Abstract
Ribonucleotide Reductase (RNR) is an enzyme responsible for the reduction of ribonucleotides to their corresponding Deoxyribonucleotides (DNA), which is a building block for DNA replication and repair mechanisms. The key role of RNR in DNA synthesis and control in cell growth has made this an important target for anticancer therapy. Increased RNR activity has been associated with malignant transformation and tumor cell growth. In recent years, several RNR inhibitors, including Triapine, Gemcitabine and GTI-2040, have entered the clinical trials. Our current work focuses on an attempted to dock this inhibitors Flavin and Phenosafranine to curtail the action of human RNR2. The docked inhibitor Flavin and Phenosafranine binds at the active site with THR176, which are essential for free radical formation. The inhibitor must be a radical scavenger to destroy the tyrosyl radical or iron metal scavenger. The iron or radical site of R2 protein can react with one-electron reductants, whereby the tyrosyl radical is converted to a normal tyrosine residue. However, compounds such as Flavin and Phenosafranine were used in most of the cases to reduce the radical activity. The docking study was performed for the crystal structure of human RNR with the radical scavengers Flavin and Phenosafranine to inhibit the human RNR2. This helps to understand the functional aspects and also aids in the development of novel inhibitors for the human RNR2.
Keywords: Ribonucleotide reductase, Flavin, Phenosafranine, radical scavenger, inhibitors
Background
Ribonucleotide Reductase (RNR) is a ubiquitous cytosolic enzyme in the cell, responsible for converting ribonucleotides into deoxyribonucleotides, the eventual substrates for DNA polymerase [1–3], and also repair DNA in all living cells [4]. In mammalian cells, this enzyme contains two dissimilar protein components, R1 and R2, which are encoded by two different genes located on different chromosomes [5]. Protein R1 is a homodimeric structure, with a molecular mass of 168kDa, and has substrate and allosteric effector sites that control enzyme activity and substrate specificity [3]. Protein R2 is a homodimer, with a molecular mass of 88kDa, And forms two equivalent dinuclear iron centers that stabilize a tyrosyl free radical, required for the initiation of electron transformation during catalysis. R1 and R2 proteins interact at their C-terminal ends to form an active holoenzyme [3].
Amidox (3, 4- dihydroxybenzamidoxime), a new polyhydroxysubstituted benzoic acid derivative, is a potent inhibitor of the enzyme ribonucleotide reductase (RR), which catalyses the de novo synthesis of DNA. RR is considered to be an excellent target for cancer chemotherapy. In the present study we investigated the antineoplastic effects of Amidox alone and in combination with Arabinofuranosylcytosine (Ara-c) in HL-60 human promyelocytic leukemia cells [6]. Ribonucleotide reductase (RR) is the rate- limiting enzyme of de novo DNA synthesis and has been shown to be up regulated linked with proliferation and malignant transformation. It was therefore identified as an excellent target for ant tumor therapy [7]. Daily oral or intravenous administration of the ribonucleotiode diphosphate reductase inhibitor, (E)-2‘-(fluromethylene) cytidine (MDL 101, 731), to nude mice caused rapid regression of colon and prostrate xenografts. Studies were performed to optimize dosing schedule and route of administration [8].
The cell division makes it a potential target for designing drug to inhibit cell growth, applications in cancer therapy, and the production of anti- malaria and trypanosome drugs, antibiotics and anti-viral agents against those viruses that have their own RNRs. An increased interest in RNR as a target for cancer therapy is seen ever since the human ribonucleotide reductase of a new type was identified which is regulated by p53. The p53 actively suppresses tumor formation but on mutation several forms of cancer are developed. As much as over 80% of the human tumors have been found to contain mutations in p53 or in the pathway that directly regulates it. Mammalian RNR-R2 is located in the cytoplasm and regulated by the cell cycle. The new R2 gene product is called p53R2 and found to be located in the nucleus. The p53 binds to a sequence in the first intron of p53R2 gene and is required for directly activating its transcription. It was reported recently that p53 enzyme binds both R2 and p53R2 subunits in testing cells but upon exposure to UV radiation, they dissociate from p53 and bind to R1. Perhaps the regulation of RNR activity by p53 is more complex than activation of P53R2 [4]
Methodology
Receptor and ligand data
Radical scavengers from R2 protein are very essential for inhibiting the RNR activity and DNA replication. The crystal structure of human RNR2 of a single subunit (PDB ID: 2IYH) was used for the current studies. Based on brief Literature survey [4,9], the potential radical scavengers Flavin and Phenosafranine molecules are constructed using Insight [10] and subsequently these small molecules were subjected to energy minimization to bring the energy level of the system to global minima, structurally stable and free from steric clashes.
