PDZ (PSD-95/DLG/ZO-1) domains constitute one of the most abundant protein interaction modules in eukaryotic cells. These domains principally bind C-terminal motifs of target proteins through formation of conserved interactions involving the ligand residues at positions 0 (hydrophobic side chain and terminal carboxylate group) and −2 (side chain). The latter interaction is commonly used to distinguish the domains into classes (S/T-X-Ψ for class I, Φ-X-Ψ for class II and E/D-X-Ψ for class III, withΨ and Φ representing aliphatic and hydrophobic amino acids respectively).1 Ligand specificity is further defined by contributions from additional residues. PDZ domains are generally found in association with other interaction domains in proteins located at cell junctions. They play a major role in the assembly and localization of macromolecular complexes involved in signal transduction pathways.2–4 With affinities in the low micromolar range and a propensity for promiscuous binding,2,5 transient PDZ domain-mediated interactions largely contribute to the plasticity of these complexes. While PDZ domains have been studied extensively, understanding the various modes of regulation and functional implications of these domains remains a major challenge that would greatly benefit from new tools for addressing the dynamics of these interactions.
Herein we report the development of a general strategy for the design, synthesis and evaluation of fluorogenic probes for PDZ domains. These probes are based in part on natural ligand sequences and incorporate an environment-sensitive fluorophore to report specific binding events. With appropriate placement of the solvatochromic group within the ligand sequence, changes in the local environment due to interactions with cognate domains result in significant modifications of the fluorescence properties (Figure 1). Chromophores of the dimethylaminophthalimide family (4-DMAP,6 6-DMN7 and 4-DMN8) exemplify these properties and have previously been applied to the development of sensors for small protein domains such as SH2 (phosphotyrosine-binding) domains. In this case, placement of the fluorophore adjacent to the conserved binding determinants has afforded sensors with modest signal enhancement (~10-fold). Alternatively, solvatochromic fluorophore-based probes have been developed for 14-3-3,6,9 calmodulin8 and class II MHC proteins,10 by replacement of a conserved hydrophobic aromatic ligand residue, which is known to interact with a defined site on the cognate binding protein. However, not all protein interaction domains allow for such a replacement approach to be utilized, as the domains may either not rely upon defined hydrophobic interactions or, alternatively, the native hydrophobic residues may constitute critical specificity determinants that cannot be modified. PDZ domains typically fall into both of these categories and therefore necessitate the development of a different, more general strategy to generate optimized fluorescent probes.
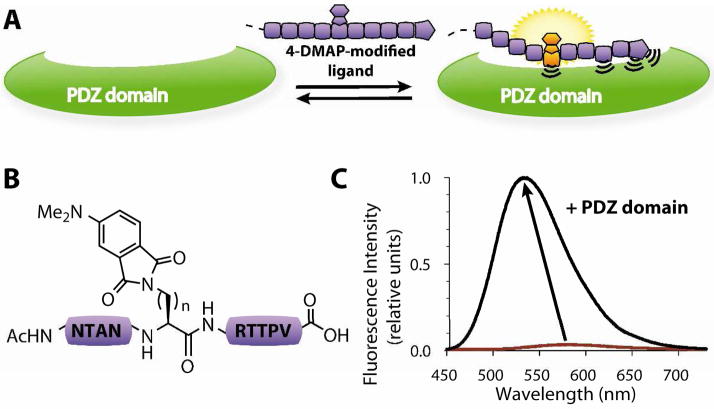
Figure 1.
Sensing PDZ domain-mediated interactions. (A) Optimized fluorescent ligands report binding to partner PDZ domains by dramatic changes in emission intensity and maximum λem. (B) Representative structures of probes 5–7 with the 4-DMAP fluorophore (n=1, Dap, 5; n=2, Dab, 6; n=3, Orn, 7). (C) Fluorescence emission spectra of probe 6 in the unbound (red) and PSD95-3-bound (black) states.
