Abstract
Genetic changes that help explain the differences between two individuals might include those that create or disrupt sites complementary to microRNAs1,2, but the extent to which such polymorphic microRNA sites mediate repression is unknown. Here, we develop a method to measure mRNA allelic imbalances associated with a regulatory site found in mRNA from one allele but not in that from the other. Applying this method, called allelic-imbalance sequencing (AI-Seq), to sites for three microRNAs (miR-1, miR-133, and miR-122) provided quantitative measurements of repression in vivo, without altering either the microRNAs or their targets. A significant fraction of polymorphic sites mediated repression in tissues that expressed the cognate microRNA, with downregulation depending on site type and site context. Extrapolating these results to the other broadly conserved microRNAs suggests that when comparing two mouse strains (or two human individuals), polymorphic microRNA sites cause expression of many (often hundreds) of genes to differ.
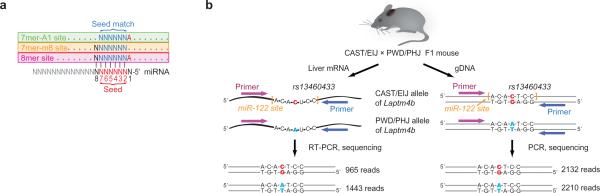
MicroRNAs (miRNAs) are ~23-nt endogenous RNAs that pair to messages of protein-coding genes to direct the posttranscriptional repression of these mRNAs3. To explore miRNA regulatory diversity within a single species, we considered miRNA complementary sites that are created or disrupted by single-nucleotide polymorphisms (SNPs) in mice. Our study centered on three types of complementary sites that previous computational and experimental results indicate can mediate miRNA recognition4. Each of these three sites includes perfect Watson-Crick pairing to the miRNA seed (miRNA positions 2-7); one is a 7-nt site, referred to here as the “7mer-m8 site,” for which seed pairing is supplemented by a Watson-Crick match to miRNA nucleotide 8 (refs. 5-7); another is the “7mer-A1 site,” for which seed pairing is supplemented by an A across from miRNA nucleotide 1 (ref. 5), and the third is the “8mer site,” which has both the m8 match and the A across from position 1 (ref. 5; Fig. 1a). We focused on sites to three miRNAs, miR-1 (for our purposes synonymous with its paralog, miR-206), miR-133, and miR-122, because of the strong, tissue-specific expression of these miRNAs in relatively homogenous and accessible tissues, muscle (miR-1 and miR-133) or liver (miR-122)8,9. In agreement with previous reports1,10-12, a search of the SNP databases13,14, focusing on 3' untranslated regions (UTRs) because these regions of the messages are most hospitable to miRNA targeting15, revealed many polymorphisms that when compared to the reference sequence create or disrupt sites for one of the three miRNAs. Because miRNAs often destabilize their target mRNAs16, we reasoned that if these sites were functional in the tissue expressing the cognate miRNA, then less RNA might accumulate from the allele with the site. Moreover, in mice heterozygous for the SNPs, destabilization of mRNA from the target allele, but not that from the non-target allele, would contribute to allelic imbalance in mRNA steady-state level. Hence, we developed a method called allelic-imbalance sequencing (AI-Seq) to measure such imbalances, reasoning that any imbalances would identify and quantify miRNA regulatory diversity within a species, and provide a unique opportunity to examine molecular consequences of miRNA-mediated repression in vivo without perturbing either the miRNA or its targets.
Figure 1.
Measurement of mRNA allelic imbalances associated with heterozygous miRNA target sites. (a) Canonical 7-8-nt miRNA target sites. (b) AI-Seq, illustrated for the SNP rs13460433, which generates a heterozygous miR-122 target site in the Laptm4b gene of CAST/EIJ × PWD/PHJ F1 mice. When using liver mRNA, the 965 and 1443 reads obtained from the target and the non-target allele, respectively, imply an allelic imbalance of 0.67 (= 965 / 1443). Because allele-specific PCR bias might have influenced this ratio, amplification and sequencing was performed in parallel with the same primers but using genomic DNA (gDNA) instead of mRNA. The gDNA template produced a target:non-target ratio of 0.96 (= 2132 / 2210), enabling the raw allelic imbalance to be corrected to 0.70 (= 0.67 / 0.96) or -0.51 in log2 scale. This ratio implied that in liver, a tissue expressing miR-122, the mRNA abundance of the target allele is 70% of that of the non-target allele.