Docking
The molecules were further subjected to Auto dock 3.0.5 to perform the docking studies which use Lamarckian Genetic Algorithm (LGA) for about 50 iterations by placing the grid arbitrarily at the active site of the crystal structure [11,12]. To reduce the complexities in the binding studies the hetero groups were removed from the crystal structure earlier. The ligand bound complexes were further analyzed for the binding affinity and HBond interaction studies using the online server HBPLUS.
Interaction identification
The free energy co efficient for the vander waals for the docked receptor and the ligand is 0.1560 and the electrostatic term for the docked receptor and the ligand is 0.1465. It is said earliar that Ribonucleotide reductase (RNR) activity is necessary for DNA replication and inhibition of this enzyme will inhibits cell division. The inhibitor must be ready to destroy the Tyrosyl radical. And thus the inhibitors such as Flavin and Phenosafranine bind at the active site Tyrosine 176 to reduce the radical activity and inhibit cell division.
Discussion
Structural features of human RNR2 subunit
The human RNR2 structure belongs to all-helical class of protein. R2 subunit contains a dinuclear ion center and a tyrosyl radical site (Tyr- 176 in human R2), which are essential for initiation of the nucleotide reduction process-taking place at the active site in R1. The di-ion center of R2 can exist in different oxidation states. If molecular oxygen is allowed to react with the diferrous R2 (Fe (II) Fe (II)), it will spontaneously oxidize the di-ion center through a series of intermediate states, leading to a μ-oxo- bridged diferric ion cluster (Fe (III) Fe (III)) and a stable tyrosyl radical [13]. The dinuclear ion center is located within a four-helix bundle and is coordinated by Glutamine 169,266,232 and Histidine 269. The R2 subunit of RNR generates, as mentioned above, a tyrosyl radical essential for the reduction of ribonucleotides to deoxyribonucleotides in eukaryotes. It is the first and second coordination sphere of the di-ion center in each protein that determines the outcome of the reaction with oxygen. Therefore, it is of great interest. The crystal structure of human RNR2 (PDB ID: 2IYH) is shown in Figure 1.
Figure 1.
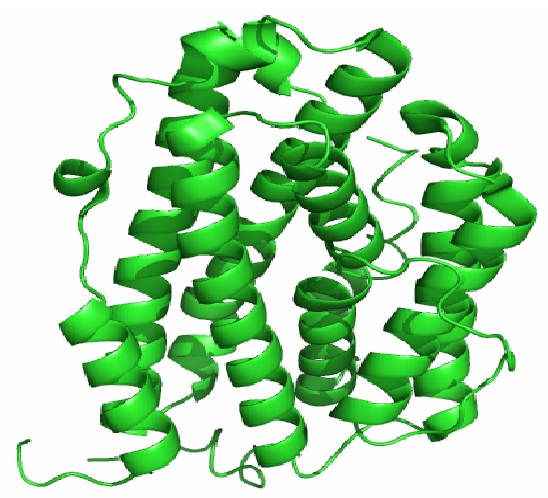
The Cartoon representation of crystal structure of human RNR R2 subunit which displays the α-helix and the loop region in distinct manner. The structure is rendered using PyMol.
Docking studies
In this paper we have attempted to dock the inhibitor Flavin and Riboflavin to curtail the action of human RNR2. Auto DOCK program version available at the web site http://www.scripps.edu/pub/olsonweb/doc/autodock/, was used for this study. This program utilizes Lamarckian Genetic Algorithm for configurationally search and evaluates the energy using grid-based molecular affinity potential. Fifty best configurations of the protein-inhibitors complexes were retrieved on binding energy. The hydrogen bond and non-bonded contacts for the complexes were calculated using the program HBPLUS.
