Abstract
Ginger (Zingiber officinale Rosc) (Family: Zingiberaceae) is a herbaceous perennial, the rhizomes of which are used as a spice. Ginger is a plant which is well known for its medicinal applications. Recently EST-derived SNPs are a free by-product of the currently expanding EST (Expressed Sequence Tag) databases. The development of high-throughput methods for the detection of SNPs (Single Nucleotide Polymorphism) and small indels (insertion/deletion) has led to a revolution in their use as molecular markers. Available (38139) Ginger EST sequences were mined from dbEST of NCBI. CAP3 program was used to assemble EST sequences into contigs. Candidate SNPs and Indel polymorphisms were detected using the perl script AutoSNP version 1.0 which has used 31905 ESTs for detecting SNPs and Indel sites. We found 64026 SNP sites and 7034 indel polymorphisms with frequency of 0.84 SNPs / 100 bp. Among the three tissues from which the EST libraries had been generated, Rhizomes had high frequency of 1.08 SNPs/indels per 100 bp whereas the leaves had lowest frequency of 0.63 per 100 bp and root is showing relative frequency 0.82/100bp. Transitions and transversion ratio is 0.90. In overall detected SNP, transversion is high when compare to transition. These detected SNPs can be used as markers for genetic studies.
Availability
The results of the present study hosted in our webserver www.spices.res.in/spicesnip
Keywords: Zingiber officinaleRosc, Expressed Sequence Tag, in silico, Ginger, SNPs, Indel
Background
Ginger (Zingiber officinale) is a perennial plant in the family Zingiberaceae ‐ its rhizome is commonly used as a cooking spice throughout the world. The ginger plant has a long history of cultivation known to originate in China and then spread to India, Southeast Asia, West Africa, and the Caribbean. India is a leading producer of ginger in the world. Ginger is cultivated in most of the states in India. Kerala and Meghalaya are major ginger growing states in the country. The rhizomes and stems of ginger have assumed significant roles in Chinese, Japanese, and Indian medicine since the 1500s [1].The oleoresin of ginger is often contained in digestive, antitussive, antiflatulent, laxative, and antacid compounds [2]. Ginger has a large genome of 23618 Mbp distributed in 2n=22 chromosomes. The phytochemistry and pharmacology of this is well studied but the molecular biological process involved in this is not yet studied. Singlepass sequencing of the 5' and/or 3' ends of randomly selected cDNA clones, is an effective approach to provide genetic information of an organism. These sequences can serve as markers or tags for transcripts, and have been used in the development of SNP markers for reference genetic map and recovery of full-length cDNA and genomic sequences. Expressed sequence tags (ESTs) are also useful for the discovery of novel genes, investigation of genes of unknown function, comparative genomic study, and recognition of exon/intron boundaries. Currently, there are 38139 available ginger sequences in the GenBank, and majority of these sequences are ESTs which had been deposited at NCBI (dbEST) http://www.ncbi.nlm.nih.gov/dbEST/. The lack of sequence information has limited the progress of gene discovery and characterization, global transcript profiling, probe design for development of gene arrays, and generation of molecular markers for Ginger. In this study, we have categorized 38139 ESTs in to three tissue libraries leaves 13274, rhizomes 12763 and roots 12092 ESTs. The availability of these EST sequences will allow comparative genomic studies between ginger and other monocotyledonous and dicotyledonous plants, development of molecular markers for the establishment of reference genetic map, design and construction of cDNA microarray for global gene expression profiling. Single nucleotide polymorphisms (SNPs) are a second class of genetic markers that can be mined from sequence data and are useful for characterizing allelic variation, genome-wide mapping, and as a tool for marker-assisted selection. In the field of human genetics, SNPs are a major focus of efforts to increase the efficiency of mapping [3–6] and are already being used for detection and mapping of a variety of diseases [7–9]. In many crop plants, SNPs are present with sufficient frequency to offer an alternative for genetic mapping and markerassisted selection. Although SNPs can be identified by sequencing selected DNA fragments, a practical limitation to this approach for ginger follows from the fact that the sequencing error rate is often higher than the polymorphism rate. The cost of SNP discovery through sequencing amplified fragments is therefore high even with reductions in the cost of sequencing. The objectives of the research described in this paper were to assess the potential of existing public databases for the discovery of single nucleotide polymorphisms. We have mined updated EST tissue libraries of zingiber officinale for this analysis to find the SNP / Indel polymorphisms. SNP detecting perl scripts AutoSNP version 1.0 is used indentify the SNP / Indel polymorphisms, DNA substitution like Transversion vs Transition and Indel [10]. There are some other SNP detecting software such as SEAN [11] PolyPhred [12] PolyBayes [13] TRACE_DIFF [14] and HarvEST (http://harvest.ucr.edu) but AutoSNP provides user friendly approach and interpretable results as html file. Thus there are ten kinds of SNP/indel (two types of transition and four types of transversion and four groups of indels) are possible in the SNP/indel sites in EST libraries. We have used three tissue libraries [15,16] of Zingiber officinale.