Because lab strains lack the heterozygosity found in natural populations, we performed five inter-strain crosses to generate mice heterozygous for the parental alleles. Approximately 300 annotated SNPs that create/disrupt target sites for one of the three miRNAs were heterozygous in at least one of the five F1 hybrids. We chose a subset of these, preferring those that create/disrupt 8mers, those in messages with evidence of expression in the tissues expressing the miRNAs, and those that were not linked to many nearby polymorphisms. Allelic imbalance was measured for 67 target sites (28 for miR-122, 28 for miR-1, and 11 for miR-133) in the tissue expressing the cognate miRNA.
For AI-Seq, mRNA fragments containing the SNPs were first reverse transcribed (RT) and amplified (PCR), and then the amplicon was subjected to high-throughput pyrosequencing17 (Fig. 1b, Supplementary Discussion and Supplementary Fig. 1 online). To economize on sequencing, amplicons derived from different primers were pooled. Because the primers flanking the SNPs used for RT-PCR were gene-specific but not allelespecific, both alleles of the same gene were amplified by the same reaction, and their relative abundance could be inferred from the number of sequencing reads representing each allele.
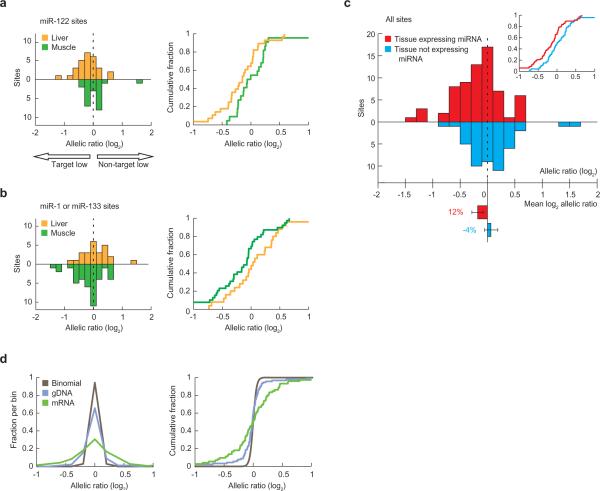
If none of the intact miRNA sites imparted repression, the distribution of log ratios would center on zero, with individual measurements deviating from zero because of experimental noise and allelic imbalance independent of repression from the heterozygous site. However, the distribution of the log ratios for the 28 miR-122 sites measured using liver, centered below zero, consistent with the hypothesis that mRNA from some of the alleles with target sites was destabilized (Fig. 2a). If miR-122 caused this destabilization, then the shift from zero would depend on the presence of miR-122. To test this dependency, we measured ratios for 22 of the 28 miR-122 sites in muscle, which does not express miR-122. The remaining six sites were missing from the muscle-measured ratios because four of the six were in messages not expressed in muscle, and the other two were unusual in that the SNP disrupting the miR-122 site (CACTCCA and ACACTCC, SNP underlined) simultaneously created a site for miR-1 (CATTCCA and ACATTCC), a miRNA expressed in muscle, thereby precluding the muscle-measured ratios as negative controls. As expected for a miRNA-mediated effect, the shift from zero disappeared in muscle (Fig. 2a). When analyzing the ratios for the 39 sites for miR-1 or miR-133, which are expressed in the muscle but not in the liver, the reciprocal pattern was observed, in which the distribution of the liver-measured ratios was centered on zero and that of the muscle-measured ratios was skewed to the left (Fig. 2b).