We are interested in probing PDZ domain-mediated interactions of postsynaptic density scaffold proteins in neurons. More specifically, the target proteins in this study are PSD-95 and Shank3, representative members of the MAGUK/SAP11 and Shank/ProSAP12 families respectively. Both proteins contain one or several class I PDZ domains, which were cloned as recombinant GST-fusion proteins (PSD95-12/3 and Shank3, Figure S3). In contrast to previous studies, where a defined hydrophobic pocket in the cognate domains immediately suggested an ideal position for insertion of the environment-sensitive fluorophores,6,8–10 PDZ domains accommodate a wide variety of ligands conforming to specific consensus sequences defined by residues at positions 0 and −2 and do not provide conserved residues to substitute without affecting critical specificity determinants. Furthermore, apart from the hydrophobic pocket, which accommodates the C-terminal aliphatic ligand residue, PDZ domains lack any obvious non-polar site flanking the binding groove. Hence a systematic approach was utilized to identify probes that efficiently report binding. Our design strategy included: (1) development of a peptide library to screen for the optimal fluorophore placement; (2) screening of probes with the fixed fluorophore position for improved affinity and specificity.
In the first phase, we screened a library of peptides derived from the C-terminal sequence of Stargazin and incorporating the 4-DMAP fluorophore (Table 1). Stargazin, an auxiliary sub-unit of AMPA receptors, was chosen for its capacity to interact with the three domains of PSD-95.13 Furthermore, given the general promiscuity of PDZ domains, it was anticipated that a library incorporating the consensus motif T-X-V would also enable screening for other class I PDZ domains such as Shank3. Although structural studies indicate that key interactions with PDZ domains primarily involve the last four ligand residues, some data suggest that additional residues may also be important.2 Thus we chose to use the terminal ten residues of Stargazin in order to best preserve native specificity. Optimal fluorophore positioning was assessed in a step-wise process, where native residues were first replaced with 4-DMAP-derived amino acids (except for positions 0 and −2, which are essential for canonical binding) and evaluated for fluorescence increases. In the second iteration, the fluorescence increase was further optimized by adjusting length of the diamino acid linker bearing 4-DMAP in the most sensitive positions (Dap, n=1; Dab, n=2 or Orn, n=3). Of note, the 4-DMAP fluorophore was initially chosen for its minimal size and the ease of synthesis of a peptide library by on-resin derivatization; hence, at this stage other environment-sensitive fluorophores, 6-DMN and 4-DMN, were also tested at the optimal position with similar and shorter linkers to compensate for ring extension, albeit with lower relative fluorescence increases (data not shown).
Table 1.
Fluorescence increases of the C-terminal Stargazin-derived peptide library with PSD-95 and Shank3 PDZ domains.
| Entry | Peptide Sequencea | Relative Fluorescence Increase | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −9 | −5 | 0 | PSD95-3 | PSD95-12 | Shank3 | ||||||||
| 1 | AC-N | T | A | N | R | R | T | T | α | V-COOH | 1.9 | 2.5 | 1.0 |
| 2 | AC-N | T | A | N | R | R | α | T | P | V-COOH | 6.6 | 3.4 | 1.1 |
| 3 | AC-N | T | A | N | R | α | T | T | P | V-COOH | 1.7 | 1.5 | 1.0 |
| 4 | AC-N | T | A | N | R | β | T | T | P | V-COOH | 8.0 | 1.5 | 1.5 |
| 5 | AC-N | T | A | N | α | R | T | T | P | V-COOH | 36.1 | 1.8 | 2.0 |
| 6 | AC-N | T | A | N | β | R | T | T | P | V-COOH | 89.5 | 3.9 | 26.7 |
| 7 | AC-N | T | A | N | γ | R | T | T | P | V-COOH | 37.6 | 2.6 | 3.0 |
| 8 | AC-N | T | A | β | R | R | T | T | P | V-COOH | 19.8 | 1.9 | 1.2 |
| 9 | AC-N | T | β | N | R | R | T | T | P | V-COOH | 28.7 | 1.7 | 1.3 |
| 10 | AC-N | β | A | N | R | R | T | T | P | V-COOH | 25.6 | 1.9 | 1.3 |
| 11 | AC-β | T | A | N | R | R | T | T | P | V-COOH | 17.4 | 2.0 | 1.3 |
α = Dap(4-DMAP), β = Dab(4-DMAP), γ= Orn(4-DMAP).