Methodology
EST database of NCBI (dbEST release 092509) contains 38139 Zingiber officinale Express sequence tag data. We have mined 38139 EST sequences consist of three tissue libraries of leaves 13274 (DV544275-ES560515), rhizomes 12763 (DY350707-DY363469) and roots 12092 (DY363470-DY375561). CAP3 program is used to assemble the EST sequence in to contigs. The SNP detection tool AutoSNP version.1.0 was used to find the candidate SNPs from these libraries. AutoSNP required input as ace or fasta format. But the perl script edited manually to analyse fasta or ace format. Sequence assembly program CAP3 is integrated in AUTOSNP to make fasta files in to contigs (http://bioweb.pasteur.fr/seqanal/interfaces/cap3.html) [17]. The DNA substitution like transition (Ts) versus transversion (Tv) ratio of all the libraries in Ginger genome was also calculated.
Discussion
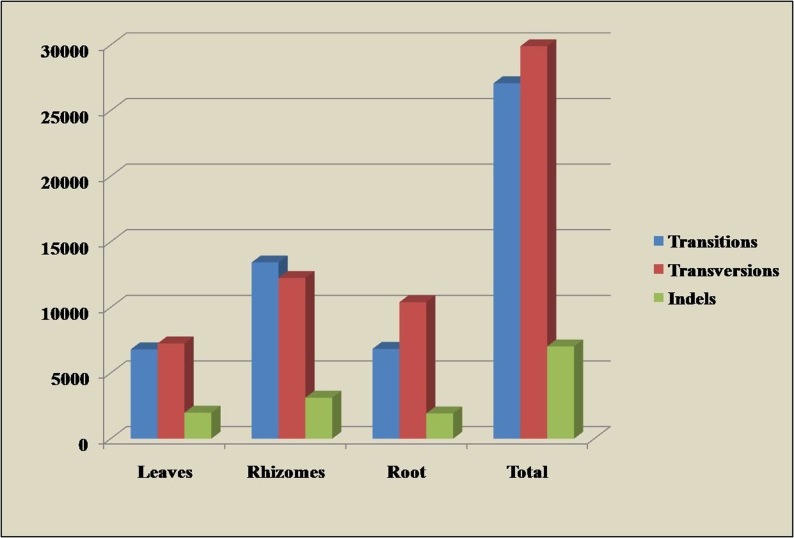
In this study it is discovered that total of 64026 SNP sites and 7034 indel polymorphisms in 38139 ESTs analyzed with an average frequency of 0.84 SNPs / 100 bp. Results of the tissue wise SNP and indel discovery are listed in Table 1 (see supplementary material) and Figure 1. In Ginger leaves tissue libraries showing high indels 1983 while comparing other tissues. Rhizome tissues showing the high SNP frequency 1SNP in 100bp. In Ginger a total of 27083 transitions, 29909 transversions and 7034 indels were found while analysis. But we found in tissue wise manner, rhizome transitions are high in number 13433. Rhizome tissues having more SNPs than others. Rhizome part is more expressed than other tissues. While discovering all SNP with DNA substitution overall transitions and transversions ratio is 0.90. When compared to ginger with others, the studies on the occurrence and nature of SNPs are beginning to receive considerable attention, particularly Arabidopsis where over 37,000 SNPs have been identified through the comparison of two accessions [18]. It has been reported in maize that there occurs a frequency of one non-coding SNP per 31 bp and 1 coding SNP per 124bp in 18 maize genes assayed in 36 inbred lines [19]. Moreover the recent evidence has indicated that SNPs appear to be even more abundant in plant systems than in the human genome. Germano and Klein [20] identified five SNPs in 1 kb of cDNA of Picea rubens and Picea mariana, and also discovered SNPs in the chloroplasts of these species. Recently, in soyabean (Glycine max), two SNPs found approximately 400 bp [21]. In maize (Zea mays), SNP has been detected even more frequently, with one SNP approximately every 48 bp and every 130 bp in 3' untranslated regions and coding regions, respectively [22,23]. The SNP analysis on Apple (Malus domestica) ESTs the Bi-allelic SNPs were on an average of every 706 bp [24] and the study in Maize ESTs [25] also showed the relative increase of over transversion and transition sites. This in silico analysis will help ginger researchers about the single nucleotide polymorphism related study and nucleotide substitution in this important crop.
Figure 1.