Figure 2.
Impact of heterozygous target sites on mRNA allelic imbalance. (a) Distribution of allelic ratios (target:non-target, log2) measured for miR-122 heterozygous sites using mRNA from either liver (orange, n = 28) or muscle (green, n = 22), plotted as a histogram (left, 0.2-unit bins) and cumulative distribution (right). (b) Distribution of allelic ratios measured for miR-1 and miR-133 heterozygous sites using mRNA from either liver (orange, n = 25) or muscle (green, n = 39), plotted as in panel a. (c) Distribution of allelic ratios, pooling ratios from panels a and b, measured using either mRNA from the tissue expressing the cognate miRNA (red, n = 67), or mRNA from the tissue not expressing the cognate miRNA (blue, n = 47). In the inset are the cumulative distributions, plotted as in panels a and b. Below the histogram is the mean offset from zero for the two distributions, with error bars indicating 95% confidence intervals (bootstrapping) for the mean, and the percentages indicating the average downregulation of target alleles compared to non-target alleles. (d) Components of variability in allelic ratios, depicted with standard (left, 0.2-unit bins) and cumulative (right) distributions. Total variability not attributable to the cognate miRNAs was measured using mRNA with heterozygous sites not predicted to be regulated by the cognate miRNAs (n = 72). Of the 72 ratios determined, 47 were from mRNAs of tissues lacking the cognate miRNA, and 25 were from mRNAs without predicted potential for allele-specific repression mediated by the three miRNAs. (These 72 ratios were not normalized to corresponding gDNA ratios.) PCR variability was measured using gDNA (n = 90). Stochastic counting error was simulated using the binomial model (n = 9,000, 100 simulations per gDNA measurement), with total counts for each simulated amplicon chosen to match those of the gDNA measurements. The differences between each of the three possible pairs of distributions were statistically significant (p < 0.01 for each comparison, two-sided KS test).
To increase sample size and thereby achieve statistical significance, datasets were combined such that the ratios measured in the presence of the cognate miRNA were analyzed together and compared to those measured in the absence of the miRNA (Fig. 2c). A significantly large fraction of the log ratios were smaller than zero in the presence of the miRNA (p < 0.01, one-sided exact binomial test), but not in the absence of the miRNA (p = 0.6), and the difference between the two distributions also was significant (p = 0.02, one-sided Kolmogorov-Smirnov [KS] test). Thus, at least a subset of the interrogated target sites mediated repression.
On average, the polymorphic sites were associated with mRNA downregulation of 12% (Fig. 2c; 95% confidence interval of 5-18%, bootstrapping). Actual downregulation was likely greater because the signal could have been diluted by both nuclear mRNA and mRNA from cells that do not express the cognate miRNA, such as those from blood or vasculature. Effects of functional sites also might have been diluted by inclusion of nonfunctional sites. Nonfunctional sites presumably were enriched among the set of sites interrogated in this study because natural selection selects against polymorphisms that either disrupt beneficial functional sites or generate functional sites in messages that should not be repressed10,18,19.
A lower bound for the fraction of functional sites was estimated by analyzing the maximal vertical displacement of the cumulative distribution curves (correcting for the bumpiness of the distributions15), and was found to be 19%. This estimate was a conservative lower bound for the true fraction; simulations incorporating the variability observed from the tissues lacking the miRNA showed that if 100% of sites mediated target repression by 20%, only 32% would be detected functional by our analyses. Therefore, at least 19% and perhaps over half the examined polymorphic sites mediated repression.