The peptide library was screened for ability to report binding, which was evaluated by comparison of 4-DMAP fluorescence emission intensities in the absence and presence of the target PDZ domains (PSD95-12/3 and Shank3, Table 1). The maximum relative fluorescence increase for all PDZ constructs was obtained for peptide 6; thus, the optimal position and linker length are −5 and Dab (n=2), respectively. This probe was particularly sensitive to PSD95-3; more than an eighty-fold increase in emission intensity was observed. Interestingly, similar overall results to PSD-95 were obtained with other MAGUK PDZ domains, which share a high sequence homology with PSD95-12/3 (SAP102-12/3 and SAP97-12/3, Figure S3 and S4) and can accommodate the same ligands.5,11 This suggests that MAGUK PDZ domains share common structural features that can be sensed by the probes. Control experiments were employed to further validate the screening approach. As expected, no significant fluorescence increase was observed for the Stargazin peptides with class II PDZ domains from PICK1 and GRIP1 (Figure S4). This confirmed that the probe binding is driven by specific PDZ domain-cognate ligand interactions. Next, the relative fluorescence increase was measured for full-length PSD-95 and compared to its constituent PDZ domains, PSD95-12 and -3 (Figure S6) using peptide 6. The similarity of the fluorescence output for the native protein and the domain constructs indicated the relevance of the results obtained with isolated domains and validated the screening approach.
In the second step of the design process, our goal was to generate probes with improved signaling properties by addressing affinity and specificity of the ligands for each domain. Therefore, C-terminal sequences of CRIPT,5,14 NR2a5 and GluR1,15 known binders for PSD95-3, PSD95-12 and Shank, respectively, were selected as ligands for the second series of probes. The series also included negative controls such as a class II ligand, derived from mGluR7a,16 and a Stargazin sequence in which critical residues were replaced by Arg (at 0 and −2). Applying the results of the library screen, the 4-DMAP fluorophore was appended to a Dab residue at the −5 positon, and probes were tested with each construct. The affinities of the new sensors were obtained by fluorescence-based titrations and are reported in Table 2, together with relative fluorescence increases. The studies reveal that 12 and 14 are the best probes for PSD95-3, 13 for PSD95-12, and 15 for Shank3. Remarkably, probe 15 exhibited a 265-fold fluorescence increase with the Shank3 PDZ domain. Control sequences, 16 and 17, exhibited negligible fluorescence increases (and poor affinities when measured) for the targeted domains, thus confirming class specificity of the probes. Together, these results indicate that selectivity of the native sequences was preserved after insertion of the fluorophore. Furthermore, for each domain, KD values of ligands identically derived with 4-DMAP (both position and linker) correlated with their relative fluorescence increases when screened under the same conditions. Next, competition experiments with non-fluorescent peptides were carried out on probes 12 and 15 with their respective target PDZ domains. As shown in Figure 2, gradual displacement of probe 12 from PSD95-3 by a non-fluorescent peptide derived from Stargazin (18) can be readily observed by decrease of the fluorescence signal. Affinity constants of the unlabeled peptides could be calculated from these competitive titrations, showing a stronger overall affinity of the labeled peptides (Figure 2 for 18 with PSD95-3 and 5.80 μM for 19 with Shank3). While further demonstrating the reversibility and specificity of the fluorescent probe-PDZ domain interactions, these experiments also illustrate the potential of these tools for the screening of high affinity ligands for target PDZ domains.
Table 2.
