergani P, Malur A, Hooghe-Peters EL, Hooghe RJ. The transcription factor Pit-1/GHF-1 is expressed in hemopoietic and lymphoid tissues. European Journal of Immunology. 1993;23:951–955. doi: 10.1002/eji.1830230428. [DOI] [PubMed] [Google Scholar]
- Freeman ME, Kanyicska B, Lerant A, Nagy G. Prolactin: structure, function, and regulation of secretion. Physiological Reviews. 2000;80:1523–1631. doi: 10.1152/physrev.2000.80.4.1523. [DOI] [PubMed] [Google Scholar]
- Fuh G, Wells JA. Prolactin receptor antagonists that inhibit the growth of breast cancer cell lines. Journal of Biological Chemistry. 1995;270:13133–13137. doi: 10.1074/jbc.270.22.13133. [DOI] [PubMed] [Google Scholar]
- Gaiddon C, de Tapia M, Loeffler JP. The tissue-specific transcription factor Pit-1/GHF-1 binds to the c-fos serum response element and activates c-fos transcription. Molecular Endocrinology. 1999;13:742–751. doi: 10.1210/mend.13.5.0275. [DOI] [PubMed] [Google Scholar]
- Gellersen B, Kempf R, Telgmann R, DiMattia GE. Nonpituitary human prolactin gene transcription is independent of Pit-1 and differentially controlled in lymphocytes and in endometrial stroma. Molecular Endocrinology. 1994;8:356–373. doi: 10.1210/mend.8.3.8015553. [DOI] [PubMed] [Google Scholar]
- Gellersen B, Kempf R, Telgmann R, DiMattia GE. Pituitary-type transcription of the human prolactin gene in the absence of Pit-1. Molecular Endocrinology. 1995;9:887–901. doi: 10.1210/mend.9.7.7476971. [DOI] [PubMed] [Google Scholar]
- Gil-Puig C, Blanco M, Garcia-Caballero T, Segura C, Perez-Fernandez R. Pit-1/GHF-1 and GH expression in the MCF-7 human breast adenocarcinoma cell line. Journal of Endocrinology. 2002;173:161–167. doi: 10.1677/joe.0.1730161. [DOI] [PubMed] [Google Scholar]
- Gil-Puig C, Seoane S, Blanco M, Macia M, Garcia-Caballero T, Segura C, Perez-Fernandez R. Pit-1 is expressed in normal and tumoral human breast and regulates growth hormone secretion and cell proliferation. European Journal of Endocrinology. 2005;153:335–344. doi: 10.1530/eje.1.01962. [DOI] [PubMed] [Google Scholar]
- Ginsburg E, Vonderhaar BK. Prolactin synthesis and secretion by human breast cancer cells. Cancer Research. 1995;55:2591–2595. [PubMed] [Google Scholar]
- Kurotani R, Yoshimura S, Iwasaki Y, Inoue K, Teramoto A, Osamura RY. Exogenous expression of Pit-1 in AtT-20 corticotropic cells induces endogenous growth hormone gene transcription. Journal of Endocrinology. 2002;172:477–487. doi: 10.1677/joe.0.1720477. [DOI] [PubMed] [Google Scholar]
- Lee EJ, Russell T, Hurley L, Jameson JL. Pituitary transcription factor-1 induces transient differentiation of adult hepatic stem cells into prolactin-producing cells in vivo. Molecular Endocrinology. 2005;19:964–971. doi: 10.1210/me.2004-0034. [DOI] [PubMed] [Google Scholar]
- Lefevre C, Imagawa M, Dana S, Grindlay J, Bodner M, Karin M. Tissue specific expression of the human growth hormone gene is conferred in part by the binding of a specific trans-acting factor. EMBO Journal. 1987;6:971–981. doi: 10.1002/j.1460-2075.1987.tb04847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Crenshaw EB, Rawson EJ, Simmons DM, Swanson LW, Rosenfeld MG. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene Pit-1. Nature. 1990;347:528–533. doi: 10.1038/347528a0. [DOI] [PubMed] [Google Scholar]
- Llovera M, Pichard C, Bernichtein S, Jeay S, Touraine P, Kelly PA, Goffin V. Human prolactin (hPRL) antagonists inhibit hPRL-activated signalling pathways involved in breast cancer cell proliferation. Oncogene. 2000;19:4695–4705. doi: 10.1038/sj.onc.1203846. [DOI] [PubMed] [Google Scholar]