The experimental variability was composed of multiple components. One was stochastic sampling error inherent to counting sequencing reads, which was modeled by the binomial distribution (Fig. 2d). A second component was PCR variability, which was estimated as the excess variability in the allelic ratios measured using gDNA, compared to the stochastic variability (Fig. 2d; difference between gDNA and binomial distributions). Another component was the biological noise, which could stem from a variety of sources, including differential epigenetic states of the two alleles or allelic differences in linked cisregulatory elements, either of which might differentially regulate the two alleles. To begin to estimate the biological noise, we examined the distribution of the allelic ratios measured using both the mRNAs from tissues lacking the cognate miRNA and other mRNAs without predicted potential for allelic-specific repression mediated by the three miRNAs. Allelic ratios from these control mRNAs were substantially more variable than those from gDNA, suggesting frequent allelic imbalance not attributable to the sites under investigation (Fig. 2d). However, we were unable to quantify the frequency or magnitude of this potentially widespread allelic imbalance because of the possibility that RT variability also contributed to the added variability of the mRNA controls compared to that of the gDNA controls.
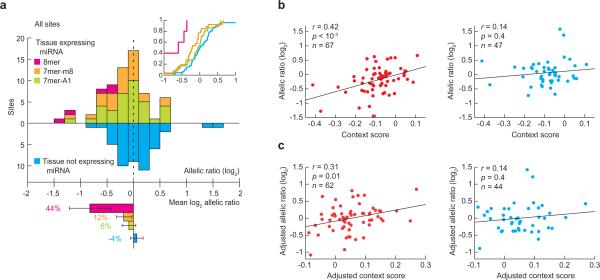
Our quantitative readout of site efficacy in vivo, without perturbation of either the miRNAs or their targets, provided a fresh opportunity to examine the influence of site type and site context. The 8mer sites performed significantly better than did 7mer-m8 or 7mer-A1 sites (Fig. 3a; p = 0.005 and p = 0.001 respectively, one-sided KS test), and 7mer-m8 sites tended to perform slightly better than 7mer-A1 sites, although this difference was not statistically significant (p = 0.1, one-sided KS test). Overall efficacy rank order of the three types was consistent with previous observations from experiments that ectopically expressed or deleted miRNAs15,20-22.
Figure 3.
Dependence of target site efficacy on site type and context. (a) Efficacy of target sites of different types (8mer, n = 5; 7mer-m8, n = 26; 7mer-A1, n = 36), plotted as in Figure 2c. (b) Relationship between allelic ratio and context score in the tissue expressing (left) and not expressing (right) the miRNA. Lines are the least-square fit to the data (r, Pearson correlation coefficient, with p value estimated by two-sided Pearson correlation test). When considering only the 47 sites in the left panel that were measured also in the absence of the cognate miRNA (right panel), the correlation remained significant (r = 0.58 and p < 10-4). (c) Relationship between adjusted allelic ratio and adjusted context score for 7-nt sites, plotted as in panel b. Context score and log ratio were each adjusted by subtracting the portion contributed by site type and the mean log ratio of sites of the same type, respectively. When considering only the 44 sites in the left panel that were measured also in the absence of the cognate miRNA (right panel), the correlation remained significant (r = 0.53 and p < 10-3).
To consider the influence of site context, the “context score” was calculated for each polymorphic site. Context scores quantitatively evaluate site type and three features of site context (surrounding AU content, position within the 3'UTR, and pairing to 3'-region of miRNA) to predict site efficacy15. Context scores significantly correlated with target downregulation in the presence of the cognate miRNA, but not in the absence of the miRNA (Fig. 3b). Significant correlation was retained in the presence of the miRNA even after the contribution of site type had been factored out, thereby indicating that site context, as scored by this model, influences the efficacy of polymorphic sites (Fig. 3c).
Our experiments focused on three of the 87 miRNA families conserved to chicken or beyond23. Expanding our SNP database search to the other 84 broadly conserved miRNA families and the ~8 million SNPs annotated in 15 mouse strains13 showed that two strains have an average of 2,430 distinct polymorphic sites (bottom and top 2.5 percentile, 810-4,600; median, 1,470) and 1,510 genes with at least one polymorphic miRNA site (bottom and top 2.5 percentile, 520-2,790; median, 950). These numbers would increase with consideration of sites to the hundreds of additional annotated miRNAs. However, because species-specific miRNAs and those conserved only within mammals tend to be expressed at lower levels, their 7-8mer sites are thought to be less frequently sufficient for mediating repression4. Therefore, to guard against overstating the impact of polymorphic sites, we did not consider these additional miRNAs.