Docking of Flavin and Phenosafranine
The free energy of binding (calculated by adding the final intermolecular energy and torsional free energy), docked energy (calculated by adding the final intermolecular energy of ligand) and the inhibition constant (Ki) for the complex configurations can be used to find the best configuration. The docked inhibitor Flavin binds the active site region of the human RNR2. The Flavin interacts with the active site tyrosine 176, which is essential for free radical formation. The binding of Flavin and phenosafranine with human RNR2 are shown in secondary structure representation in Figure 2 and Figure 3. The diagram represented by displaying the THR176 in stick representation and flavin and phenosafranine in wireframe representation. The fifty best configuration of RNR1 and Phenosafranine complex is chosen on the basis of free energy of binding, docked energy and inhibition constant. The Phenosafranine binds the active site of RNR2. The Phenosafranine also interacts with the active site tyrosine 176.
Figure 2.
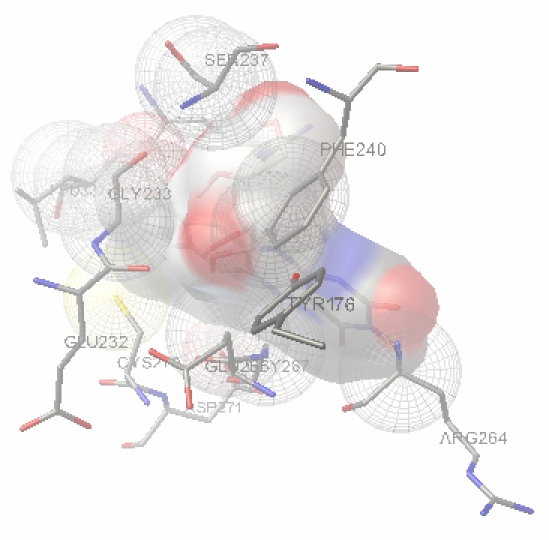
The interaction of Flavin with human RNR was performed using Auto Dock 3.0.5 which displays the residue THR176 interacts with the inhibitor Flavin. THR176 of Human RNR and the inhibitor molecule Flavin were found intact at the end of the docking.
Figure 3.
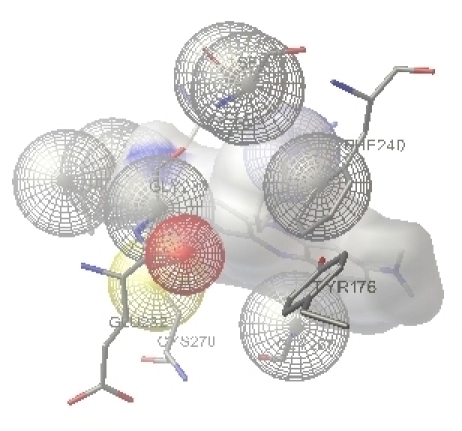
The interaction of phenosafranine with human RNR was performed using Auto Dock 3.0.5 which displays the residue THR176 interacts with the inhibitor phenosafranine. THR176 of Human RNR and the inhibitor molecule phenosafranine were found intact at the end of the docking.
Flavin action
Upon anaerobic incubation of R2 protein with reduced flavin and the reduction of the tyrosyl radical result in the formation of metR2. This reaction occurs in the absence of ferrous ion. In their presence, the Fe (III) center is reduced to Fe (II), and reduced R2 becomes the final product. Admission of oxygen then results in the reformation of active R2. The enzyme provides the reduced flavin, which under anaerobic conditions transforms R2 into metR2 by scavenging the tyrosyl radical [14]. In a second step, together with Fe (II) ions, the flavin reductase catalyzes the formation of reduced R2, with its Fe2+ center. Oxygen then closes the circle by reforming the radical Fe3+ center of protein R2. In the presence of oxygen, the tyrosyl radical is formed; in its absence the radical is removed [9].
Mechanism of radical scavenging by Flavin molecule
In anaerobic condition, the first step, the oxido oxygen atom of ion (III) cluster scavenge the hydrogen radical from the acidic hydrogen of flavin and form the hydroxyl Fe (III). Another Fe (III) was reduced to Fe (II) and that the reductive step precedes oxygen requiring radical formation [5,9]. According to this model a form of R2 containing reduced ion (“reduced R2”) should be an intermediate in the reaction. Simultaneously the unstable flavin radical (IIa & IIb) hydrogen bonded [flavin N-H (of Uracil moiety)] with oxygen atom of oxido-Fe (III) and serve as a driving force for the oxido-Fe (III) bond cleavage. To attain the stable aromatic nature, the II b flavin radical eliminates one more electron from the piperazine ring and form the yellow color oxidized flavin (IV) in the second step. Hydroxyl Fe (III) reacts with second hydrogen radical and form one more reduced Fe (II) and water.