- Manfroid I, Van de Weerdt C, Baudhuin A, Martial JA, Muller M. EGF stimulates Pit-1 independent transcription of the human prolactin pituitary promoter in human breast cancer SK-BR-3 cells through its proximal AP-1 response element. Molecular and Cellular Endocrinology. 2005;229:127–139. doi: 10.1016/j.mce.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Markoff E, Lee DW. Glycosylated prolactin is a major circulating variant in human serum. Journal of Clinical Endocrinology and Metabolism. 1987;65:1102–1106. doi: 10.1210/jcem-65-6-1102. [DOI] [PubMed] [Google Scholar]
- Mertani HC, Garcia-Caballero T, Lambert A, Gerard F, Palayer C, Boutin JM, Vonderhaar BK, Waters MJ, Lobie PE, Morel G. Cellular expression of growth hormone and prolactin receptors in human breast disorders. International Journal of Cancer. 1998;79:202–211. doi: 10.1002/(sici)1097-0215(19980417)79:2<202::aid-ijc17>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Nelson C, Albert VR, Elsholtz HP, LI-W Lu, Rosenfeld MG. Activation of cell-specific expression of rat growth hormone and prolactin genes by a common transcription factor. Science. 1988;239:1400–1405. doi: 10.1126/science.2831625. [DOI] [PubMed] [Google Scholar]
- Peers B, Monget P, Nalda A, Voz ML, Berwaer M, Belayew A, Martial JA. Transcriptional induction of the human prolactin gene by cAMP requires two cis-acting elements and at least the pituitary-specific factor Pit-1. Journal of Biological Chemistry. 1991;266:18127–18134. [PubMed] [Google Scholar]
- Seoane S, Perez-Fernandez R. The vitamin D receptor represses transcription of the pituitary transcription factor Pit-1 gene without involvement of the retinoid X receptor. Molecular Endocrinology. 2006;20:735–748. doi: 10.1210/me.2005-0253. [DOI] [PubMed] [Google Scholar]
- Seoane S, Ben I, Centeno V, Perez-Fernandez R. Cellular expression levels of the vitamin D receptor are critical to its transcriptional regulation by the pituitary transcription factor Pit-1. Molecular Endocrinology. 2007;21:1513–1525. doi: 10.1210/me.2006-0554. [DOI] [PubMed] [Google Scholar]
- Shaw-Bruha CM, Pirrucello SJ, Shull JD. Expression of the prolactin gene in normal and neoplastic human breast tissues and human mammary cell lines: promoter usage and alternative mRNA splicing. Breast Cancer Research and Treatment. 1997;44:243–253. doi: 10.1023/a:1005879103367. [DOI] [PubMed] [Google Scholar]
- Shinha YN. Structural variants of prolactin: occurrence and physiological significance. Endocrine Reviews. 1995;16:354–369. doi: 10.1210/edrv-16-3-354. [DOI] [PubMed] [Google Scholar]
- Tworoger SS, Eliassen AH, Sluss P, Hankinson SE. A prospective study of plasma prolactin concentrations and risk of premenopausal and postmenopausal breast cancer. Journal of Clinical Oncology. 2007;25:1482–1488. doi: 10.1200/JCO.2006.07.6356. [DOI] [PubMed] [Google Scholar]
- Vonderhaar BK. Prolactin involvement in breast cancer. Endocrine-Related Cancer. 1999;6:389–404. doi: 10.1677/erc.0.0060389. [DOI] [PubMed] [Google Scholar]
- Voss JW, Yao T-P, Rosenfeld MG. Alternative translation initiation site usage results in two structurally distinct forms of Pit-1. Journal of Biological Chemistry. 1991;266:12832–12835. [PubMed] [Google Scholar]
- Welsch CW, Nagasawa H. Prolactin and murine mammary tumorogenesis: a review. Cancer Research. 1977;37:951–963. [PubMed] [Google Scholar]
- Wennbo H, Törnell J. The role of prolactin and growth hormone in breast cancer. Oncogene. 2000;19:2744–2751. doi: 10.1038/sj.onc.1203349. [DOI] [PubMed] [Google Scholar]
- Wennbo H, Gebre-Medhin M, Gritli-Linde A, Ohlsson C, Isaksson OG, Tornell J. Activation of the prolactin receptor but not the growth hormone receptor is important for induction of mammary tumors in transgenic mice. Journal of Clinical Investigation. 1997;100:2744–2751. doi: 10.1172/JCI119820. [DOI] [PMC free article] [PubMed] [Google Scholar]