DNA substitution and indel polymorphism of SNPs in Ginger EST libraries.
Large-scale sequencing of Expressed Sequence Tags and complete genomes offers information of use to plant breeding programs. With the completion of the first crop genome sequencing projects [26,27] the potential for plant breeding to be impacted by new technology has never been greater. In ginger, sequencing projects offer a potential solution to the scarcity of markers that can be used in elite breeding populations. Of special interest is the ability to discover DNA polymorphisms by mining sequence data [28,29] . The frequency of single nucleotide polymorphisms that we detected is considerably lower than reported for maize, wheat, barley, and soybean. Not surprisingly it is also lower than the one SNP per approximately 100 bases that was detected in some of tissue libraries [30]. There was a relative increase in the proportion of transition (6805) over transversion (7258) in Ginger ESTs except in leaves libraries (Figure 1). C / T transition was found to be high in ginger (Table 1 in supplementary material). High frequency of the C to T mutation is usually seen due to methylation. We also used the Shannon information index to analyze the proportion of ten possible types of SNP/indels. ESTs from tissues of root showed highest values of indices (0.164) whereas leaves had the least value (0.150) and rhizome showed relatively increased value (0.152). Our study on higher number and Shannon index of SNP/indel sites in root tissue than other tissues also gives the additional information about in genomic variation in genes expressed specifically in root tissue. Ratio of transition to transversion (Ts/Tv) was very useful to compare the genotypes of hepatitis virus C and also differences among the mitochondrial genomes of animals. Our study gives a method, which compares the ten possible types of SNP/ indels in a single index. The results of detected SNPs were accessed through online at www.spices.res.in/spicesnip/
Conclusion
In total, we have identified over 64026 candidate SNP polymorphisms with frequency of 0.84 SNPs/100bp in Ginger EST sequence data, along with two measures of confidence for each predicted polymorphism. Segregation of these SNPs with haplotype along with validation demonstrates that candidate SNPs with high redundancy and co-segregation confidence scores are likely to represent true SNPs. The transition to transversion ratio and indel size frequencies correspond to those observed by the analysis methods of SNP discovery and suggest that the majority of predicted SNPs and indel identified using this approach represent true genetic variation in ginger. Overall transversion is high because ginger is vegetative propagated through rhizome. This in silico analysis on ginger shows the potential SNP markers for use in ginger breeding and the online information we created would help to designing new primers and develop more markers and to saturate the linkage maps.
Supplementary material
Acknowledgments
We are grateful to Department of Biotechnology Government of India for the financial support.