Our results on the performance of polymorphic sites matching three miRNAs, when extrapolated to the sites of all broadly conserved miRNAs, provided the first glimpse of the direct impact of such sites on gene-expression variation within a species. For the 67 sites examined, we observed average downregulation of 12%, with at least 19% of the sites responsible for the observed downregulation. Correcting for our preference in choosing 8mers for analysis slightly lowered these percentages to 10% and 18%, as our best estimates, respectively, for all 7-8mer polymorphic sites in mRNAs expressed in cognate tissues. If 50% of the genes with the sites are expressed in any cell type of the body that express the cognate miRNA and if the actual fraction of functional sites matches our observed lower limit of 18%, then at least 9% (= 0.5 × 0.18) of the genes with polymorphic sites will be differentially regulated between two strains. Thus, between most mouse strains, over a hundred messages are likely to be differentially regulated through polymorphic sites, with average mRNA downregulation for these messages exceeding 55% (average downregulation of all sites of 10%, divided by 0.18). In a more likely scenario in which half the sites in cognate tissues are functional, proportionally more messages would be downregulated, with average downregulation of functional sites still approximating 20%. Because miRNAs also influence translation, effects on the proteome are presumed even greater. Overall, it is hard to escape the conclusion that polymorphic miRNA regulatory sites have a substantial impact on gene-expression variation within a species.
Our results in mice, considered together with SNP frequencies in humans, indicate that any two unrelated humans are likely to have more than a hundred genes differentially regulated due to polymorphic miRNA targeting. Assuming that some of these could explain differences in disease risk among individuals, our results suggest that, as more genome-wide association studies are conducted with improved coverage in 3'UTRs, more miRNA target site polymorphisms will be associated with clinical conditions and individual traits2.
Despite the success in mammalian tissues, miRNA effects were not detected in a HapMap24 panel of lymphoblastoid cell lines when we used AI-Seq to measure the allelic imbalance of 56 heterozygous target sites for nine miRNA families most highly expressed in these cell lines (data not shown). Cell lines exhibit variable degrees of clonality with random monoallelic expression in each clone25, and the resulting random allelic imbalance might have overwhelmed any imbalance resulting from polymorphic regulatory elements of interest. Moreover, the process of establishing lymphoblastoid cell lines, which involved Epstein-Barr-virus infection and subsequent transformation of B-cells, might have downregulated miRNA expression26.
Experiments examining the influence of miRNA knockouts on the transcriptome and proteome have been very informative for inferring effects of both conserved and nonconserved sites that are not polymorphic21,27,28. Our results complement those of these previous studies, revealing the frequent influence of polymorphic sites, with the added advantage of detecting miRNA regulatory impact in vivo without perturbing either the miRNAs or their targets. Upregulation of a target following miRNA knockout can trigger offsetting feedback regulation that reduces the observed effect of the miRNA. Our AI-Seq results are less likely to be confounded by such a response, because the feedback regulation is usually not allele-specific and therefore is unlikely to change the relative expression of the target compared to the non-target allele. Our approach can be extended to characterization of other cis-regulatory elements that might influence mRNA level. As the capacity of high-throughput sequencing increases, we anticipate that RNA-Seq coverage will expand to the point that directed amplification of specific loci will no longer be required to accurately detect many allelic imbalances and that our approach of correlating imbalances with predicted regulatory sites can be applied transcriptome-wide to reveal many of the polymorphic regulatory sites contributing to these imbalances.