Binding constants and fluorescence increases for peptides with the optimally positioned 4-DMAP fluorophore.
| Entry | Sequence origin | Sequencea | PSD95-3 | PSD95-12 | Shank3 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KD (μM) | RFIb | KDapp (μM)c | RFIb | KD (μM) | RFIb | |||||||||||||||
| 12 | Stargazin | AC-N | R | R | N | T | A | N | β | R | T | T | P | V-COOH | 0.57 ± 0.23 | 62.6 | 5.99 ± 0.76 | 6.9 | 10.00 ± 2.44 | 27.6 |
| 13 | NR2a | AC-N | R | R | K | K | λ | P | β | I | E | S | D | V-COOH | 1.66 ± 0.18 | 35.1 | 1.50 ± 0.26 | 8.6 | 23.10 ± 0.68 | 4.9 |
| 14 | CRIPT | AC-D | T | K | N | β | K | Q | T | S | V-COOH | 1.21 ± 0.20 | 48.8 | 9.63 ± 1.34 | 4.5 | 12.40 ± 4.30 | 21.0 | |||
| 15 | GluR1 | AC-N | R | R | S | G | λ | P | β | G | A | T | G | V-COOH | 89.5 ± 20.6 | 4.6 | 20.45 ± 3.17 | 2.0 | 0.17 ± 0.02 | 265.0 |
| 16 | Control | AC-N | R | R | N | T | A | N | β | R | T | R | P | R-COOH | ≫50 | 1.4 | n.d.d | 1.7 | n.d.d | 1.5 |
| 17 | mGluR7a | AC-A | K | K | K | Y | V | S | β | N | N | L | V | I-COOH | n.d.d | 2.7 | n.d.d | 1.7 | n.d.d | 1.9 |
β = Dab(4-DMAP), λ= Norleucine, NRR- added to improve solubility.
Relative fluorescence increase.
Apparent KD.
not determined.
Figure 2.
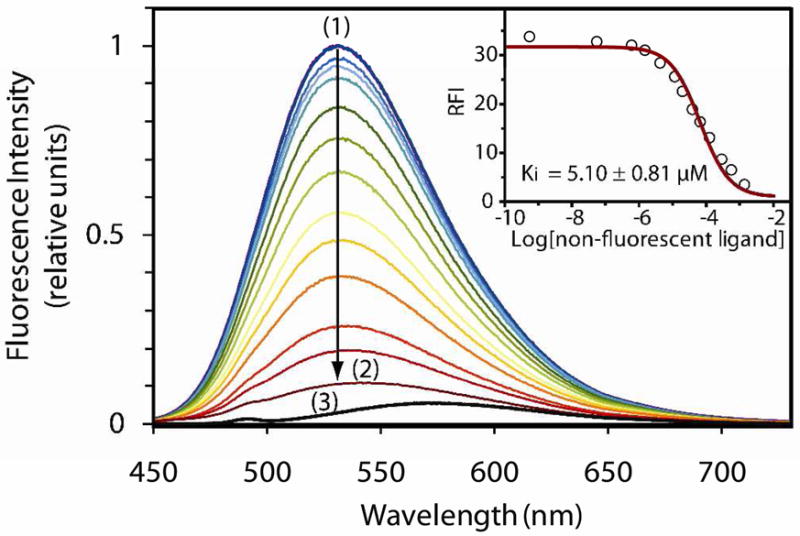
Competitive titration of unlabeled Stargazin peptide vs probe 12/PSD95-3 complex. Main graph: Fluorescence emission spectra of a 5 μM solution of peptide 12 and PSD95-3 after sequential additions of 0 (1) to 200 equivalents (2) of the non-fluorescent peptide; (3) 5 μM solution of peptide 12. Insert: Relative fluorescence increases at corresponding concentrations of unlabeled competing peptide (data, fit and Ki).