Footnotes
Citation:Chandrasekar et al, Bioinformation 4(3): 119-122 (2009)
References
- 1.Purseglove JW, et al. Spice. 1981;2 [Google Scholar]
- 2.Govindarajan VS, et al. Crit Rev Food Sci Nutr. 1982;17:1. doi: 10.1080/10408398209527343. [DOI] [PubMed] [Google Scholar]
- 3.International SNP Map Working Group Nature. 2001;409:928. [Google Scholar]
- 4.Aerts J, et al. Human Mutation. 2002;20:162. doi: 10.1002/humu.10107. [DOI] [PubMed] [Google Scholar]
- 5.Balasubramanian B, et al. Pharmacogenomics. 2002;3:393. doi: 10.1517/14622416.3.3.393. [DOI] [PubMed] [Google Scholar]
- 6.Chen LYY, et al. Genome Research. 2002;12:1106. doi: 10.1101/gr.224502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verhage BAJ, et al. Int J Cancer. 2002;100:683. doi: 10.1002/ijc.10541. [DOI] [PubMed] [Google Scholar]
- 8.Sugimoto Y, et al. Journal of Viral Hepatitis. 2002;9:377. doi: 10.1046/j.1365-2893.2002.00365.x. [DOI] [PubMed] [Google Scholar]
- 9.Margiotti K, et al. The Prostate. 2002;53:65. doi: 10.1002/pros.10134. [DOI] [PubMed] [Google Scholar]
- 10.Barker G, et al. Bioinformatics. 2003;19:421. doi: 10.1093/bioinformatics/btf881. [DOI] [PubMed] [Google Scholar]
- 11.Huntley D, et al. Bioinformatics. 2006;22:495. doi: 10.1093/bioinformatics/btk006. [DOI] [PubMed] [Google Scholar]
- 12.Nickerson DA, et al. Nucleic Acids Res. 1997;25:2745. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marth GT, et al. Nat Genet. 1999;23:452. doi: 10.1038/70570. [DOI] [PubMed] [Google Scholar]
- 14.Bonfield JK, et al. Nucleic Acids Res. 1998;26:3404. doi: 10.1093/nar/26.14.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jouannic S, et al. FEBS. 2005;579:2709. doi: 10.1016/j.febslet.2005.03.093. [DOI] [PubMed] [Google Scholar]
- 16.Ho CL, et al. BMC Genomics. 2007;8:381. doi: 10.1186/1471-2164-8-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang X, et al. Genom Res. 1999;9:868. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jander G, et al. Plant Physiol. 2002;129:440. doi: 10.1104/pp.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ching A, et al. BMC Genet. 2002;3:19. doi: 10.1186/1471-2156-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Germano J, et al. Theor Appl Genet. 1999;99:37. [Google Scholar]
- 21.Coryell VH, et al. Theor Appl Genet. 1999;101:1291. [Google Scholar]
- 22.Tenaillon MI, et al. Proc Natl Acad Sci. 2001;98:9161. doi: 10.1073/pnas.151244298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rafalski A, et al. Curr Opin Plant Biol. 2002;5:94. doi: 10.1016/s1369-5266(02)00240-6. [DOI] [PubMed] [Google Scholar]
- 24.Newcomb RD, et al. Plant Physiology. 2006;141:147. doi: 10.1104/pp.105.076208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batley J, et al. Plant Physiology. 2003;132:84. doi: 10.1104/pp.102.019422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goff SA, et al. Science. 2002;296:92. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 27.Yu J, et al. Science. 2002;296:79. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 28.Smulders MJM, et al. Theor Appl Genet. 1997;94:264. [Google Scholar]
- 29.Bredemeijer GMM, et al. Theor Appl Genet. 2002;105:1019. doi: 10.1007/s00122-002-1038-6. [DOI] [PubMed] [Google Scholar]
- 30.Suliman-Pollatschek S, et al. Cellular and Molecular Biology Letters. 2002;7:583. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.