METHODS
Mouse tissues and preparation of cDNA and gDNA
The study was approved by the MIT Committee on Animal Care. The Jackson laboratory performed five inter-strain crosses (CAST/EIJ × PWD/PHJ, FVB/NJ × PWD/PHJ, A/J × C57BL/6J, WSB/EiJ × MOLF/EiJ, A/J × DBA/2J) and dissected liver and skeletal muscle from two 4-week-old F1 littermates of each cross. For each cross and tissue, ~0.6 g tissue (~0.3 g from each littermate) was homogenized for RNA extraction (RNeasy Maxi kit, Qiagen), and cDNA was synthesized from total RNA in RT reactions (Superscript III, Invitrogen) primed with random hexamers. For each cross, gDNA was isolated from ~50 mg of either liver or muscle from either littermate (DNeasy Blood and Tissue kit, Qiagen).
Computational identification of polymorphic sites
Genomic coordinates of known mouse SNPs on the July 2007 genome assembly (mm9) were obtained from NCBI dbSNP build 12814. Genotypes of mouse strains were obtained from dbSNP build 128, mm9 genomic sequence (for C57BL/6J strain), and the Perlegen data (http://mouse.perlegen.com)13. Gene annotation on mm9 was obtained from UCSC genome browser29. We identified SNPs that generate heterozygous sites for miR-122, miR-1 or miR-133 in at least one of the five crosses (123 SNPs for miR-122, 109 SNPs for miR-1, and 74 SNPs for miR-133; 7-nt sites to 8-nt sites ratio, 10.3), excluding those that modify the sites, e.g., by converting a 7mer to 8mer, or vice verse.
Site selection and DNA amplification
Polymorphic sites located <15 nt from the stop codon were excluded15. Also excluded were those in the genes expressed, according to the mouse expression atlas30, at a level lower than 90% of all genes in the tissue that expresses the cognate miRNA; sites in genes without an expression measurement were not excluded. Out of the remaining sites, all polymorphic 8mers were chosen. A subset of the 7-nt polymorphic sites were chosen somewhat arbitrarily, preferring those with fewer additional SNPs in flanking regions, which could potentially interfere with primer annealing. A total of 138 SNPs were carried forward for primer design, performed with the aid of PRIMER3 and suitable primers were found for 124 of those, creating 136 polymorphic sites (7-nt sites to 8-nt sites ratio, 5.5). PCR amplification reactions of each of the SNPs were performed individually (Phusion Hot Start polymerase, NEB). All PCR reactions done with cDNA were accompanied by a matching no-RT control. Fragments that failed to be amplified at sufficient yield from either gDNA or cDNA were discarded. For each successful amplification, the procedure was repeated using cDNA from the noncognate tissue. As additional controls to examine variability of allelic imbalances that were not attributable to miRNA targeting, SNPs that do not generate polymorphic miRNA sites for the three miRNAs were identified in the ORFs of 27 genes that had polymorphic sites in the 3'UTRs, and these 27 SNPs were amplified using F1 hybrids that were heterozygous for the ORF SNP but homozygous for the 3'UTR SNP. In total, 240 amplicons (70 3'UTR SNPs from cDNA of the tissue with miRNA expression, 49 3'UTR SNPs from cDNA of the tissue without miRNA expression, 27 ORF SNPs from cDNA of either tissue, and 94 SNPs from gDNA) were prepared for sequencing.
Mixing and purifying PCR products for pyrosequencing
Because gDNA-templated, liver-mRNA-templated, and muscle-mRNA-templated amplicons all shared the same primers, they needed to be sequenced in separate pools, so that they could be distinguished from one another. Because one pyrosequencing plate can be divided into four segments without contamination between segments, the 240 amplicons were mixed into 4 pools. Each pool had ~60 amplicons, mixed in equimolar amounts following determination of each amplicon concentration (Bioanalyzer, Agilent Technologies). Each pool was deproteinated (phenol, chloroform with iso-amyl-alcohol), purified by native PAGE gel, taking precautions to avoid denaturing the double-stranded PCR products, and submitted to the 454 Life Sciences for sequencing. Three sequencing runs were performed.