Finally, the binding affinities were also independently evaluated by using a non-fluorescence-based method, isothermal titration calorimetry (ITC). In this case, the aims were to validate the fluorescence-based results and also to determine the potential influence of the fluorophore on the probe affinity. The binding constant of peptide 12 for PSD95-3, determined by ITC, was similar to that obtained by fluorescence titration, while the KDs of non-fluorescent peptides derived from CRIPT and Stargazin were in agreement with reported data (Table S8). Overall, these results confirmed our method and revealed an improved affinity of peptides 12 and 14 in comparison to the native peptide sequences.
The affinity improvement was attributed to interactions of the fluorophore with a non-polar patch of the PDZ domains that, when combined with the other ligand interactions, translates into high fluorescence increases and stronger affinities. However, attempts at correlating these results with structural information did not allow identification of obvious shared elements for the three sets of domains that would account for the observed response from the 4-DMAP chromophore at the −5 position. By considering the X-ray crystallographic structure of PSD95-3 complexed with a ligand (PDB entry 1TP5) as a starting point and modeling the last 6 residues of probe 12, we could define the regions accessible by 4-DMAP on the PDZ domain surface (Figure 3). Surprisingly, the fluorophore is modeled to be located at the edge of the PDZ domain binding groove and does not seem to be in the vicinity of any clearly defined hydrophobic patch (similar observations were made for probe 15 with Shank3, Figure S9). In this context, it is worth noting that another PSD95-3 structure complexed with a 9-residue CRIPT peptide17 (PDB entry 1BE9) failed to show sufficient electron density to resolve the location of the ligand −5 Tyr residue, thus stressing the lack of a predefined strong interaction of the PDZ domain with an aromatic ligand residue at this position. Current structural analysis is underway to elucidate the origin of the remarkable fluorescence increases observed with the 4-DMAP-based peptide probes. Overall, these structural considerations underscore the importance and efficiency of the general screening approach in the design of environment-sensitive probes for interaction domains such as PDZ domains that do not allow for a rational ligand residue replacement strategy.
Figure 3.
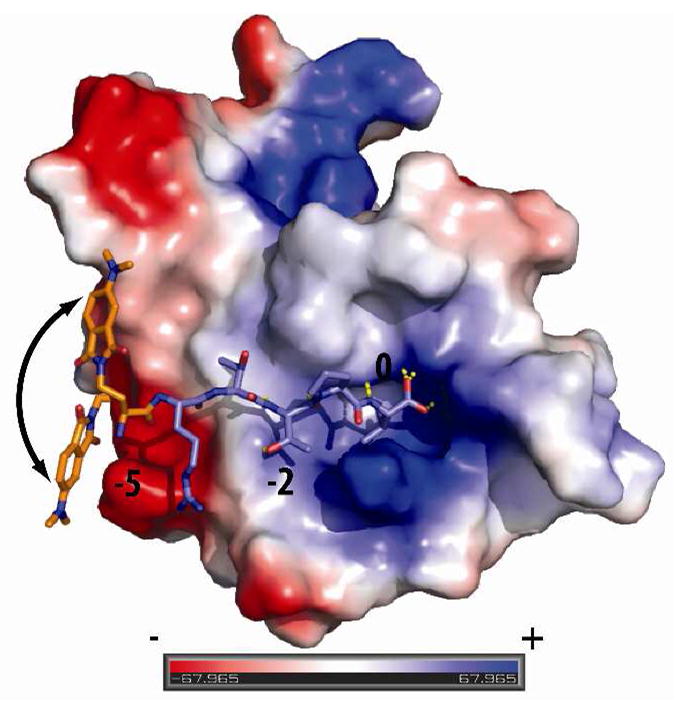
Defining the potential sites of interaction between the 4-DMAP chromophore and PSD95-3 for probe 12. The six C-terminal amino acids (βRTTPV, stick representation) of peptide 12 were docked into PSD95-3 structure (electrostatic potential surface representation, PDB accession code 1TP5). Rotation of the chromophore and the linker around the fixed ligand backbone allows for identification of proximal PDZ domain residues. Structure rendered using PyMOL v1.1 (DeLano Scientific LLC).