Action of Phenosafranine molecule
In the anaerobic environment with the presence of Phenosafranine (I), the reduction of both the free radical and the ion center takes place. Phenosafranine cation first loses one of its protons and forms the neural phenosafranine (II) and hydroxyl Fe- (III). One Fe (III) gets reduced to Fe (II) at the first step. In the second step, dimethyl amine nitrogen donates a pair of electrons to make aromaticity in the molecule and the piperazine moiety expels the hydrogen anion from N-H. This anion reacts with HO-Fe (II)-R to form water (IV) and reduced to Fe (II). After the radical reduction of Fe center, air was introduced into the sample and a partial regeneration of both the iron center and the radical occurs.
Non bonded interaction
The internal non bonded interactions of the Ribonucleotide reductase with Flavin before weeding is 861 and after weeding is 550 and that of phenosafranine before interaction is 630 and after weeding is 385. The internal energy of the unbound extended state of Flavin was computed to be -0.313kcal/mol and that of Phenosafranine is - 0.213kcal/mol. It is noted that the Auto dock internal energy of the “extended” state for both the ligands Flavin and Phenosafranine denotes Negative.
Conclusion
The Human RNR activity is necessary for DNA replication and inhibition of this enzyme helps to inhibit cell division. Therefore an understanding of the molecular mechanism of RNR is essential for the design of new cytostatic drugs. In this paper, the docked inhibitors Flavin and Phenosafranine binds at the active cite TYR176, which are essential for free radical formation. This mechanism helps to understand the functional aspects of human RNR2 and leads to the development of novel inhibitors. An increased level of RNR produces more DNA, and thus drives the cells through DNA synthesis. In a hypoxic environment where the activity of RNR is hampered due to lack of oxygen, the level of RNR regulates the production of DNA and drives the cells faster through DNA synthesis. This implies that the replication machinery in the cells is able to detect the level of DNA, when exogenous DNA is supplied. This helps to explore more about the structure of Human RNR and its interaction with its inhibitors. Based on our current results, the work can be further narrowed towards novel leads molecule synthesis by structure based Drug discovery approach.
Footnotes
Citation:Priya & Shanmughavel, Bioinformation 4(3): 123-126 (2009)
References
- 1.Lewis WH, et al. Cell Physiol. 1978;97:87. doi: 10.1002/jcp.1040970109. [DOI] [PubMed] [Google Scholar]
- 2.Reichard P. Science. 1993;260:1773. doi: 10.1126/science.8511586. [DOI] [PubMed] [Google Scholar]
- 3.Wright JA, et al. Biochem Cell Biol. 1990;68:1364. doi: 10.1139/o90-199. [DOI] [PubMed] [Google Scholar]
- 4.Eklund H, et al. Prog Biophys Mol Biol. 2001;77:177. doi: 10.1016/s0079-6107(01)00014-1. [DOI] [PubMed] [Google Scholar]
- 5.Cocking JM, et al. Somat cell mol Genet. 1987;13:221. doi: 10.1007/BF01535204. [DOI] [PubMed] [Google Scholar]
- 6.Bauer W, et al. Amidox, an inhibitor of RNR. 2004;23:1541. doi: 10.1081/NCN-200027766. [DOI] [PubMed] [Google Scholar]
- 7.Horvath Z, et al. Synergistis cytotoxicity. 2004;54:139. [Google Scholar]
- 8.Bitonti AJ, et al. Response of human colon. 1995;15:1179. [PubMed] [Google Scholar]
- 9.Fontecave M, et al. J Bio Chem. 1989;264:9164. [PubMed] [Google Scholar]
- 10.Biosym, Insight II user guide. San Diego, USA: Biosym MSI; 1995. [Google Scholar]
- 11.Brooijimans N, Kundz ID. Annu Rev Biophys Biomol Struct. 2003;32:335. doi: 10.1146/annurev.biophys.32.110601.142532. [DOI] [PubMed] [Google Scholar]
- 12.Jordan A, Reichard P. Ann Rev Biochem. 1998;67:71. doi: 10.1146/annurev.biochem.67.1.71. [DOI] [PubMed] [Google Scholar]
- 13.Reichard P, Ehrenberg A. Science. 1983;221:514. doi: 10.1126/science.6306767. [DOI] [PubMed] [Google Scholar]
- 14.Stubbe J, Vander Donk WA. Chem Rev. 1998;98:705. [Google Scholar]