Analysis of sequencing reads
Of the ~1.194 million reads acquired, ~1.085 million (~91%) correctly mapped to the 3'-terminal 10-nt fragments of the unique primer pairs. Of the 240 amplicons, 9 were excluded for at least one of the following reasons: (1) the number of reads obtained per amplicon was less than 300, (2) a SNP was not detected, (3) a severe allelic bias was observed with gDNA. The remaining 231 amplicons had a median of 3,978 reads (range, 541-18,714) and corresponded to 65 3'UTR SNPs and 25 ORF SNPs. Information and results for each amplicon are provided (Supplementary Tables 1,2 online). Although most of the sites were polymorphic for only one of the three miRNAs, five exceptional SNPs allowed one allele to have a miR-122 site and the other allele to have a miR-1 or miR-133 site. Three of the five were in mRNA expressed only in muscle, but the remaining two (rs IDs: 36333425, 30114270) were expressed in both tissues, allowing the measurement taken from liver to report on the miR-122 site and that from muscle to report on miR-1 site. Another exception was the two polymorphic sites in the same miRNA (rs IDs: 32325030, 32323893) that exist in the same cross and collectively allow one Snap29 allele to have two miR-122 sites and the other allele to have none. For these, the allelic ratio was measured separately in liver for each site, but because each ratio was likely to reflect the effect of two target sites, each was analyzed after reducing by half the log2 value.
Statistical analyses
MATLAB was used for all statistical analyses. To calculate the statistical significance for the observed number of log allelic ratios with values less than zero, the one-sided exact binomial test was used, in which the p value was the probability that a random variable following binomial distribution (parameters: p = 0.5, n = [the total number of sites]) was equal to or larger than the observed number. To calculate the significance of difference between two distributions, the KS test was chosen over the Wilcoxon rank sum test (Mann-Whitney U test) or t-test, because the KS test is based on fewer assumption on the data and almost always provided the most conservative p value compared to the other two tests. When estimating the lower bound for the number of active polymorphic sites, the contribution of experimental noise (illustrated by the bumpiness of the cumulative distributions) in increasing the maximal offset between the cognate and control distributions needed to be subtracted. To estimate the contribution of this noise, we merged the two distributions and generated 1000 pairs of distributions by random sampling, with replacement, maintaining the sizes of the original distributions. Then we calculated the maximum difference in cumulative fraction for each pair of simulated distributions, and subtracted the median of the 1,000 values from the observed maximum offset. The observed average downregulation of all examined polymorphic sites was corrected for our preferential choice of 8mers for analysis by recalculating the mean log ratio of all sites after reducing the contribution from 8mers by 1.87 fold (= 10.3 / 5.5), which was the enrichment of 8mers among the polymorphic sites analyzed. The observed lower bound for the fraction of functional sites was similarly adjusted.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tim Harkins and the 454 Sequencing Facility for high-throughput sequencing, members of the laboratory for helpful comments on this manuscript, and T. DiCesare for illustration. Supported by a grant from the NIH. D.B. is an investigator of the Howard Hughes Medical Institute.
Footnotes
COMPETING INTERESTS The authors declare no competing interests.
References
- 1.Clop A, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–8. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 2.Sethupathy P, Collins FS. MicroRNA target site polymorphisms and human disease. Trends Genet. 2008;24:489–97. doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 8.Lagos-Quintana M, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–9. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 9.Sempere LF, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38:1452–6. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 11.Bao L, et al. PolymiRTS Database: linking polymorphisms in microRNA target sites with complex traits. Nucleic Acids Res. 2007;35:D51–4. doi: 10.1093/nar/gkl797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A. 2007;104:3300–5. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frazer KA, et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–3. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- 14.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 17.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farh KK, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–21. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 19.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution. Cell. 2005;123:1133–46. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen CB, et al. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 23.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2008 doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plagnol V, et al. Extreme clonality in lymphoblastoid cell lines with implications for allele specific expression analyses. PLoS ONE. 2008;3:e2966. doi: 10.1371/journal.pone.0002966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–17. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 29.Karolchik D, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–9. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.