In conclusion, we have developed and successfully applied a systematic library approach to the design of fluorogenic peptide-based probes for PDZ domains. The method can be applied to other classes of domains and can be readily implemented for the screening of sensors as well as high affinity competing ligands. The new probes exhibit large fluorescence increases while conserving native sequence specificity. Thus, they constitute valuable tools for in vitro studies of PDZ domain-mediated interactions by enabling a direct readout of binding. In on-going work, we are extending this approach to other PDZ domains and evolving our probes to report these interactions in native conditions within the cellular environment.
Supplementary Material
Acknowledgments
This research was supported by HFSP (RGP0007/2006, B.I.), the Cell Migration Consortium (GM064346) and the Marie Curie postdoctoral Fellowship (PICK-CPP, M.S.).
Footnotes
Supporting Information Available: Experimental procedures, peptides characterization and additional figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Science. 1997;275:73–7. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 2.Hung AY, Sheng M. J Biol Chem. 2002;277:5699–5702. doi: 10.1074/jbc.R100065200. [DOI] [PubMed] [Google Scholar]
- 3.Kim E, Sheng M. Nat Rev Neurosci. 2004;5:771–81. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- 4.Nourry C, Grant SG, Borg JP. Sci STKE. 2003;2003:RE7. doi: 10.1126/stke.2003.179.re7. [DOI] [PubMed] [Google Scholar]
- 5.Lim IA, Hall DD, Hell JW. J Biol Chem. 2002;277:21697–21711. doi: 10.1074/jbc.M112339200. [DOI] [PubMed] [Google Scholar]
- 6.Vazquez ME, Rothman DM, Imperiali B. Org Biomol Chem. 2004;2:1965–1966. doi: 10.1039/b408001g. [DOI] [PubMed] [Google Scholar]
- 7.Vazquez ME, Blanco JB, Imperiali B. J Am Chem Soc. 2005;127:1300–1306. doi: 10.1021/ja0449168. [DOI] [PubMed] [Google Scholar]
- 8.Loving G, Imperiali B. J Am Chem Soc. 2008;130:13630–13638. doi: 10.1021/ja804754y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vazquez ME, Nitz M, Stehn J, Yaffe MB, Imperiali B. J Am Chem Soc. 2003;125:10150–10151. doi: 10.1021/ja0351847. [DOI] [PubMed] [Google Scholar]
- 10.Venkatraman P, Nguyen TT, Sainlos M, Bilsel O, Chitta S, Imperiali B, Stern LJ. Nat Chem Biol. 2007;3:222–228. doi: 10.1038/nchembio868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elias GM, Nicoll RA. Trends Cell Biol. 2007;17:343–52. doi: 10.1016/j.tcb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Sheng M, Kim E. J Cell Sci. 2000;113:1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- 13.Dakoji S, Tomita S, Karimzadegan S, Nicoll RA, Bredt DS. Neuropharmacology. 2003;45:849–856. doi: 10.1016/s0028-3908(03)00267-3. [DOI] [PubMed] [Google Scholar]
- 14.Niethammer M, Valtschanoff JG, Kapoor TM, Allison DW, Weinberg RJ, Craig AM, Sheng M. Neuron. 1998;20:693–707. doi: 10.1016/s0896-6273(00)81009-0. [DOI] [PubMed] [Google Scholar]
- 15.Uchino S, Wada H, Honda S, Nakamura Y, Ondo Y, Uchiyama T, Tsutsumi M, Suzuki E, Hirasawa T, Kohsaka S. J Neurochem. 2006;97:1203–1214. doi: 10.1111/j.1471-4159.2006.03831.x. [DOI] [PubMed] [Google Scholar]
- 16.Hirbec H. J Biol Chem. 2002;277:15221–15224. doi: 10.1074/jbc.C200112200. [DOI] [PubMed] [Google Scholar]
- 17.Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Cell. 1996;85:1067–76. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.